Abstract
Modern cell biology is reliant on light and fluorescence microscopy for analysis of cells, tissues and protein localisation. However, these powerful techniques are ultimately limited in resolution by the wavelength of light. Electron microscopes offer much greater resolution due to the shorter effective wavelength of electrons, allowing direct imaging of sub-cellular architecture. The harsh environment of the electron microscope chamber and the properties of the electron beam have led to complex chemical and mechanical preparation techniques, which distance biological samples from their native state and complicate data interpretation. Here we describe recent advances in sample preparation and instrumentation, which push the boundaries of high-resolution imaging. Cryopreparation, cryoelectron microscopy and environmental scanning electron microscopy strive to image samples in near native state. Advances in correlative microscopy and markers enable high-resolution localisation of proteins. Innovation in microscope design has pushed the boundaries of resolution to atomic scale, whilst automatic acquisition of high-resolution electron microscopy data through large volumes is finally able to place ultrastructure in biological context.
Keywords: Transmission electron microscopy, Scanning electron microscopy, Artifacts, Native state, Cryopreparation, Cryo-EM, ESEM, Correlative, Volume EM, FIB/SEM, SBF/SEM
Overview
The properties of the electron beam dictate both the environment of the electron microscope chamber and the physical properties of the specimen to be imaged. Electrons are easily scattered by the molecules in air, and so electron microscopes operate under a vacuum. Biological samples are mainly composed of light elements (carbon, hydrogen and oxygen), which have low electron density and therefore low contrast in the electron beam. Additionally, in the transmission electron microscope (TEM) the sample must be thin enough for the electron beam to penetrate in order to form an image on a detector below (Fig. 1a), whereas in the scanning electron microscope (SEM) the sample must be conductive in order for the electron beam to scan the surface layer without charge build-up or excessive heating (Fig. 1b). Traditional sample preparation techniques were designed to address these challenges (Fig. 2).
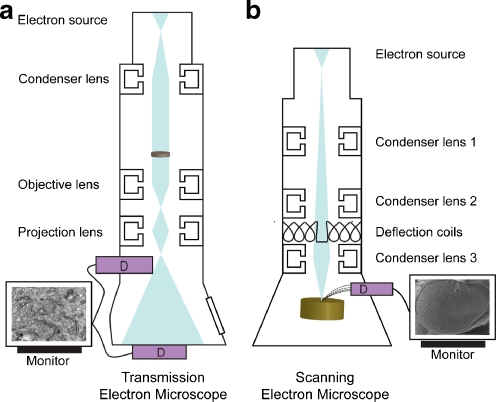
Fig. 1.
Schematic overview of transmission and scanning electron microscopes. a Samples must be cut into ultrathin sections in order for the electron beam to transmit and form an image on the detector in the TEM (Micrograph - stacked membranes of the Golgi apparatus). b In the SEM, the electron beam is scanned over the surface of the sample to produce topographical or compositional information from the surface layer only (Micrograph - Drosophila melanogaster compound eye). D detector
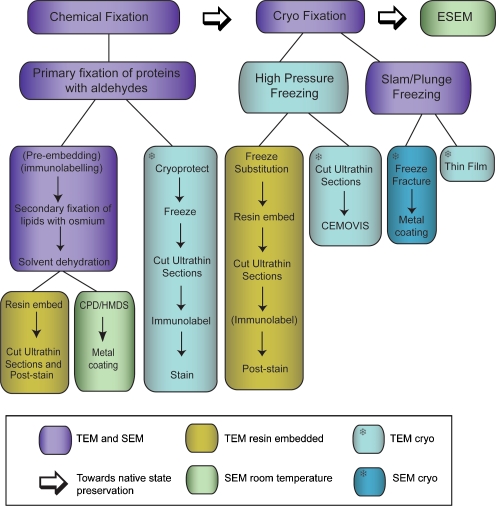
Fig. 2.
Flow diagram of sample preparation techniques for electron microscopy. Traditional chemical fixation, staining and resin embedding procedures protect biological samples against the harsh environment of the EM chamber but induce processing artifacts. Developments in cryopreservation and cryo-EM minimise processing and preserve samples closer to their native state. Environmental SEMs take high-resolution imaging a step closer to native state using hydrated samples at ambient temperature
The first step in preparing a biological sample for electron microscopy (EM) is to stabilise or ‘fix’ the macromolecular structure. Primary fixation for routine biological EM is achieved by chemical cross-linking of proteins using aldehydes [52]. Secondary fixation with osmium tetroxide reduces extraction of lipids and introduces contrast due to deposition of the heavy metal onto membranes [68, 89]. Tannic acid [69] and uranyl acetate [44, 87] may be incorporated as secondary or tertiary fixatives to improve membrane contrast. However, infiltration of chemicals can be slow and limits sample size to approximately 1 mm3.
Microwave-accelerated immobilisation [100] has been used to increase the penetration rate of chemicals into samples and improve preservation through volumes. Cell monolayers can be fixed in a sub-minute timescale, improving preservation of cytoskeleton and raising the possibility of studying dynamic processes [81]. Plant material can be notoriously difficult to infiltrate due to the thick cell wall, but using microwave technology sample preparation times can be reduced from more than 3 days to just 5 h [105]. However, the use of microwaves in cell biology EM is in its infancy, and further development of protocols and investigation of microwave-induced artifacts is required [102].
Samples must then be protected against structural collapse in the vacuum of the EM chamber. In conventional processing for TEM this is achieved by embedding the sample in a liquid resin and polymerising to create a hard block. Most resins are not miscible with water so the sample first needs to be dehydrated using solvents, which can cause artifacts due to shrinkage. There are many commercially available resins, the most common being the epoxy resins, which polymerise uniformly, suffer negligible shrinkage during polymerisation and are relatively stable under the electron beam making them a popular embedding medium for routine TEM [70]. Once polymerised, the block is cut into sections thin enough for the electron beam to penetrate (typically 50–200 nm) using an ultramicrotome and a glass or diamond knife. This process can introduce sampling artifacts as an ultrathin section may represent only 0.5% of the thickness of a single cell, as well as mechanical artifacts in the form of knife marks, compression and chatter. The sections are floated onto a water bath, picked up on EM grids and post-stained with heavy metals to enhance the contrast of various sub-cellular structures. Common post-embedding stains include lead citrate [83, 99] and uranyl acetate [49, 99]. Although necessary for good contrast in the final image, heavy metals can cause additional artifacts in the form of precipitation.
In SEM, samples are typically protected against structural collapse in the vacuum by full dehydration. This can be achieved using simple chemical evaporation from hexamethyldisilazane or by critical point drying (CPD). In both cases the aim is to avoid the damaging effects of surface tension on ultrastructure. CPD is carried out in a temperature- and pressure-controlled chamber that is able to reach a critical point at which there is no apparent difference between the liquid and the gas state of the transitional medium (in this case carbon dioxide) giving zero surface tension. However, in both techniques dehydration may lead to shrinkage artifacts [12, 13]. Once fully dehydrated the sample is mounted onto a stub and coated with a conductive material (most commonly gold or platinum) to enhance surface detail and ameliorate distortion and excessive heating caused by charging of non-conductive samples during electron beam imaging.
Routine TEM and SEM can produce informative and visually arresting images of biological samples, but at each step in processing, the samples are subjected to harsh treatments that can induce artifacts. These may be accepted and tolerated by the experienced electron microscopist but can easily lead to misinterpretation or masking of the data. Recent advances in sample preparation have focused on imaging proteins, cells and tissues as close to their native state as possible in the demanding environment of the electron microscope.
Advances in sample preparation techniques
Currently, the most effective alternative to chemical fixation is cryofixation (Fig. 2). All freezing methods have the common aim of preserving the sample in as life-like a state as possible. This is achieved by removing heat at such a rapid rate that water molecules form amorphous vitreous ice, thus avoiding the destructive effects of crystalline ice on cellular ultrastructure. When the vitrification temperature is attained by rapid freezing, viscosity reaches a level which prevents movement, thus immobilising all the molecules in a cell within milliseconds [72].
The limitation of most freezing methods is the depth of vitrification. Several microns of good freezing can be achieved by rapidly slamming the sample onto a cooled metal block or plunging into a cooled cryogen (usually liquid ethane, propane or nitrogen). This method was first used in observations of a viral suspension embedded in a vitrified layer of water [1].
Alternatively, high-pressure freezing (HPF) at around 2,000 bar using a jet of liquid nitrogen can vitrify to a depth of around 200 μm (Fig. 2) [93]. HPF is particularly suited to fixation of samples with thick cell walls such as yeast [74], Caenorhabditis elegans [73, 101] and plant tissues [16] as well as fast fixation of volatile cellular components including cytoskeleton and coat proteins [84]. Intriguingly, a recent report suggests that plunge freezing of specimens in sealed capillary tubes may achieve comparable vitrification to HPF due to pressure build-up in the confined space [57].
In order to image the frozen sample in a standard EM, the sample must be brought to room temperature and embedded in resin. An organic solvent, usually acetone or methanol, is substituted for the vitreous ice at temperatures below −90 °C in a process known as freeze substitution (FS) [10]. The exact temperature at which fixation takes place during FS is not entirely clear but is believed to be somewhere between −50 and −30 °C [90]. At these low temperatures protein and lipid molecules do not have sufficient thermal energy to move so many of the fixation artifacts seen at room temperature that are avoided [54]. A variety of FS protocols have been reported in the literature, optimised for a wide diversity of biological samples, many involving the addition of gluteraldehyde, osmium or tannic acid as fixatives or membrane stains [65]. To date, no single protocol or group of protocols has emerged as a ‘standard’ for processing biological samples by FS.
HPF has proved to be critical in studies of organelle biogenesis. Weibel–Palade bodies (WPBs) are organelles found in endothelial cells, which play a central role in the initiation of inflammation and haemostasis. Whilst WPBs can be imaged in aldehyde-fixed cells (Fig. 3a), volatile elements, including coat proteins and the content protein Von Willebrand factor (VWF), are only preserved well by HPF (Fig. 3b) [106]. Delineating the formation of WPBs and the folding of VWF is crucial in understanding the role of these organelles in platelet recruitment during wound healing. HPF has also been invaluable in the study of foot-and-mouth disease virus (FMD) where FMD virions cannot be detected within infected cells prepared by routine processing methods. By contrast, high-pressure freezing of cells revealed clusters of viral particles in association with membrane-bound vesicles allowing further study of FMD infection [71].
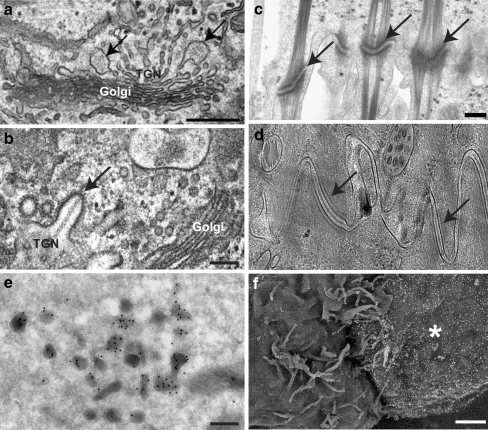
Fig. 3.
Comparison of different sample preparation techniques for electron microscopy. a TEM image of a routinely processed HUVEC showing WPBs (arrows) forming at the trans-Golgi network (TGN), compared with b TEM image of a high-pressure frozen and freeze substituted HUVEC showing improved preservation of volatile elements including coats and protein tubules in forming WPBs (arrow). c TEM image of desmosomes (arrows) in routinely processed tissue culture cells compared with d TEM image of desmosomes (arrows) prepared by CEMOVIS showing improved resolution and near native state preservation. e Immunogold localisation of secretogranin in dense granules in PC12 cells, imaged by TEM. f Immunogold labelling of the plasma membrane of an immune cell (asterisk) demonstrating membrane transfer across the immune synapse, imaged by backscattered electron imaging in the SEM. Bars = 200 nm except f = 1 μm. a, b Adapted with permission from [106]. d Image courtesy of Dr. Ashraf Al-Amoudi, EMBL
Towards native-state imaging in the electron microscope
Development of liquid nitrogen and liquid helium-cooled cryo-EM stages has allowed frozen samples to be imaged without the need for chemical fixation, dehydration, heavy metal staining or FS. In cryo-TEM, imaging is again limited by the ability of the electron beam to penetrate the sample and by the need to reduce the interaction of the electron beam with the specimen. Frozen hydrated samples larger than 1 μm must be sectioned at low temperature (typically −140 °C) in a cryoultramicrotome prior to imaging [24]. Cryoelectron microscopy of vitreous sections (CEMOVIS) [1, 4, 11, 25] requires advanced technical and interpretative skills [26] and despite removing chemical artifacts, mechanical artifacts including knife marks and crevasses remain [5]. However, the results can be quite exceptional and with no chemical fixation or staining there is, at the present time, no comparable EM technique for viewing biological samples in near native state and at unprecedented resolution (Fig. 3c, d) [5, 11, 107]. This technique has led to the discovery of a novel branched tubular structure containing lamellar material and a new ribosome complex-like structure not previously seen by classical EM techniques [76].
In addition, imaging protein structure itself is a major element of modern electron microscopy. Negative staining with ammonium molybdate, uranyl acetate, uranyl formate, phosphotungstic acid, osmium tetroxide or osmium ferricyanide has traditionally been used to image ultrasmall biological samples including viruses, bacteria, membranes and proteins at high resolution [36, 37]. However, drying of the macromolecules and subsequent coating with heavy metals induces artifacts as already described for large samples. Cryo-TEM of macromolecules is now becoming routine and has the advantage that specimens are observed without chemical fixation, staining or coating whilst minimising radiation damage. Problems may arise due to the inherently low contrast of biological macromolecules, and although new techniques combine negative stains with cryofixation to increase contrast, stain-based artifacts remain an issue [2, 17].
Many laboratories now use energy-filtering TEM (EFTEM) to overcome the challenges of viewing frozen samples with inherently low contrast (Fig. 4). EFTEM can be used in two ways to enhance information from an unstained cryo-TEM sample. Firstly, devices can be adjusted to select for zero-loss electrons (zero-loss filtering), which can significantly increase the signal-to-noise ratio to improve contrast on unstained samples. Secondly, in electron energy loss spectroscopy (EELS), electrons which have lost a specific amount of energy can be selected using an adjustable slit to build an image of elemental distributions within a sample. In the 1980s, EELS was frequently used by biologists for elemental mapping, with particular application to the composition of regulated secretory granules and elemental distribution along the regulated secretory pathway [27, 56].
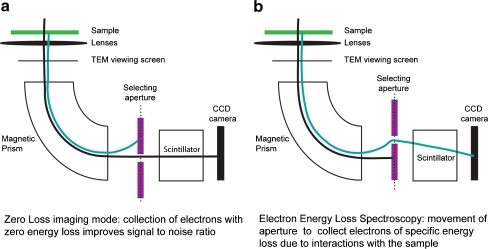
Fig. 4.
Comparison of zero-loss imaging and electron energy loss spectroscopy. a In zero-loss imaging mode, the selecting aperture allows only transmitted electrons to reach the detector. This increases contrast and improves resolution for thick sections due to the elimination of scattered electrons. b In EELS, the selecting aperture is moved, and the accelerating voltage of the TEM is increased to select for electrons that were slowed down by their interactions with a specific element
Using EFTEM, beam damage to the frozen sample is minimised by electron dose control, allowing cryoelectron tomography (CET) to be used in the study of heterogeneous or non-symmetrical samples at high resolution. In CET, images are collected over a tilt series, usually ±70°, and reconstructed into a high-resolution three-dimensional (3D) volume through the depth of the sample [35, 60]. CET has emerged as a 3D imaging technique to bridge the information gap between X-ray crystallography and optical microscopy methods. CET allows investigation of the structure–function relationship of molecular complexes and supramolecular assemblies in their cellular environments without fixation, dehydration, embedding or sectioning artifacts.
Similarly, many of the artifacts associated with sample preparation for SEM can be avoided by imaging frozen samples on a cryo-SEM stage. Sucrose may be used as a cryoprotectant when imaging the internal structure of large specimens by freeze fracture, whereas plunge freezing into nitrogen slush (approximately −210 °C) usually provides a sufficient depth of vitrification when imaging the sample surface. Cryo-SEM also carries the additional advantage that samples sensitive to dehydration may be imaged [47]. Specimens are preserved much closer to their fully hydrated native state, as demonstrated in work by Walther [98] where 10-nm-diameter filaments attached to the inner membrane of the nuclear envelope of Xenopus laevis stage VI oocytes were shown to be composed of distinct globular sub-units of approximately 5 nm in diameter, arranged in a helical manner with right-handed periodicity. This could not be seen using room temperature preparation protocols and analysis and enabled identification of the filaments as F-actin.
The ultimate aim of native-state imaging is to view fully hydrated, chemically unmodified biological samples at high resolution in the electron microscope. A promising solution is wet SEM, where a hydrated sample is placed in an electron transparent, pressure-resistant capsule and then imaged in a conventional SEM using a backscattered electron detector. This technique has yet to be fully exploited but has achieved resolutions between 10 and 100 nm and is suitable for samples that are adversely affected by dehydration using organic solvents, including lipid-rich structures and hydrated gels [31].
Innovations in microscope design have led to environmental or variable pressure SEM (ESEM or VPSEM), which allows a gaseous environment in the chamber [91]. Fully hydrated insulating biological samples can be imaged at room temperature and without the need for coating with conducting elements. The atmosphere in the chamber is carefully controlled to ensure that water evaporation or condensation does not take place, although this condition may be deliberately manipulated in order to follow the natural state of a sample as it undergoes the hydration and dehydration process. Although processing artifacts have been reduced to an absolute minimum, as with all EM imaging, there is still a possibility of electron beam damage to the sample. Also, as there is no fixation step, there is the possibility of movement of molecules within cells, cell degradation and eventually cell death.
A comprehensive study was carried out on yeast cells to monitor radiation damage under high and low vacuum conditions, with and without electron beam exposure in ESEM. Fully hydrated yeast cells survived longer, but were more severely affected by electron-induced radiation damage, under low vacuum conditions [82]. A second report showed that it is possible to view unfixed, fully hydrated and uncoated cells with no dehydration effects for extended periods (in some cases even hours) but the user must have good knowledge of their sample. The osmotic pressure and rate of water loss from the specimen needs to be known to achieve this task [75, 92].
Although the potential of ESEM for imaging biological samples is yet to be fully realised, application to in situ tissue engineering studying endothelial cell retention and differentiation on biomaterial surfaces in hydrated conditions has been demonstrated [8].
Locating areas of interest
Fluorescence microscopy revolutionised cell biology by allowing localisation of proteins in cells, thus generating vital information as to their function. In general, protein localisation may be studied either in intact cells using clonable reagents like green fluorescent protein (GFP) or by detergent permeabilisation of cells followed by immunolabelling with fluorescent marker-conjugated antibodies. Even considering recent breakthroughs in super resolution light microscopy which circumvent the diffraction limit [42, 45, 48], electron microscopy is required to image protein localisation in direct relation to cell ultrastructure.
Protein localisation in EM is challenging. Good preservation of sample morphology is often at odds with localisation of target molecules—antigens may be destroyed or masked by harsh fixation and staining conditions, while detergent permeabilisation disrupts membrane ultrastructure.
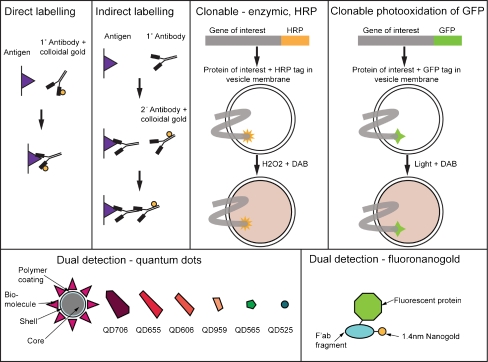
Markers must be electron dense and discrete from the contrasting agents used to visualise sample morphology (Fig. 5). Clonable markers include GFP, which can be detected using photooxidation [32], and the enzyme horseradish peroxidase (HRP), which produces an electron dense reaction product when treated with diaminobenzidine and hydrogen peroxide (Fig. 5) [46]. This strategy is optimised when the tag resides within the lumen of a sub-cellular compartment so that the reaction product is concentrated in a small volume. Recent work has led to the development of a new clonable marker based on the metal-binding protein metallothionein, which binds directly to gold salts during freeze substitution to produce electron dense particles, although endogenous metallothioneins may cause background labelling [22, 67].
Fig. 5.
Examples of strategies for localisation of proteins in cells and tissues for EM. Direct and indirect immunolabelling uses antibodies specific for the protein of interest, conjugated to an electron dense marker (usually colloidal gold) for pre-embedding labelling of permeabilised cells, post-embedding labelling onto hydrophilic resins and immunolabelling of cryosections. Clonable markers, including HRP and green fluorescent protein (GFP), can be converted to electron dense products that can be detected in the EM. CLEM is most efficient when used with markers, which can be detected in both the light and electron microscope including quantum dots and fluoronanogold. Quantum dots are illustrated to show approximate shape in EM as well as colour in the fluorescence microscope
Cells can be permeabilised for pre-embedding immunolabelling with weak detergents such as digitonin or saponin, which maintain recognisable cell ultrastructure but lead to loss of cytosolic constituents. Antigens on the cytosolic side of membrane-bound compartments become accessible to direct or indirect detection using antibodies conjugated to electron dense particles, usually colloidal gold with a diameter of 5–20 nm [20]. Samples may also be immunolabelled post-embedding using a hydrophilic methacrylate resin, although detection efficiency is often significantly reduced as only those antigens available at the resin surface will be labelled.
Cryosectioning and immunolabelling is the most elegant method currently available for protein localisation although it is labour intensive, requires a high level of skill and is critically dependent on good primary antibodies. Aldehydes, in particular glutaraldehyde, can cause antigen denaturation by cross-linking proteins internally. For this reason a light fix of paraformaldehyde only is usually used with some sacrifice of ultrastructural preservation [34]. Support is provided by gelatin, and the sample is cryoprotected in 2.3 M sucrose, mounted on pins, frozen in liquid nitrogen and sectioned using a cryoultramicrotome [94, 95]. Sections are picked up on a solution of sucrose and methyl cellulose [58], placed onto a formvar-coated EM grid and immunolabelled. In the absence of harsh chemicals and resins, antigens are better preserved and more accessible to immunolabelling (Figs. 3e and 5). Recently, cryosectioning has been used in combination with HPF to localise proteins in cells, tissues [96] and yeast whilst maintaining excellent morphological preservation [33].
Correlative light electron microscopy (CLEM) is becoming more popular in the quest to link fluorescence data to high-resolution morphology. Simple correlation can be made by growing cells on glass coverslips photo-etched with a grid. Relocation of the cell of interest post-embedding is critically dependent on careful mapping of cell position by light microscopy. Fluorescence data can then be overlaid onto the EM images to correlate the area of interest and identify the sub-cellular location of the fluorescent marker [79]. This technique is most informative when using dual detection markers which appear in both fluorescence microscopes and EM, for example fluoronanogold with silver enhancement for EM detection [15] and quantum dots [19, 30] (Fig. 5).
Recent advances in HPF equipment and accessories [65] have made it possible to record a live event by fluorescence video microscopy, transfer the sample to the high-pressure freezer for cryofixation within 4 s and image the same event in EM with excellent preservation of ultrastructure. This has been applied to the study of multivesicular bodies (MVBs), which are highly dynamic sub-cellular organelles involved in protein sorting to the degradative and recycling cellular pathways. Fluorescence video microscopy was used to image MVBs until a membrane was observed budding from one structure and fusing with an adjacent structure, at which point the sample was immediately high-pressure frozen. The area of interest was relocated post-embedding and identified as a tubular profile attached to the MVB via an open connection, suggesting a mechanism for protein sorting [97]. The development of a cryofluorescence microscope enables visualisation of the fluorophore after HPF and CEMOVIS and prior to imaging in cryo-EM for correlation of protein localisation with near native-state preservation [85, 86]. Direct imaging of fluorescent molecules in situ in an EM is the ultimate realisation of CLEM. Integrated laser electron microscopy has taken the first steps towards this by feeding a laser line into the goniometer of a TEM [3] to image UV-induced nuclear structures in apoptotic cells [53].
Similarly, cell surface antigens may be immunolabelled for analysis in the SEM. Backscattered electron imaging detects markers like colloidal gold due to high atomic mass. Detection of 5–20-nm particles has been made possible by advances in high-resolution field emission SEMs and high-resolution ion beam coaters. SEM imaging of immune cells with 20-nm gold particles decorating the cell surface has been used to demonstrate intercellular transfer of membrane between natural killer cells and target cells across the immune synapse (Fig. 3f) [104], believed to be a key step in immune cell communication during immune surveillance.
Resolution versus sample size
In all imaging techniques the sample volume has an inverse relationship to the achievable resolution. This is due to physical laws governing the penetration of light and electrons through the sample. In order to achieve highest resolution in any imaging modality the sample size must therefore be physically reduced. However, this introduces sampling artifacts and masks interactions between cells and their environment in 3D space. This is a particular problem when dealing with cells, tissues and whole organisms, which are complex and heterogenous.
Even in the most powerful scanning TEMs, the maximum section thickness that can be imaged is approximately 1 μm [59]. A section thickness larger than the mean free path of the electrons leads to a large contribution from inelastically scattered electrons causing blurring in the final image. Energy filtering can offer some remedy: When operated in a zero-loss mode the inelastically scattered electrons can be removed, and contrast is improved even in an EM operating at conventional voltages (80–120 keV) [35] (Fig. 4). For thick biological specimens, due to the wide maximum spectrum observed (100–300 eV) many researchers now use most probable loss filtering [50, 80].
Electron tomography is also severely limited by section thickness—due to geometry a specimen with a thickness of only 200 nm at 0° tilt will increase in thickness to 585 nm at 70° tilt. Tomography also suffers from several artifacts including the ‘missing wedge’. This can be partially overcome by using a dual axis holder, leading to a reduced ‘missing pyramid’ of information [6, 62] or by post-acquisition software calculations [9]. Recent innovations include a 360° holder, which can collect a full tilt series from a cylinder of material produced by ion milling [51].
Serial sectioning and imaging of the sample may be used to collect data through volumes in excess of 1 μm. This technique is achievable in a standard biological EM laboratory but is time consuming and has many difficult and rate limiting steps. Artifacts include chatter and thickness variation during sectioning, folds or tears in the sections, staining artifacts and differences in orientation between sections and consecutive grids. Images must be realigned using software and corrections made for differences in brightness and contrast, orientation and stretching or contraction between sections, often leading to a reduced field of view in the final reconstruction [38]. A serial tomography strategy [88] has been successfully used to collect data from serial semi-thin sections to analyse the Golgi apparatus in a pancreatic beta cell line [61]. Nevertheless tomography, serial sectioning and serial tomography studies tend to be limited to a thickness of less than 5 μm, which is frequently less than the thickness of a single cell. Where studies of larger volumes have been undertaken, they have required many electron microscopists working over many years to cut, image and analyse thousands of sections, as in the reconstruction of neurons in C. elegans [103].
Recent innovations have led to a paradigm shift in high-resolution imaging of large volumes of biological samples. These techniques exploit different sectioning methods to cut and image slices of material automatically, thus minimising artifacts and speeding up the process whilst freeing the operator to perform other work. In general, samples are prepared as for traditional TEM. Much of the work in this area has been driven by neurobiology, where traditional EM cannot cope with the conflict between volume imaging of complex neural networks and high-resolution imaging of neuronal connections [41].
Atomic force microscopy (AFM) has been integrated with ultramicrotomy to sequentially scan the block face of resin-embedded samples at high resolution following each sectioning cycle. AFM phase contrast is generated by local variation of the viscoelastic properties of the sample so that heavy metal staining is no longer required, although interpretation of ultrastructure is therefore more complex. As sections are collected after cutting, correlative AFM/TEM can be used to link macromolecular information to more traditional high-resolution images [29, 63, 64].
Automatic tape-collecting lathe ultramicrotomy (ATLUM) automatically sections and collects embedded tissue on a long carbon-coated tape for imaging in an SEM. Volumes as large as tens of millimetres can be collected at 50-nm section thickness and imaged with a lateral resolution of 5 nm [40]. The development of ATLUM has been targeted towards producing a map of the entire neural circuitry (connectome) of the human brain.
Two additional techniques section the sample in situ within the chamber of an SEM. In focused ion beam SEM (FIB/SEM) [18, 43, 55] and serial block face SEM (SBF/SEM) [14, 21, 77] a slice of material is removed, and the revealed surface is imaged using the scanning electron beam. This cutting and imaging process is repeated sequentially to automatically collect a stack of high-resolution images through the sample (Fig. 6). In the FIB/SEM a gallium ion beam, which is inherently destructive, is used to ‘mill’ or ‘sputter’ away material, whereas the SBF/SEM uses a modified ultramicrotome inside the SEM chamber to remove material using a diamond knife. A backscattered electron detector is used to image the heavy metal staining of the tissue [18, 66].
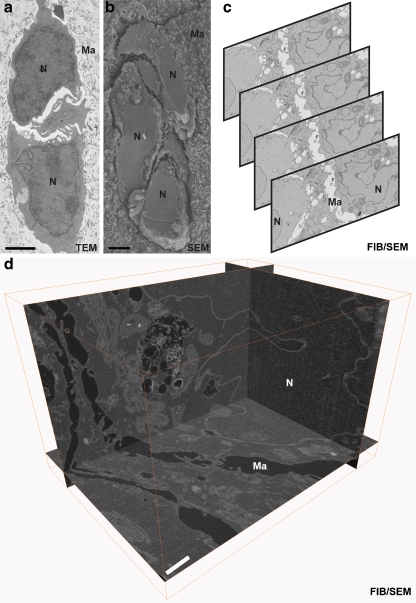
Fig. 6.
Comparison of traditional TEM and SEM with volume EM. a–d Tumour cells invading in response to stromal fibroblasts in a collagen I/matrigel matrix (samples courtesy of Dr. Erik Sahai, Cancer Research UK London Research Institute). a TEM image of cells gives high-resolution information but represents only an ultrathin section through a large sample. N cell nucleus, Ma matrix. b SEM image of a freeze fracture plane through the sample, which gives 3D information from the surface but lacks high-resolution information through the volume. c Serial images from a dataset of 219 images, automatically collected by focused ion beam/scanning electron microscopy (FIB/SEM). The contrast has been inverted to resemble traditional TEM images. d 3D reconstruction (Amira software, Visage Imaging Inc.) of the full FIB/SEM dataset showing x, y and z orthoslices, giving high-resolution information through the volume of the sample to aid analysis of cell–cell contacts (data acquisition in collaboration with Dr. Andreas Schertel, Carl Zeiss NTS Gmbh). Bar (a, b) 2 μm, (d) 1 μm
Although FIB/SEM and SBF/SEM give similar results, the mechanism of in situ sectioning means that the two techniques have distinct but complementary applications. Ion beam milling is currently limited by speed and surface area but can be targeted to specific areas, mill slice thicknesses down to ~10 nm and cut through materials with different hardnesses in a single sample. In contrast, the diamond knife can cut larger surface areas and the speed of cutting is high and independent of surface area, but section thickness is limited to ~25 nm. Lateral resolutions of 4 nm2/pixel have been achieved in the FIB/SEM [55]; however the slight sacrifice in resolution compared to TEM is likely to be outweighed by the increased volume of data when studying large biological samples. SBF/SEM and FIB/SEM have recently been used in series to locate a minute area of interest within a whole organism. Live confocal microscopy was used to follow fluorescent developing blood vessels in a transgenic zebrafish to the point of fusion between two adjacent vessels. SBF/SEM was then used to create an atlas of the microanatomy of the organism, and FIB/SEM was targeted to area of interest to image the ultrastructure of the fusion event [6], representing a volume of just 3 μm3 in a total volume of ~12,600,000 μm3—the proverbial ‘needle in the haystack’.
At present the artifacts arising from FIB/SEM of biological samples remain largely uncharacterised. Computer-based modelling can be used to analyse the ion beam/material interaction for simple crystalline materials [78] but biological materials stained with heavy metal and embedded in resin are heterogenous and complex. Effects due to gallium ion implantation, anisotropic milling, curtaining and the interaction volume of the electron beam must be taken into account [23]. Even so, FIB/SEM is the most flexible of the volume EM techniques. The ion beam is capable of cutting through materials of different hardness, a property which has been exploited to map the interaction of soft biological tissues with titanium-based biomedical implants [28]. Areas of interest can be identified and excised as thin wafers or cylinders for subsequent tomographic analysis in a TEM by in situ lift-out [43]. FIB/SEM of frozen hydrated biological materials is under development, and although this is technically challenging it finally offers the possibility of linking high-resolution volume and near native-state electron microscopy [28, 39].
Conclusions and future outlook
Excellent sample preparation is absolutely critical in cell biological electron microscopy for delivery of meaningful, accurate results. Major technical breakthroughs in cryopreparation, cryomicroscopy, high-resolution and volume EM are pushing the boundaries of imaging. However, with each new microscope advance and preparation technique comes a new raft of artifacts, which must be carefully characterised.
A current major challenge lies in the quantity of data produced by automated high-resolution and volume EM. Thousands of images of proteins, cells and tissues can be collected in a matter of hours or days. A concerted effort is now required to improve data storage, processing, analysis and interpretation through developments in computer software and hardware to make best use of the information generated.
Nevertheless, high-resolution ultrastructural imaging is coming ever closer to visualising proteins, cells and tissues in their native state at unprecendented resolution and in true biological context. Application of different imaging media to biological specimens, from light and electrons to new microscopes utilising X-rays and ions, will allow multiple properties of a single sample to be mapped across 3D space and time.
Acknowledgement
The authors’ research was supported by Cancer Research UK.
References
- 1.Adrian M, Dubochet J, Lepault J, McDowall AW. Cryo-electron microscopy of viruses. Nature. 1984;308:32–36. doi: 10.1038/308032a0. [DOI] [PubMed] [Google Scholar]
- 2.Adrian M, Dubochet J, Fuller SD, Harris JR. Cryo-negative staining. Micron. 1998;29:145–160. doi: 10.1016/s0968-4328(97)00068-1. [DOI] [PubMed] [Google Scholar]
- 3.Agronskaia AV, Valentijn JA, van Driel LF, Schneijdenberg CTWM, Humbel BM, van Bergen En Henegouwen PMP, Verkleij AJ, Koster AJ, Gerritsen HC (2008) Integrated fluorescence and transmission electron microscopy. J Struct Biol 164:183–189 [DOI] [PubMed]
- 4.Al-Amoudi A, Norlen LP, Dubochet J. Cryo-electron microscopy of vitreous sections of native biological cells and tissues. J Struct Biol. 2004;148:131–135. doi: 10.1016/j.jsb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Al-Amoudi A, Studer D, Dubochet J. Cutting artefacts and cutting process in vitreous sections for cryo-electron microscopy. J Struct Biol. 2005;150:109–121. doi: 10.1016/j.jsb.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Armer HEJ, Mariggi G, Png KMY, Genoud C, Monteith AG, Bushby AJ, Gerhardt H, Collinson LM (2009) Imaging transient blood vessel fusion events in zebrafish by correlative volume electron microscopy. PLoS ONE 4: e7716 [DOI] [PMC free article] [PubMed]
- 7.Arslan I, Tong JR, Midgley PA. Reducing the missing wedge: high-resolution dual axis tomography of inorganic materials. Ultramicroscopy. 2006;106:994–1000. doi: 10.1016/j.ultramic.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Baguneid M, Murray D, Salacinski HJ, Fuller B, Hamilton G, Walker M, Seifalian AM. Shear-stress preconditioning and tissue-engineering-based paradigms for generating arterial substitutes. Biotechnol Appl Biochem. 2004;39:151–157. doi: 10.1042/BA20030148. [DOI] [PubMed] [Google Scholar]
- 9.Bartesaghi A, Sprechmann P, Liu J, Randall G, Sapiro G, Subramaniam S. Classification and 3D averaging with missing wedge correction in biological electron tomography. J Struct Biol. 2008;162:436–450. doi: 10.1016/j.jsb.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bohrmann B, Kellenberger E. Cryosubstitution of frozen biological specimens in electron microscopy: use and application as an alternative to chemical fixation. Micron. 2001;32:11–19. doi: 10.1016/s0968-4328(00)00029-9. [DOI] [PubMed] [Google Scholar]
- 11.Bouchet-Marquis C, Fakan S. Cryoelectron microscopy of vitreous sections: a step further towards the native state. Methods Mol Biol. 2009;464:425–439. doi: 10.1007/978-1-60327-461-6_24. [DOI] [PubMed] [Google Scholar]
- 12.Braet F, Zanger R, Wisse E. Drying cells for SEM, AFM and TEM by hexamethyldisilazane: a study on hepatic endothelial cells. J Microsc. 1997;186:84–87. doi: 10.1046/j.1365-2818.1997.1940755.x. [DOI] [PubMed] [Google Scholar]
- 13.Bray DF, Bagu J, Koegler P. Comparison of hexamethyldisilazane (HMDS), Peldri II, and critical-point drying methods for scanning electron microscopy of biological specimens. Microsc Res Tech. 1993;26:489–495. doi: 10.1002/jemt.1070260603. [DOI] [PubMed] [Google Scholar]
- 14.Briggman KL, Denk W. Towards neural circuit reconstruction with volume electron microscopy techniques. Curr Opin Neurobiol. 2006;16:562–570. doi: 10.1016/j.conb.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Cheutin T, Sauvage C, Tchelidze P, O’Donohue MF, Kaplan H, Beorchia A, Ploton D. Visualizing macromolecules with fluoronanogold: from photon microscopy to electron tomography. Methods Cell Biol. 2007;79:559–574. doi: 10.1016/S0091-679X(06)79022-7. [DOI] [PubMed] [Google Scholar]
- 16.Craig S, Staehelin LA. High pressure freezing of intact plant tissues. Evaluation and characterization of novel features of the endoplasmic reticulum and associated membrane systems. Eur J Cell Biol. 1988;46:81–93. [PubMed] [Google Scholar]
- 17.Carlo S, El-Bez C, Alvarez-Rua C, Borge J, Dubochet J. Cryo-negative staining reduces electron-beam sensitivity of vitrified biological particles. J Struct Biol. 2002;138:216–226. doi: 10.1016/s1047-8477(02)00035-7. [DOI] [PubMed] [Google Scholar]
- 18.Winter DAM, Schneijdenberg C, Lebbink MN, Lich B, Verkleij AJ, Drury MR, Humbel BM. Tomography of insulating biological and geological materials using focused ion beam (FIB) sectioning and low-kV BSE imaging. Journal of Microscopy-Oxford. 2009;233:372–383. doi: 10.1111/j.1365-2818.2009.03139.x. [DOI] [PubMed] [Google Scholar]
- 19.Deerinck TJ, Giepmans BN, Smarr BL, Martone ME, Ellisman MH. Light and electron microscopic localization of multiple proteins using quantum dots. Methods Mol Biol. 2007;374:43–53. doi: 10.1385/1-59745-369-2:43. [DOI] [PubMed] [Google Scholar]
- 20.Deinhardt K, Berninghausen O, Willison HJ, Hopkins CR, Schiavo G. Tetanus toxin is internalized by a sequential clathrin-dependent mechanism initiated within lipid microdomains and independent of epsin1. J Cell Biol. 2006;174:459–471. doi: 10.1083/jcb.200508170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diestra E, Fontana J, Guichard P, Marco S, Risco C. Visualization of proteins in intact cells with a clonable tag for electron microscopy. J Struct Biol. 2009;165:157–168. doi: 10.1016/j.jsb.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 23.Drobne D, Milani M, Leser V, Tatti F. Surface damage induced by FIB milling and imaging of biological samples is controllable. Microsc Res Tech. 2007;70:895–903. doi: 10.1002/jemt.20494. [DOI] [PubMed] [Google Scholar]
- 24.Dubochet J, McDowall AW, Menge B, Schmid EN, Lickfeld KG. Electron microscopy of frozen-hydrated bacteria. J Bacteriol. 1983;155:381–390. doi: 10.1128/jb.155.1.381-390.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 26.Dubochet J, Zuber B, Eltsov M, Bouchet-Marquis C, Al-Amoudi A, Livolant F. How to “read” a vitreous section. Methods Cell Biol. 2007;79:385–406. doi: 10.1016/S0091-679X(06)79015-X. [DOI] [PubMed] [Google Scholar]
- 27.Dudek RW, Boyne AF. An excursion through the ultrastructural world of quick-frozen pancreatic islets. Am J Anat. 1986;175(217–43):354. doi: 10.1002/aja.1001750209. [DOI] [PubMed] [Google Scholar]
- 28.Edwards HK, Fay MW, Anderson SI, Scotchford CA, Grant DM, Brown PD. An appraisal of ultramicrotomy, FIBSEM and cryogenic FIBSEM techniques for the sectioning of biological cells on titanium substrates for TEM investigation. J Microsc. 2009;234:16–25. doi: 10.1111/j.1365-2818.2009.03152.x. [DOI] [PubMed] [Google Scholar]
- 29.Efimov AE, Tonevitsky AG, Dittrich M, Matsko NB. Atomic force microscope (AFM) combined with the ultramicrotome: a novel device for the serial section tomography and AFM/TEM complementary structural analysis of biological and polymer samples. J Microsc. 2007;226:207–217. doi: 10.1111/j.1365-2818.2007.01773.x. [DOI] [PubMed] [Google Scholar]
- 30.Giepmans BN, Deerinck TJ, Smarr BL, Jones YZ, Ellisman MH. Correlated light and electron microscopic imaging of multiple endogenous proteins using quantum dots. Nat Methods. 2005;2:743–749. doi: 10.1038/nmeth791. [DOI] [PubMed] [Google Scholar]
- 31.Gileadi O, Sabban A. Squid sperm to clam eggs: imaging wet samples in a scanning electron microscope. Biol Bull. 2003;205:177–179. doi: 10.2307/1543239. [DOI] [PubMed] [Google Scholar]
- 32.Grabenbauer M, Geerts WJ, Fernadez-Rodriguez J, Hoenger A, Koster AJ, Nilsson T. Correlative microscopy and electron tomography of GFP through photooxidation. Nat Methods. 2005;2:857–862. doi: 10.1038/nmeth806. [DOI] [PubMed] [Google Scholar]
- 33.Griffith J, Mari M, Maziere A, Reggiori F. A cryosectioning procedure for the ultrastructural analysis and the immunogold labelling of yeast Saccharomyces cerevisiae. Traffic. 2008;9:1060–1072. doi: 10.1111/j.1600-0854.2008.00753.x. [DOI] [PubMed] [Google Scholar]
- 34.Griffith JM, Posthuma G. A reliable and convenient method to store ultrathin thawed cryosections prior to immunolabeling. J Histochem Cytochem. 2002;50:57–62. doi: 10.1177/002215540205000106. [DOI] [PubMed] [Google Scholar]
- 35.Grimm R, Barmann M, Hackl W, Typke D, Sackmann E, Baumeister W. Energy filtered electron tomography of ice-embedded actin and vesicles. Biophys J. 1997;72:482–489. doi: 10.1016/S0006-3495(97)78689-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harris JR. Negative staining of thinly spread biological particulates. Methods Mol Biol. 1999;117:13–30. doi: 10.1385/1-59259-201-5:13. [DOI] [PubMed] [Google Scholar]
- 37.Harris JR, Scheffler D. Routine preparation of air-dried negatively stained and unstained specimens on holey carbon support films: a review of applications. Micron. 2002;33:461–480. doi: 10.1016/s0968-4328(01)00039-7. [DOI] [PubMed] [Google Scholar]
- 38.Harris KM, Perry E, Bourne J, Feinberg M, Ostroff L, Hurlburt J. Uniform serial sectioning for transmission electron microscopy. J Neurosci. 2006;26:12101–12103. doi: 10.1523/JNEUROSCI.3994-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayles MF, Stokes DJ, Phifer D, Findlay KC. A technique for improved focused ion beam milling of cryo-prepared life science specimens. Journal of Microscopy-Oxford. 2007;226:263–269. doi: 10.1111/j.1365-2818.2007.01775.x. [DOI] [PubMed] [Google Scholar]
- 40.Hayworth K, Kasthuri N, Schalek R, Lichtman J. Automating the collection of ultrathin serial sections for large volume TEM reconstructions. Microsc Microanal. 2006;12:86–87. [Google Scholar]
- 41.Helmstaedter M, Briggman KL, Denk W. 3D structural imaging of the brain with photons and electrons. Curr Opin Neurobiol. 2008;18:633–641. doi: 10.1016/j.conb.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 42.Henriques R, Mhlanga MM. PALM and STORM: what hides beyond the Rayleigh limit? Biotechnol J. 2009;4:846–857. doi: 10.1002/biot.200900024. [DOI] [PubMed] [Google Scholar]
- 43.Heymann JA, Hayles M, Gestmann I, Giannuzzi LA, Lich B, Subramaniam S. Site-specific 3D imaging of cells and tissues with a dual beam microscope. J Struct Biol. 2006;155:63–73. doi: 10.1016/j.jsb.2006.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hirsch JG, Fedorko ME. Ultrastructure of human leukocytes after simultaneous fixation with glutaraldehyde and osmium tetroxide and “postfixation” in uranyl acetate. J Cell Biol. 1968;38:615–627. doi: 10.1083/jcb.38.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirvonen LM, Wicker K, Mandula O, Heintzmann R. Structured illumination microscopy of a living cell. Eur Biophys J. 2009;38:807–812. doi: 10.1007/s00249-009-0501-6. [DOI] [PubMed] [Google Scholar]
- 46.Hopkins C, Gibson A, Stinchcombe J, Futter C. Chimeric molecules employing horseradish peroxidase as reporter enzyme for protein localization in the electron microscope. Methods Enzymol. 2000;327:35–45. doi: 10.1016/s0076-6879(00)27265-0. [DOI] [PubMed] [Google Scholar]
- 47.Hsu KC, Chung WH, Lai KM (2009) Histological structures of native and cooked yolks from duck egg observed by SEM and Cryo-SEM. J Agric Food Chem 57(10):4218–4223 [DOI] [PubMed]
- 48.Huang B, Bates M, Zhuang X. Super-resolution fluorescence microscopy. Annu Rev Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huxley HE, Zubay G. Preferential staining of nucleic acid-containing structures for electron microscopy. J Biophys Biochem Cytol. 1961;11:273–296. doi: 10.1083/jcb.11.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Izard J, Renken C, Hsieh CE, Desrosiers DC, Dunham-Ems S, La Vake C, Gebhardt LL, Limberger RJ, Cox DL, Marko M, Radolf JD (2009) Cryo-electron tomography elucidates the molecular architecture of Treponema pallidum, the syphilis spirochete. J Bacteriol: JB.01031–09 [DOI] [PMC free article] [PubMed]
- 51.Jarausch K, Leonard DN. Three-dimensional electron microscopy of individual nanoparticles. J Electron Microsc (Tokyo) 2009;58:175–183. doi: 10.1093/jmicro/dfn028. [DOI] [PubMed] [Google Scholar]
- 52.Karnovsky MJ. A formaldehyde-gluteraldehyde fixative of high osmolality for use in electron microscopy. J Cell Biol. 1965;27:137A. [Google Scholar]
- 53.Karreman MA, Agronskaia AV, Verkleij AJ, Cremers FF, Gerritsen HC, Humbel BM. Discovery of a new RNA-containing nuclear structure in UVC-induced apoptotic cells by integrated laser electron microscopy. Biol Cell. 2009;101:287–299. doi: 10.1042/BC20080076. [DOI] [PubMed] [Google Scholar]
- 54.Kellenberger E. The response of biological macromolecules and supramolecular structures to the physics of specimen cryopreparation. In: Steinbrecht RA, Zierold K, editors. Cryotechniques in biological electron microscopy. Berlin: Springer; 1987. pp. 35–63. [Google Scholar]
- 55.Knott G, Marchman H, Wall D, Lich B. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. J Neurosci. 2008;28:2959–2964. doi: 10.1523/JNEUROSCI.3189-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leapman RD, Ornberg RL. Quantitative electron energy loss spectroscopy in biology. Ultramicroscopy. 1988;24:251–268. doi: 10.1016/0304-3991(88)90314-2. [DOI] [PubMed] [Google Scholar]
- 57.Leunissen JL, Yi H. Self-pressurized rapid freezing (SPRF): a novel cryofixation method for specimen preparation in electron microscopy. J Microsc. 2009;235:25–35. doi: 10.1111/j.1365-2818.2009.03178.x. [DOI] [PubMed] [Google Scholar]
- 58.Liou W, Geuze HJ, Slot JW. Improving structural integrity of cryosections for immunogold labeling. Histochem Cell Biol. 1996;106:41–58. doi: 10.1007/BF02473201. [DOI] [PubMed] [Google Scholar]
- 59.Loos J, Sourty E, Lu K, Freitag B, Tang D, Wall D. Electron tomography on micrometer-thick specimens with nanometer resolution. Nano Lett. 2009;9:1704–1708. doi: 10.1021/nl900395g. [DOI] [PubMed] [Google Scholar]
- 60.Marsh BJ. Lessons from tomographic studies of the mammalian Golgi. Biochim Biophys Acta. 2005;1744:273–292. doi: 10.1016/j.bbamcr.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 61.Marsh BJ, Mastronarde DN, Buttle KF, Howell KE, McIntosh JR. Organellar relationships in the Golgi region of the pancreatic beta cell line, HIT-T15, visualized by high resolution electron tomography. Proc Natl Acad Sci U S A. 2001;98:2399–2406. doi: 10.1073/pnas.051631998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marsh BJ, Volkmann N, McIntosh JR, Howell KE. Direct continuities between cisternae at different levels of the Golgi complex in glucose-stimulated mouse islet beta cells. Proc Natl Acad Sci U S A. 2004;101:5565–5570. doi: 10.1073/pnas.0401242101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matsko N, Mueller M. AFM of biological material embedded in epoxy resin. J Struct Biol. 2004;146:334–343. doi: 10.1016/j.jsb.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 64.Matsko NB. Atomic force microscopy applied to study macromolecular content of embedded biological material. Ultramicroscopy. 2007;107:95–105. doi: 10.1016/j.ultramic.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 65.McDonald KL, Morphew M, Verkade P, Muller-Reichert T. Recent advances in high-pressure freezing: equipment- and specimen-loading methods. Methods Mol Biol. 2007;369:143–173. doi: 10.1007/978-1-59745-294-6_8. [DOI] [PubMed] [Google Scholar]
- 66.Merchan-Pèrez A, Rodriguez J-R, Alonso-Nanclares L, Schertel A, DeFelipe J. Counting synapses using FIB/SEM microscopy: a true revolution for ultrastructural volume reconstruction. Front Neuroanat. 2009;3:18. doi: 10.3389/neuro.05.018.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mercogliano CP, DeRosier DJ. Concatenated metallothionein as a clonable gold label for electron microscopy. J Struct Biol. 2007;160:70–82. doi: 10.1016/j.jsb.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Millonig G, Marinozzi V. Fixation and embedding in electron microscopy. Advances in Optical and Electron Microscopy. 1968;2:251. [Google Scholar]
- 69.Mizuhira V, Futaesaku Y. New fixation method for biological membranes using tannic acids. Acta Histochem Cytochem. 1972;5:233–236. [Google Scholar]
- 70.Mollenhauer HH. Artifacts caused by dehydration and epoxy embedding in transmission electron microscopy. Microsc Res Tech. 1993;26:496–512. doi: 10.1002/jemt.1070260604. [DOI] [PubMed] [Google Scholar]
- 71.Monaghan P, Cook H, Hawes P, Simpson J, Tomley F. High-pressure freezing in the study of animal pathogens. J Microsc. 2003;212:62–70. doi: 10.1046/j.1365-2818.2003.01245.x. [DOI] [PubMed] [Google Scholar]
- 72.Moor H. Theory and practice of high pressure freezing. In: Steinbrecht RA, Zierold K, editors. Cryotechniques in biological electron microscopy. Berlin: Springer; 1987. pp. 175–191. [Google Scholar]
- 73.Muller-Reichert T, Hohenberg H, O’Toole ET, McDonald K. Cryoimmobilization and three-dimensional visualization of C. elegans ultrastructure. J Microsc. 2003;212:71–80. doi: 10.1046/j.1365-2818.2003.01250.x. [DOI] [PubMed] [Google Scholar]
- 74.Murray S. High pressure freezing and freeze substitution of Schizosaccharomyces pombe and Saccharomyces cerevisiae for TEM. Methods Cell Biol. 2008;88:3–17. doi: 10.1016/S0091-679X(08)00401-9. [DOI] [PubMed] [Google Scholar]
- 75.Muscariello L, Rosso F, Marino G, Giordano A, Barbarisi M, Cafiero G, Barbarisi A. A critical overview of ESEM applications in the biological field. J Cell Physiol. 2005;205:328–334. doi: 10.1002/jcp.20444. [DOI] [PubMed] [Google Scholar]
- 76.Norlen L. Exploring skin structure using cryo-electron microscopy and tomography. Eur J Dermatol. 2008;18:279–284. doi: 10.1684/ejd.2008.0387. [DOI] [PubMed] [Google Scholar]
- 77.O’Connell MK, Murthy S, Phan S, Xu C, Buchanan J, Spilker R, Dalman RL, Zarins CK, Denk W, Taylor CA. The three-dimensional micro- and nanostructure of the aortic medial lamellar unit measured using 3D confocal and electron microscopy imaging. Matrix Biol. 2008;27:171–181. doi: 10.1016/j.matbio.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Prenitzer BI, Urbanik-Shannon CA, Giannuzzi LA, Brown SR, Irwin RB, Shofner TL, Stevie FA. The correlation between ion beam/material interactions and practical FIB specimen preparation. Microsc Microanal. 2003;9:216–236. doi: 10.1017/S1431927603030034. [DOI] [PubMed] [Google Scholar]
- 79.Razi M, Tooze SA. Correlative light and electron microscopy. Methods Enzymol. 2009;452:261–275. doi: 10.1016/S0076-6879(08)03617-3. [DOI] [PubMed] [Google Scholar]
- 80.Reimer H, Kohl H. Transmission electron microscopy: physics of image formation. 5. Berlin: Springer; 2008. [Google Scholar]
- 81.Reipert S, Kotisch H, Wysoudil B, Wiche G. Rapid microwave fixation of cell monolayers preserves microtubule-associated cell structures. J Histochem Cytochem. 2008;56:697–709. doi: 10.1369/jhc.7A7370.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ren Y, Donald AM, Zhang Z. Investigation of the morphology, viability and mechanical properties of yeast cells in environmental SEM. Scanning. 2008;30:435–442. doi: 10.1002/sca.20126. [DOI] [PubMed] [Google Scholar]
- 83.Reynolds ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rostaing P, Real E, Siksou L, Lechaire JP, Boudier T, Boeckers TM, Gertler F, Gundelfinger ED, Triller A, Marty S. Analysis of synaptic ultrastructure without fixative using high-pressure freezing and tomography. Eur J NeuroSci. 2006;24:3463–3474. doi: 10.1111/j.1460-9568.2006.05234.x. [DOI] [PubMed] [Google Scholar]
- 85.Sartori A, Gatz R, Beck F, Rigort A, Baumeister W, Plitzko JM. Correlative microscopy: bridging the gap between fluorescence light microscopy and cryo-electron tomography. J Struct Biol. 2007;160:135–145. doi: 10.1016/j.jsb.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 86.Schwartz CL, Sarbash VI, Ataullakhanov FI, McIntosh JR, Nicastro D. Cryo-fluorescence microscopy facilitates correlations between light and cryo-electron microscopy and reduces the rate of photobleaching. J Microsc. 2007;227:98–109. doi: 10.1111/j.1365-2818.2007.01794.x. [DOI] [PubMed] [Google Scholar]
- 87.Silva MT, Guerra FC, Magalhaes MM. The fixative action of uranyl acetate in electron microscopy. Experientia. 1968;24:1074. doi: 10.1007/BF02138757. [DOI] [PubMed] [Google Scholar]
- 88.Soto GE, Young SJ, Martone ME, Deerinck TJ, Lamont S, Carragher BO, Hama K, Ellisman MH. Serial section electron tomography: a method for three-dimensional reconstruction of large structures. Neuroimage. 1994;1:230–243. doi: 10.1006/nimg.1994.1008. [DOI] [PubMed] [Google Scholar]
- 89.Stein O, Stein Y. Light and electron microscopic radioautography of lipids: techniques and biological applications. Adv Lipid Res. 1971;9:1–72. doi: 10.1016/b978-0-12-024909-1.50008-9. [DOI] [PubMed] [Google Scholar]
- 90.Steinbrecht RA, Muller M. Freeze substitution and freeze drying. In: Steinbrecht RA, Zierold K, editors. Cryotechniques in biological electron microscopy. Berlin: Springer; 1987. pp. 87–113. [Google Scholar]
- 91.Stokes DJ. Recent advances in electron imaging, image interpretation and applications: environmental scanning electron microscopy. Philos Transact A Math Phys Eng Sci. 2003;361:2771–2787. doi: 10.1098/rsta.2003.1279. [DOI] [PubMed] [Google Scholar]
- 92.Stokes DJ, Rea SM, Best SM, Bonfield W. Electron microscopy of mammalian cells in the absence of fixing, freezing, dehydration, or specimen coating. Scanning. 2003;25:181–184. doi: 10.1002/sca.4950250404. [DOI] [PubMed] [Google Scholar]
- 93.Studer D, Humbel BM, Chiquet M. Electron microscopy of high pressure frozen samples: bridging the gap between cellular ultrastructure and atomic resolution. Histochem Cell Biol. 2008;130:877–889. doi: 10.1007/s00418-008-0500-1. [DOI] [PubMed] [Google Scholar]
- 94.Tokuyasu KT. A technique for ultracryotomy of cell suspensions and tissues. J Cell Biol. 1973;57:551–565. doi: 10.1083/jcb.57.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tokuyasu KT, Singer SJ. Improved procedures for immunoferritin labeling of ultrathin frozen sections. J Cell Biol. 1976;71:894–906. doi: 10.1083/jcb.71.3.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Donselaar E, Posthuma G, Zeuschner D, Humbel BM, Slot JW. Immunogold labeling of cryosections from high-pressure frozen cells. Traffic. 2007;8:471–485. doi: 10.1111/j.1600-0854.2007.00552.x. [DOI] [PubMed] [Google Scholar]
- 97.Verkade P. Moving EM: the Rapid Transfer System as a new tool for correlative light and electron microscopy and high throughput for high-pressure freezing. J Microsc. 2008;230:317–328. doi: 10.1111/j.1365-2818.2008.01989.x. [DOI] [PubMed] [Google Scholar]
- 98.Walther P. High-resolution cryo-SEM allows direct identification of F-actin at the inner nuclear membrane of Xenopus oocytes by virtue of its structural features. J Microsc. 2008;232:379–385. doi: 10.1111/j.1365-2818.2008.02109.x. [DOI] [PubMed] [Google Scholar]
- 99.Watson ML. Staining of tissue sections for electron microscopy with heavy metals. II. Application of solutions containing lead and barium. J Biophys Biochem Cytol. 1958;4:727–730. doi: 10.1083/jcb.4.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Webster P. Microwave-assisted processing and embedding for transmission electron microscopy. Methods Mol Biol. 2007;369:47–65. doi: 10.1007/978-1-59745-294-6_4. [DOI] [PubMed] [Google Scholar]
- 101.Weimer RM. Preservation of C. elegans tissue via high-pressure freezing and freeze-substitution for ultrastructural analysis and immunocytochemistry. Methods Mol Biol. 2006;351:203–221. doi: 10.1385/1-59745-151-7:203. [DOI] [PubMed] [Google Scholar]
- 102.Wendt KD, Jensen CA, Tindall R, Katz ML. Comparison of conventional and microwave-assisted processing of mouse retinas for transmission electron microscopy. J Microsc. 2004;214:80–88. doi: 10.1111/j.0022-2720.2004.01310.x. [DOI] [PubMed] [Google Scholar]
- 103.White J, Southgate E, Thomson J, Brenner S. The structure of the nervous system of the nematode C. elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 104.Williams GS, Collinson LM, Brzostek J, Eissmann P, Almeida CR, McCann FE, Burshtyn D, Davis DM. Membranous structures transfer cell surface proteins across NK cell immune synapses. Traffic. 2007;8:1190–1204. doi: 10.1111/j.1600-0854.2007.00603.x. [DOI] [PubMed] [Google Scholar]
- 105.Zechmann B, Zellnig G. Microwave-assisted rapid plant sample preparation for transmission electron microscopy. J Microsc. 2009;233:258–268. doi: 10.1111/j.1365-2818.2009.03116.x. [DOI] [PubMed] [Google Scholar]
- 106.Zenner HL, Collinson LM, Michaux G, Cutler DF. High-pressure freezing provides insights into Weibel–Palade body biogenesis. J Cell Sci. 2007;120:2117–2125. doi: 10.1242/jcs.007781. [DOI] [PubMed] [Google Scholar]
- 107.Zuber B, Chami M, Houssin C, Dubochet J, Griffiths G, Daffe M (2008) Direct visualization of the outer membrane of native mycobacteria and corynebacteria. J Bacteriol: JB.01919-07 [DOI] [PMC free article] [PubMed]