Abstract
Both over expression of cyclic AMP response element binding protein (CREB) in the nucleus accumbens (NAc), and intra-accumbal injection of cocaine- and amphetamine-regulated transcript (CART) peptides, have been shown to decrease cocaine reward. Also, over expression of CREB in the rat NAc increased CART mRNA and peptide levels, but it is not known if this was due to a direct action of P-CREB on the CART gene promoter. The goal of this study was to test if CREB and P-CREB bound directly to the CRE site in the CART promoter, using chromatin immunoprecipitation (ChIP) assays. ChIP assay with anti-CREB antibodies showed an enrichment of the CART promoter fragment containing the CRE region over IgG precipitated material, a non-specific control. Forskolin, which was known to increase CART mRNA levels in GH3 cells, was utilized to show that the drug increased levels of P-CREB protein and P-CREB binding to the CART promoter CRE-containing region. A region of the c-Fos promoter containing a CRE cis-regulatory element was previously shown to bind P-CREB, and it was used here as a positive control. These data suggest that the effects of CREB over expression on blunting cocaine reward could be, at least in part, attributed to the increased expression of the CART gene by direct interaction of P-CREB with the CART promoter CRE site, rather than by some indirect action.
Keywords: CART, CREB, Cocaine, Reward, ChIP
1. Introduction
Cocaine- and amphetamine-regulated transcript (CART) was discovered as an mRNA that was increased in the ventral striatum of rats after an acute injection of psychostimulants [1, 2]. While the increase was not always found [3–6], recent studies demonstrated that acute cocaine injections dose- and time-dependently increased c-Fos immunoreactivity in CART-containing cells of the rat nucleus accumbens (NAc), a key brain nucleus in the reward pathway [7]. c-Fos expression in neurons is regulated by extracellular stimuli and considered to represent neuronal activation [7, 8]; thus cocaine activated CART-containing neurons in the rat NAc. Additional studies have shown interactions between cocaine and CART. After a binge regimen of cocaine administration in rats [3, 9, 10], and after self-administration in Rhesus monkeys [11], cocaine increased CART mRNA levels in the amygdala (rat and monkey) and NAc (rat). Human cocaine overdose victims also had up regulated CART mRNA levels in the NAc [12].
CART peptides appeared to function in the NAc to counteract the behavioral effects of cocaine, amphetamine and dopamine [13–16]. For example, rat self-administration studies showed that intra-NAc injections of CART peptides reduced the breakpoint, or the amount of work rats would do in order to receive an injection of cocaine [15]. In reducing the breakpoint for cocaine self-administration, but not for food or sucrose, CART peptides blunted the reinforcing properties of cocaine. Thus it has been hypothesized that CART peptides in the NAc are homeostatic regulators of dopamine-induced activities [17].
Experiments have also found an association between cocaine and CREB. CREB activity was up-regulated in the NAc by cocaine, and its over expression resulted in increases in serine 133 phospho-CREB (P-CREB) levels that were associated with a reduction in cocaine reward [18]. Thus, a leading hypothesis is that CREB activity in the NAc regulates motivational aspects of drug addiction via homeostatic, negative feedback adaptations modulated by CREB target gene proteins which decrease reward [19]. Interestingly, the CART gene contains a CRE cis-regulatory site in its proximal promoter that was shown to bind CREB in nuclear extracts from the rat NAc in vitro [20], and, as noted above, intra-accumbal CART peptides were shown to decrease cocaine reward [14, 15] just as CREB does. In addition, over expression of CREB in the rat NAc increased CART mRNA and peptide levels [20]; thus CART is a CREB-regulated gene in the NAc and other tissues. We hypothesized that CREB blunts the rewarding properties of cocaine, at least in part, by increasing the expression of CART peptides, which also blunt behavioral responses to psychostimulants [15, 20].
The question addressed here is: are the effects of P-CREB on CART mRNA and peptide levels due to a direct action of P-CREB at the CART promoter CRE site, or to indirect actions through neuronal and subcellular mechanisms. While a direct interaction between P-CREB and the CART gene promoter has been hypothesized [6, 20–26] but not yet demonstrated, and because most CART gene regulation studies were carried out with linearized luciferase plasmid constructs driven by 1 kilobase or less of the CART promoter in vitro [21, 24, 26], we tested the hypothesis that CREB and its transcriptionally active form, P-CREB, were able to bind directly to the rat CART promoter CRE cis-regulatory site in the native chromatin of live cells. The technology used to test the hypothesis was chromatin immunoprecipitation (ChIP), a technique that identifies protein-DNA interactions within the chromatin of genomic DNA in live cells. Unfortunately, ChIP assays for brain tissue have not yet been fully optimized [27]; thus this research was performed with cultured GH3 cells previously shown to express CART mRNA and to exhibit CART promoter-driven luciferase after stimulation of the PKA-CREB pathway by forskolin [21, 24].
2. Results
Chromatin immunoprecipitation (ChIP) assays identified appropriate chromatin fragments from the CART and c-Fos genes
Several experiments were carried out to validate the feasibility of using the ChIP assay in GH3 cells. Table 1 shows the forward and reverse primers used in quantitative, real-time PCR reactions to amplify DNA enriched during ChIP assays. The table specifies the nucleotide sequences and the predicted amplicon lengths of the PCR products in ChIP assays. The c-Fos promoter was included because it was previously shown to bind P-CREB in ChIP assays [27], and the c-Fos gene was also up-regulated by forskolin treatment in GH3 cells [28]. Therefore, it was used in this study as a positive control. Figure 1 illustrates how the CART gene promoter consensus CRE DNA cis-regulatory element is located between the flanking primers. The CRE site, the TATA box, and +1 site of transcription initiation are identified in bold.
Table 1.
Primers used to amplify the CRE-containing regions of the CART and c-Fos promoters,
Table 1. Shown are: the sequences of each pair of forward and reverse primers from 5' to 3', the real-time PCR predicted amplicon sizes, and each gene's Genbank accession number. See text for additional details.
Figure 1. Genomic DNA sequence of the rat CART proximal promoter region.
Shown is the nucleotide sequence of the rat CART gene promoter (Genbank accession no. AF519794) originally identified and published by Barrett and colleagues [21] as well as the Genbank nucleotide numbering in the left-hand margin. The PCR primer forward and reverse sequences (corresponding to DNA sequences that are underlined, bolded and labeled as "5'-Start” and "3'-End", respectively) were recommended by Primer Express v3.0 software (Applied Biosystems, Foster City, CA). The CART gene promoter consensus CRE DNA cis-regulatory element is identified in bold and located between the flanking primers. That region of the promoter was amplified in PCR reactions. Both the TATA box necessary to initiate promoter-driven transcription and the +1 site of CART gene transcriptional initiation are also delineated in bold to orient the reader.
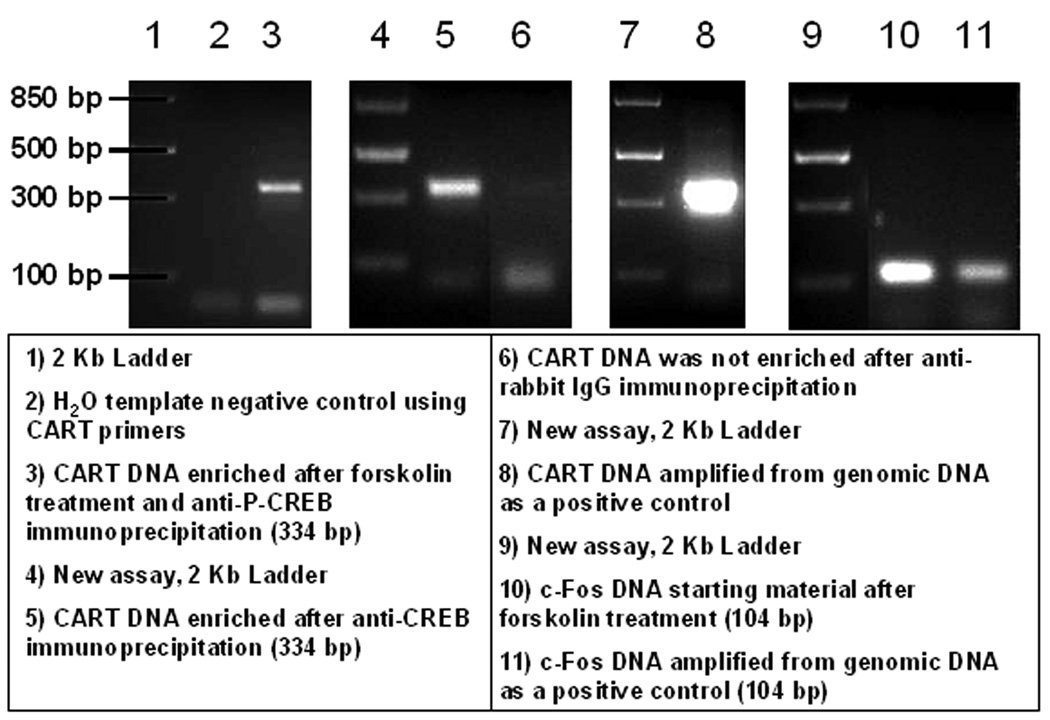
To verify that amplicons generated in PCR reactions were of the predicted sizes (334 base pairs for CART and 104 for c-Fos), PCR reaction products were loaded into 2% agarose gels supplemented with ethidium bromide and separated by electrophoresis. Figure 2 shows a composite figure of several groups of separate assays showing gels of PCR amplicons. New/separate groups of assays are partitioned in figure 2 by 2 kb DNA standards. CART promoter fragments of the predicted size of 334 base pairs (bp) were found after: forskolin treatment of GH3 cells and anti-P-CREB immunoprecipitation (IP) (lane 3); anti-CREB IP (lane 5); and utilization of GH3 genomic DNA (gDNA) isolated with a genomic DNA isolation kit from Qiagen (Valencia, CA; lane 8). The 334bp amplicons correlated with melting temperatures (Tm's) around 87.5°C (Figures 3C and 4C). Lane 6 of figure 2 did not contain a CART promoter fragment at 334bp after PCR with CART primers because anti-rabbit IgG IP was a non-specific precipitator that did not selectively target the CART promoter. c-Fos promoter fragments were of the predicted size of 104 base pairs (lanes 10 and 11), with melting temperatures around 80.5°C (Figure 5C). Thus, these data indicated that the CART and c-Fos promoter primers produced the appropriate and expected PCR amplicons.
Figure 2. Composite figure of gels of DNA enriched in chromatin immunoprecipitation (ChIP) assays by anti-CREB and -P-CREB antibodies and amplified in PCR reactions.
Four groups of lanes from agarose gels are shown and they correspond to four separate experiments. The experiments were used to verify that PCR amplicons produced by CART and c-Fos were their predicted sizes. Some non-specific amplification was present below 100 base pairs in lanes 3, 5, 6 and 8, which were likely primer dimers that arose because of low template DNA quantities. See figure and text for additional details.
Figure 3. Enrichment of the CART promoter CRE-containing region by ChIP assays after immunoprecipitation with a CREB-specific antibody.
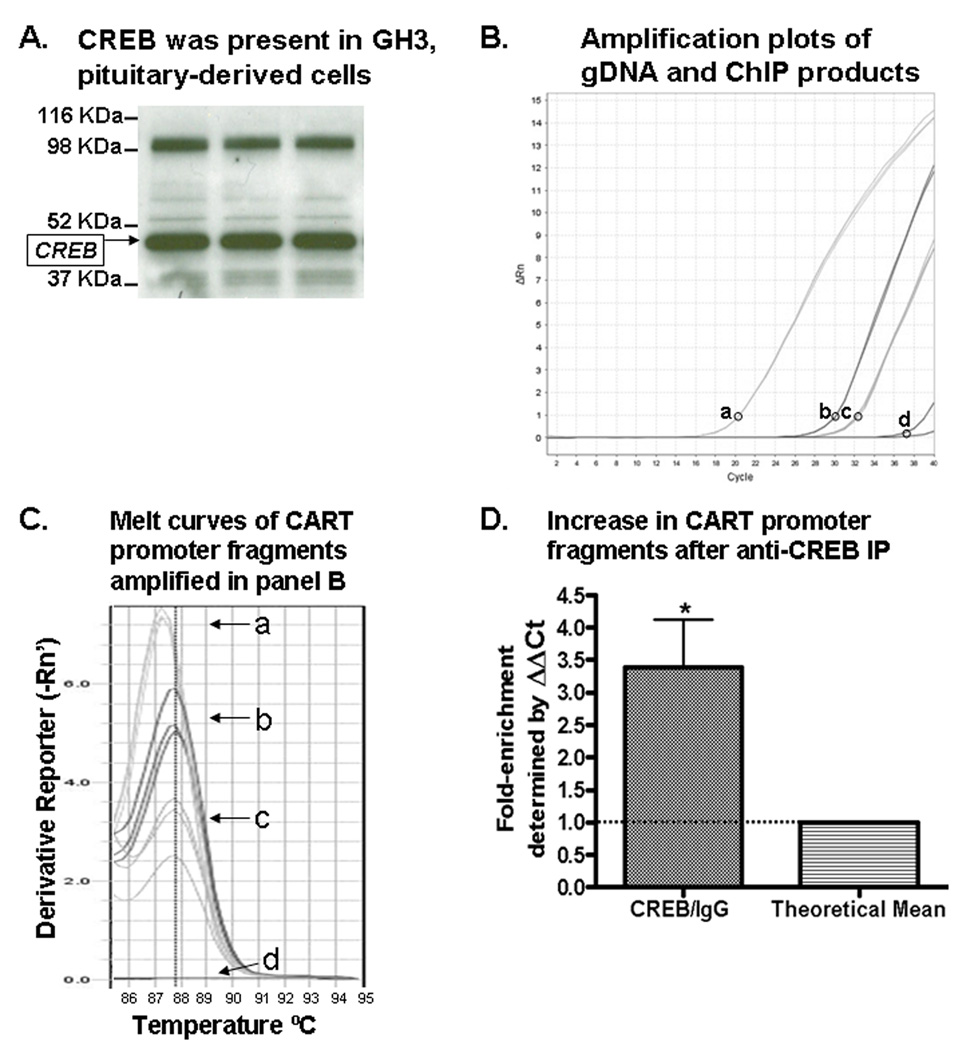
(A) Western blot analysis identified a CREB immunoreactive band from GH3 cell lysates (shown in triplicate), indicated by an arrow on the left of the gel. (B) Shown are representative PCR amplification plots of GH3 genomic DNA amplified by CART primers (a); anti-CREB IP DNA amplified by CART primers (b); IgG IP DNA amplified by CART primers (c); and H2O-template negative control PCR reactions amplified by CART primers (d). The circles next to the letters in panel B identify the y-axis position of the cycle threshold (Ct) setting used in relative quantification calculations to determine fold-enrichment values. (C) Melting temperatures (Tm's) were identified at the peak of the curves. Lettered curves correspond to lettered samples in B. (D) CART promoter fragments isolated by anti-CREB IP were more abundant than those isolated with IgG IP (used as a non-specific IP control), as expected, indicating that CREB was bound to the CART promoter region containing the CRE cis-regulatory element. See text for additional details.
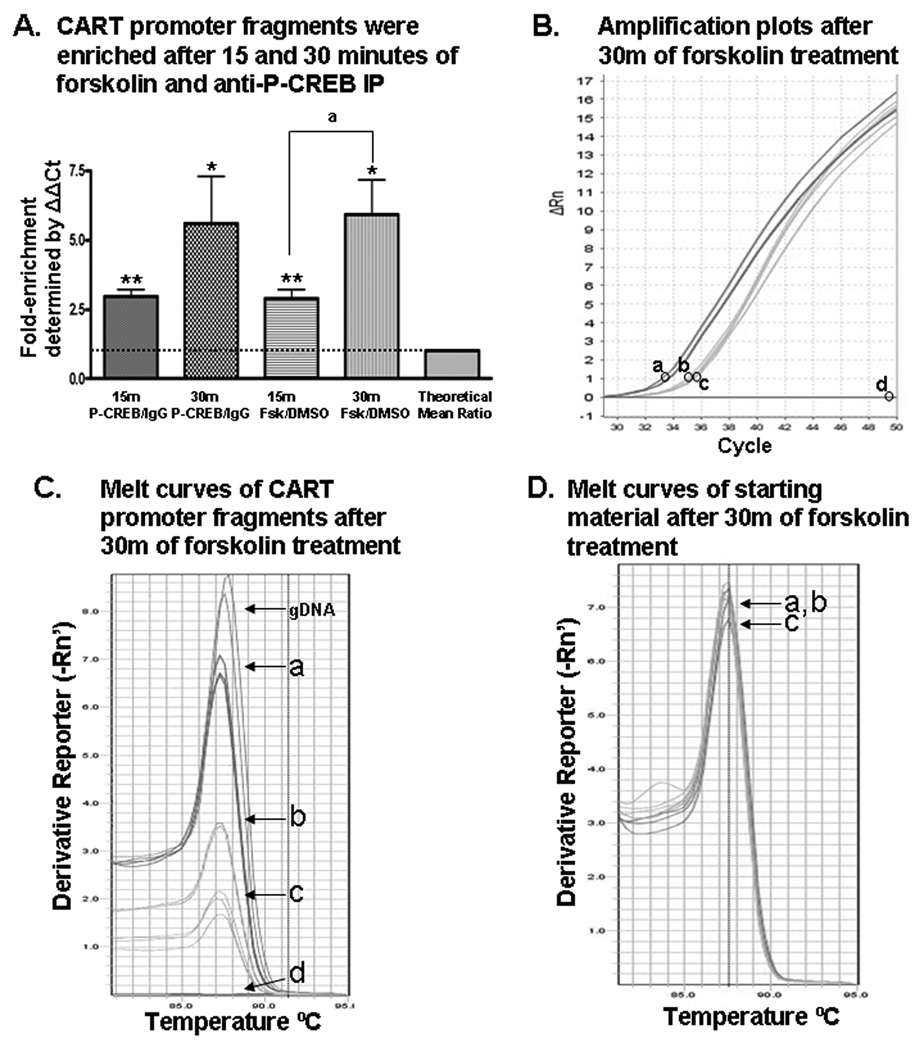
Figure 4. Forskolin stimulation enhanced P-CREB binding to the CART gene promoter region containing the CRE cis-regulatory element as determined by ChIP assays.
(A) After 15 and 30 minutes (15m and 30m) of forskolin treatment, CART promoter fragments were enriched in GH3 cell lysates after anti-P-CREB immunoprecipitations (IP) compared to DNA from cell lysates immunoprecipitated with IgG. *p < 0.05, one-sample t-test; **p < 0.001, one-sample t-test; ap < 0.05, two-sample t-test. (B) Shown is a representative PCR amplification plot of CART promoter fragments enriched after: 30 min of forskolin and anti-P-CREB IP (a); DMSO and anti-P-CREB IP (b); DMSO and IgG IP (c); and H2O-template negative control PCR reactions amplified with CART primers (d). A slight variation in one of the replicates of sample (a) makes it appear as two separate lines. The circles next to the letters in panel B indicate the y-axis position of the Ct setting used in relative quantitation calculations to determine fold-enrichment values. (C) The melt curves after 30 minutes of forskolin or DMSO treatment showed that melting temperature (Tm) values for amplicons amplified with CART primers were around 87.5°C, confirming that the PCR amplicons were the predicted size of 334 base pairs. Melt curves after 15 minutes of treatment (not shown) also revealed Tm values around 87.5°C for DNA enriched by anti-P-CREB IP. (D) In determining the fold-enrichment of CART DNA after anti-P-CREB IP compared to IgG IP, values were determined by normalizing the amount of CART promoter enriched by anti-P-CREB IP or IgG IP to the total amount of CART promoter present in those sample before antibody IP, defined here as the "starting material" (see statistical analysis for more detail). Shown are melt curves for starting material (without IP) amplified by CART primers which also had Tm's around 87.5°C, meaning the DNA fragments amplified in those PCR reactions were the predicted 334 base pairs long (a–c the same as in panel B).
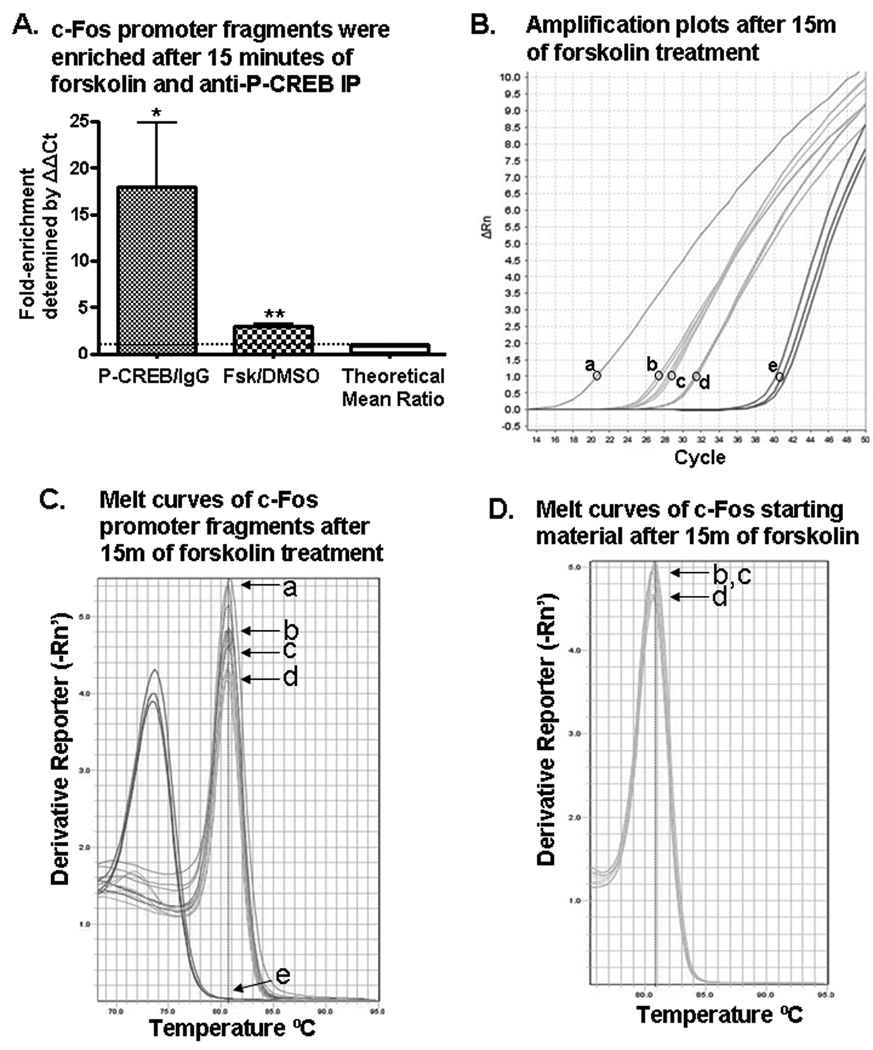
Figure 5. Forskolin stimulation (15 min) enhanced P-CREB binding to the c-Fos gene promoter region containing the CRE cis-regulatory element.
(A) After 15 minutes of forskolin treatment of GH3 cells and anti-P-CREB immunoprecipitations (IP), c-Fos promoter fragments were enriched compared to DNA from forskolin or vehicle (DMSO) treated cells immunoprecipitated with IgG. gDNA (a); forskolin treatment and anti-P-CREB IP (b); DMSO treatment and anti-P-CREB IP (c); forskolin treatment and IgG IP (d); and H2O-template negative control PCR reactions with c-Fos primers (e). (B) Shown are representative PCR amplification plots. (C) The melt curves after 15 minutes of forskolin or DMSO treatment showed that melting temperature (Tm) values for DNA isolated by antibody IP's and gDNA amplicons were around 80.5°C, confirming the PCR amplicons were the predicted size of 104 base pairs. Letters correspond to samples in 5B. (D) Starting material amplified by c-Fos promoters, which was used to normalize the quantity of DNA enriched by antibody IP’s, also had melting temperatures around 80.5°C, meaning the DNA fragments amplified in PCR reactions were the predicted 104 base pairs long (b–d the same as in panel B). Starting material was the amount of c-Fos promoter present in the GH3 cell lysate of each ChIP sample before antibody IP. See text for additional details.
Verification that GH3 cells contained CREB, P-CREB, and the CART gene promoter region containing the CRE site
To verify that GH3 cells contained CREB transcription factors, Western blot analyses were performed and CREB was identified (Figure 3A). Also, Western blots identified the presence of P-CREB using an antibody for the phosphorylated serine 133 site on the P-CREB protein (Figure 6A). To verify that GH3 cells contained the CART gene and that our assay would amplify the promoter region containing the CRE cis-regulatory site, GH3 cell gDNA was amplified in real time PCR reactions using CART primers (Figure 3B, a). The amplicons were the predicted size of 334 base pairs (Figure 2) and consistently amplified with Tm's around 87.5° C (Figures 3C and 4C), which confirmed that the Tm of 87.5°C correlated with an amplicon size of 334 base pairs and that the rat CART gene promoter was present in GH3 cells. These results justified the use of GH3 cells in the following experiments as they expressed CREB, P-CREB and the rat CART gene.
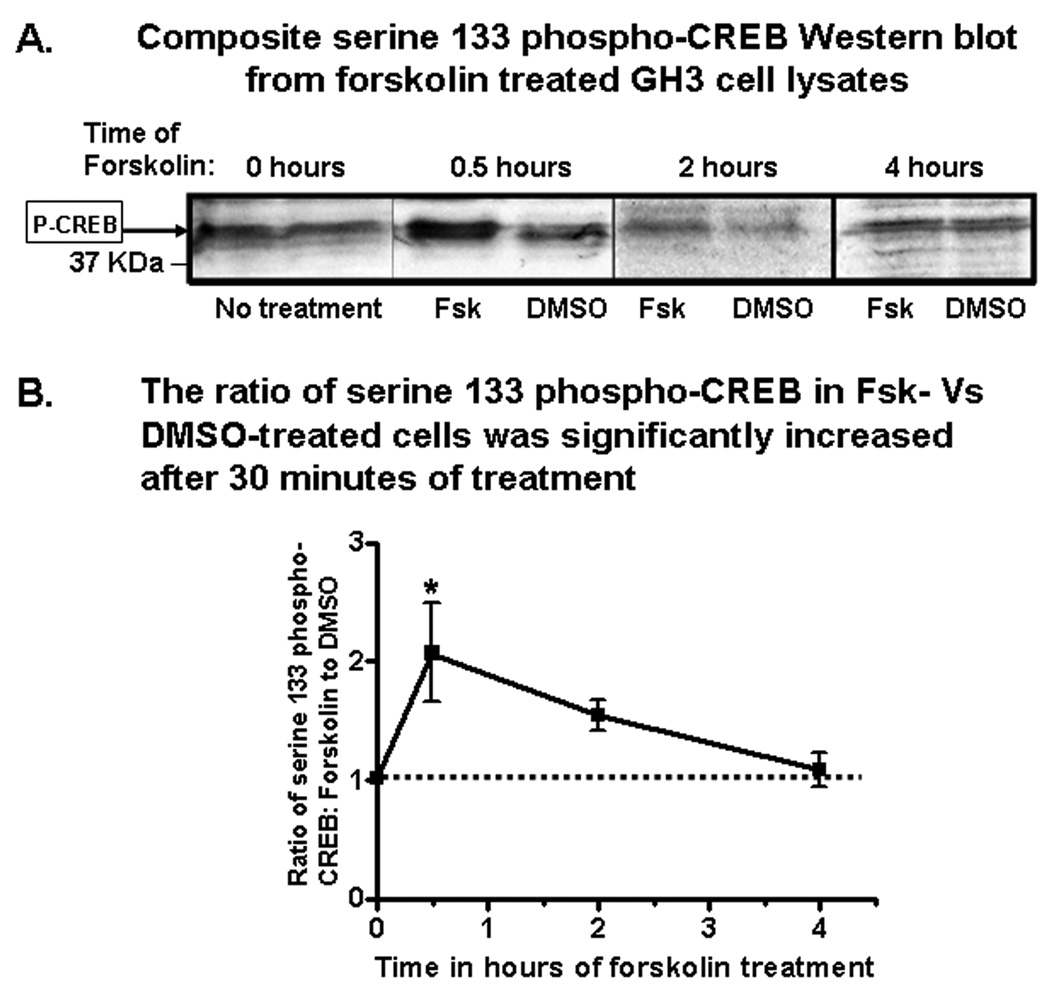
Figure 6. Forskolin time-dependently stimulated P-CREB levels in GH3 cells.
A) Shown is a composite figure of representative P-CREB Western blots from the lysates of paired plates of GH3 cells incubated with either 20µM forskolin or DMSO (vehicle). Immunoreactive P-CREB bands are indicated by an arrow in the left-hand margin of the panel. Blots from the different time points had different raw optical densities because of differences in exposure times. Thus, the ratios of forskolin to DMSO treated pairs of dishes at the individual time points were compared. The mean ratios of P-CREB levels in forskolin treated cells versus DMSO treated cells at different time points are presented graphically in panel (B) (n of 3–5 groups/time point). The asterisk indicates a significantly greater ratio. See text for additional details.
Enrichment of the CART promoter CRE-containing region in GH3 cells by Chromatin immunoprecipitation (ChIP) with a CREB-specific antibody
ChIP assays were carried out as described in Experimental Procedures. Precipitation with CREB antibodies isolated more of the CART promoter region containing the CRE cis-regulatory element than did precipitation with anti-rabbit IgG antibodies (IgG), which was a non-specific precipitator used as a control (Figure 3). The data in figure 3B show representative PCR amplification plots. The lower cycle threshold (Ct) value of sample (b) compared to sample (c) in figure 3B meant that more CART DNA was enriched by anti-CREB IP compared to IgG IP.
Melt curves in figure 3C showed that the DNA isolated by anti-CREB IP and IgG IP and amplified by CART promoter primers had Tm's of approximately 87.5°C, which were similar to the Tm of gDNA subjected to PCR using CART primers. Slight variations in Tm values ± 1°C were normal due to small differences in the general conditions of the final PCR products such as salt and DNA amplicon concentrations at the end of the PCR run (AB technical services, Carlsbad, CA, personal communication). Data in figure 2 confirmed that those Tm values around 87.5°C corresponded to the predicted CART promoter amplicon size of 334 bp. In contrast, the Tm's for some amplicons of IgG IP DNA and H2O were approximately 75°C, which meant that IgG IP did not significantly enrich CART promoter DNA and that the CART promoter primers did not amplify the CART gene from H2O, an important control for primer specificity.
Real-time PCR data were quantified by the StepOne Plus real-time PCR system software (Applied Biosciences, Carlsbad, CA) using the ΔΔCt method. In three independent ChIP assays with each antibody, the mean fold-enrichment ratio of the CART promoter CRE region after anti-CREB IP was 3.383 ± 0.735 (mean ± SEM) compared to IgG control IP (p < 0.05 one sample t-test, n = 3 ratios; Figure 3D). The fold-enrichment ratio greater than 1.0 signified that a greater amount of the CART promoter was isolated by anti-CREB IP compared to IgG IP. Those results indicated that CREB transcription factors were bound to the CART promoter region containing the CRE cis-regulatory element.
Western blot analyses verified that forskolin time-dependently increased P-CREB levels in GH3 cells
Because it was previously shown that forskolin increased CART mRNA levels in GH3 cells by a PKA-mediated pathway [24], and we hypothesized that PKA activation increased CART levels via increases in P-CREB protein levels that bound to the CART promoter, Western blot analyses were performed with antibodies specific to P-CREB on pairs of cultured cells that were treated with either 20µM forskolin or DMSO (vehicle) for various time periods (Figure 6A). The raw optical densities of P-CREB immunoreactive bands in blots from different time points were different due to different exposure times, but the ratios of pairs of samples, one treated the other untreated, were consistent and thus used to calculate the fold-stimulation of P-CREB in forskolin treated cells compared to DMSO treated cells.
After 30 minutes of treatment, the ratio of forskolin-stimulated P-CREB levels to the levels of P-CREB in DMSO-treated cells was increased compared to zero hours of treatment (F[3,31] = 3.60, p < 0.05, one-way ANOVA, Dunnette's post-hoc test). The ratios of P-CREB levels in forskolin treated cells compared to DMSO treated cells at each time point were (mean ± SEM,): 0 h = 1.03 ± 0.045 (n = 3); 0.5 h = 2.07* ± 0.416 (*p < 0.05, Dunnette's post-hoc test, n = 5); 2.0 h = 1.55 ± 0.131 (n = 4); and 4.0 h = 1.09 ± 0.140 (n = 4; Figure 6B). Thus, P-CREB levels were significantly higher after 30 minutes of forskolin treatment, but not after 2 or 4 hours. These data showed that forskolin increased the amount of P-CREB protein in GH3 cells and we hypothesized that the previously demonstrated forskolin-regulation of CART mRNA in GH3 cells [22, 24] occurred because of increased P-CREB binding directly to the CART promoter CRE site. That hypothesis was tested in the next set of experiments.
Enrichment of the CART promoter CRE-containing region in GH3 cells by ChIP with a P-CREB-specific antibody; effects of forskolin
P-CREB protein values were increased most by forskolin after 30 minutes (Figure 6). Thus, to determine if forskolin likewise stimulated P-CREB binding directly to the CART promoter region containing the CRE element, ChIP assays were performed with P-CREB-specific antibodies after 15 and 30 minutes of forskolin treatment. After 15 minutes of forskolin treatment, a mean fold-enrichment ratio (P-CREB/IgG) of 2.964 ± 0.265 (mean ± SEM, p < 0.001, one-sample t-test, n = 9 ratios; Figure 4A) was found. In addition, anti-P-CREB IP of cells treated with forskolin for 15 minutes enriched more of the CART promoter than did anti-P-CREB IP of cells treated with 15 minutes of DMSO, where the mean fold-enrichment ratio (Fsk/DMSO) was 2.899 ± 0.316 (mean ± SEM, p < 0.001. one-sample t-test, n = 9 ratios; Figure 4A). These data indicated that P-CREB was bound to the CART promoter after 15 minutes of forskolin treatment and that forskolin stimulated that binding compared to DMSO treatment alone.
The CART promoter fragment containing the CRE cis-regulatory element was also enriched by anti-P-CREB IP after 30 minutes of forskolin treatment compared to IgG IP, where the mean fold-enrichment was 5.590 ± 1.712 (mean ± SEM, p < 0.05, one-sample t-test, n = 12 ratios; Figure 4A). When CART promoter enrichment by anti-P-CREB IP from forskolin treated cells was compared to enrichment from DMSO treated cells, the mean fold-enrichment ratio was approximately the same: 5.934 ± 1.234 (mean ± SEM, p < 0.05, one-sample t-test, n = 12 ratios; Figure 4A). The data indicated that after 15 minutes and 30 minutes of forskolin treatment, forskolin stimulated P-CREB binding to the CART promoter relative to DMSO treatment.
The data in figure 4A also revealed that enrichment of the CART promoter region was time dependent. More P-CREB was bound to the CART promoter after 30 minutes of forskolin treatment than after 15 minutes of forskolin treatment, where the difference in fold-enrichment ratios was 3.3036 ± 1.400 (mean ± SEM, p < 0.05, two-sample t-test). No significant increases were observed after 2, 4 or 6 hours of treatment (data not shown).
The data in figure 4B showed representative PCR amplification plots of CART promoter fragments. The lower Ct value of sample (a) compared to sample (c) in figure 4B meant more CART DNA was enriched by anti-P-CREB IP compared to IgG IP. Furthermore, the lower Ct value of sample (a) compared to sample (b) indicated that forskolin stimulated more P-CREB binding to the CART promoter compared to DMSO-alone treatment.
Data previously shown in figure 2, lane 3 confirmed that the PCR amplicons were the predicted size of approximately 334 base pairs, corresponding to the CART promoter CRE-containing region. Data in figure 4C showed that the DNA isolated by anti-P-CREB had Tm's of approximately 87.5°C which were similar to the Tm for gDNA amplified with the same CART primers and they corresponded to the correct amplicon size. The Tm's for some IgG immunoprecipitated DNA amplicons were around 87.5°C, indicating a small amount of non-specific immunoprecipitation of the CART promoter with IgG antibodies. The data in figure 4D showed that Tm values were also 87.5°C for CART promoter fragments amplified from the starting material, which were aliquots of the total DNA present in each sample before immunoprecipitation and used to normalize the amount of DNA enriched after antibody immunoprecipitations of a–c (Figure 4B). See Experimental Procedure for details on starting material.
Enrichment of the c-Fos promoter CRE-containing region by ChIP with a P-CREB-specific antibody; effects of forskolin
c-Fos, a CREB regulated gene previously found in ChIP assays to bind P-CREB at a CRE-containing region of its promoter [27], was used as a positive control in this study. It bound P-CREB, as expected (Figure 5). After fifteen minutes of forskolin stimulation, the mean fold-enrichment ratio (P-CREB/IgG) of the c-Fos promoter by anti-P-CREB IP was 18.01 ± 6.806 (mean ± SEM) compared to IgG IP (p < 0.05, one-sample t-test, n = 8 ratios; Figure 5A). When forskolin treated cells were compared to DMSO treated cells after anti-P-CREB IP, the mean fold-enrichment ratio of c-Fos promoter isolated above DMSO was 2.957 ± 0.354 (mean ± SEM, p < 0.001, one-sample t-test, n = 8 ratios; Figure 5A).
Data in figure 5B show representative PCR amplification plots of c-Fos DNA. The lower Ct value of (b) compared to (c) and (d) indicated that anti-P-CREB IP isolated more of the c-Fos promoter after forskolin stimulation than did exposure to DMSO or IgG IP after forskolin stimulation. Thus, P-CREB was bound to the c-Fos promoter CRE-containing region.
Melt curves in figure 5C showed that the DNA isolated by anti-P-CREB IP and IgG IP had Tm's corresponding to the Tm for gDNA amplified with c-Fos primers of approximately 80.6°C and data in figure 2 previously showed example c-Fos promoter amplicons that had the same Tm that were the predicted size of 104 base pairs. Furthermore, starting material, which was a measure of the total amount of c-Fos promoter present in ChIP samples before antibody IP, had the same Tm around 80.6°C (Figure 5D).
CREB and P-CREB from the rat pituitary gland bound the rat CART promoter CRE cis-element in electrophoretic mobility shift assays and super shift assays in vitro
Having shown that CREB and P-CREB could bind to a region of the CART promoter containing a consensus CRE site in native, chromatin DNA in the nuclei of live pituitary-derived GH3 cells, we tested the hypothesis that CREB and P-CREB made in the rat pituitary itself could specifically bind to the CART CRE site from rat. The rationale was that GH3 cells were pituitary-derived and in order to extend the ChIP findings presented herein to mammalian tissues in vivo, it was important to show that CREB and P-CREB from the pituitary itself could bind to the CART gene promoter CRE site. Thus, electrophoretic mobility shift assays (EMSAs) and antibody super shifts (SS) were performed with rat pituitary nuclear extracts and a radioactively labeled oligonucleotide identical in sequence to the rat CART gene CRE cis-regulatory sequence (Figure 7).
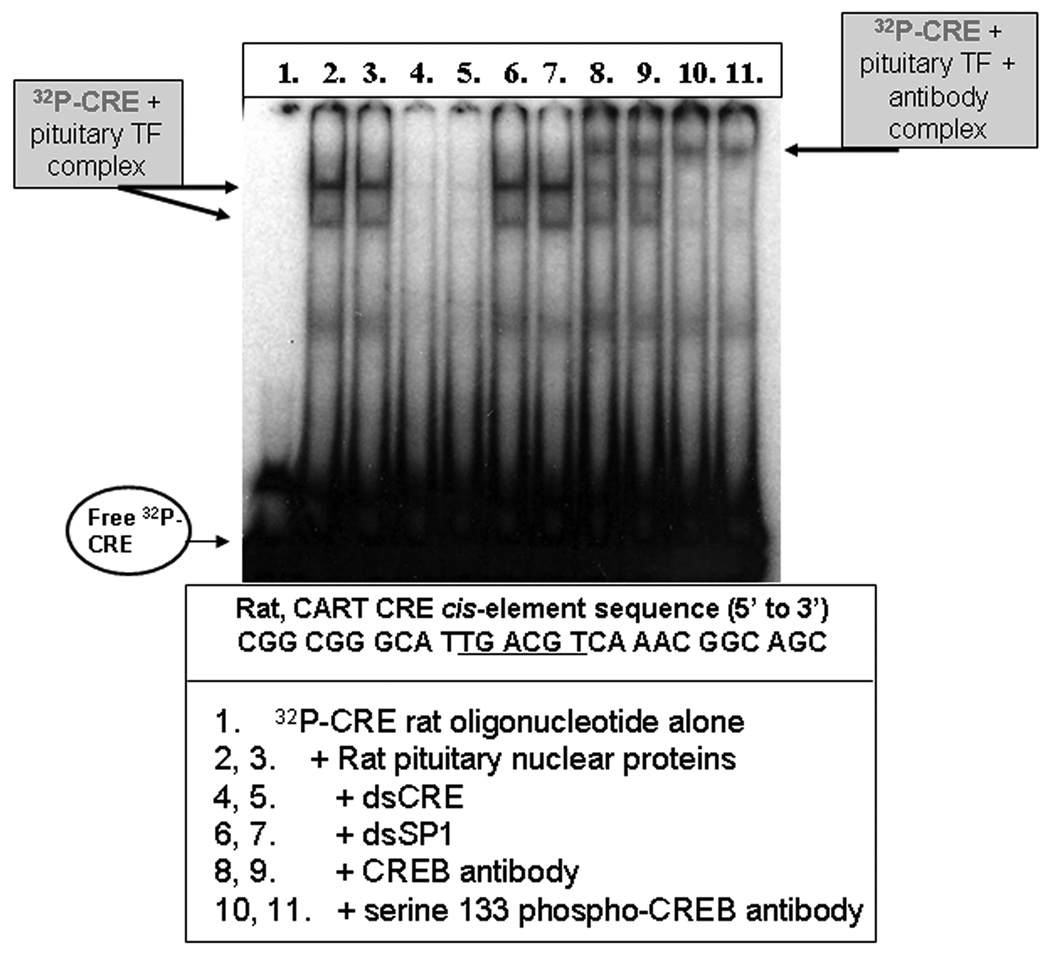
Figure 7. CREB and P-CREB from the rat pituitary gland bound the rat CART CRE cis-regulatory element in EMSA/SS assays.
Nuclear proteins were isolated from rat pituitaries, and binding to the CART CRE DNA cis-regulatory site was observed by electrophoretic mobility shift assays (EMSA) and antibody super shifts (SS) using an oligonucleotide identical in sequence to the rat CART promoter CRE site, and CREB and P-CREB antibodies. See figure and text for additional details.
Nuclear proteins were extracted from the pituitaries of rats and CREB and P-CREB binding to the CART promoter CRE site were observed. The data in figure 7 show: mobility shifts of radiolabeled CRE oligonucleotides (lanes 2 and 3), competition with 50× non-radiolabeled CRE oligonucleotides (lanes 4 and 5), no competition with 50× non-radiolabeled, SP1 oligonucleotides with a sequence unrelated to the CRE oligonucleotide (lanes 6, 7), and antibody super shifts with CREB and P-CREB antibodies (lanes 8–11). The data presented in figure 7 confirmed that the CRE site in the CART promoter was able to bind CREB and P-CREB from the pituitary, and the ChIP data presented in figures 3 and 4 bolstered the hypothesis that CREB and P-CREB may bind to the CART promoter in its histone-bound, chromatin state in the cell nucleus, in vivo.
3. Discussion
A major hypothesis for the role of CREB in drug addiction has been that CREB regulates a specific subset of genes in the brain reward pathway after drug intake, that results in the production of or down regulation of key neuropeptides and neurotransmitters, that modulate DA signaling such that the behavioral responses to those drugs are reduced [29]. CART peptides blunt the effects of psychostimulants in the NAc, and it has been suggested that the CART gene is a CREB target gene and that one mechanism by which CREB produced its anti-cocaine effects was by increasing expression of the CART gene in vivo. We previously showed that CREB can bind to the CRE site in the CART proximal promoter in vitro (Rogge et al 2009). This study, using ChIP assays, demonstrated that CREB and P-CREB interacted directly with the CART promoter in the nuclei of intact cells.
Direct CREB regulation of the CART gene in the nucleus, at the level of the chromatin, was not a given. GC-rich regions directly adjacent to core CRE regulatory sequences, global DNA topology, the presence or absence of enhancer elements and other nearby cis-elements, variations in core CRE site sequences themselves, the position of the CRE site relative to the TATA box and accessibility of the CRE site to TFs such as CREB in the midst of the nuclear chromatin, all affect the affinity of CRE cis-regulatory elements for CREB family TFs [30–32]. As one example, the CRE element is less active in locations more than 100 base pairs upstream of the TATA box and when flanked by GC-rich sequences [32]. In the rat CART gene, the consensus CRE cis-regulatory element was near a GC-rich SP1 cis-regulatory element at nucleotide position −146 from the site of transcriptional initiation [21]. Due to the complexity of CREB-mediated transcription at the level of chromatin, the observations presented in this study that CREB and P-CREB directly interacted with the CART promoter bolstered the hypothesis that CREB may be able to directly regulate CART in vivo.
The data presented in this paper examining P-CREB-DNA promoter interactions in two different CRE-containing genes, c-Fos and CART, revealed a difference in basal P-CREB binding between the two genes. In the CART gene, P-CREB binding to the CRE-containing region was stimulated by forskolin, but in the absence of forskolin, P-CREB was not significantly bound to the CART promoter CRE site. In contrast, the c-Fos gene was previously shown by ChIP to bind P-CREB at the CRE site examined after vehicle-alone treatment [27] and that result was confirmed here. The reason for these differences was not clear, and it would require additional experiments to explain them. Nevertheless, the c-Fos gene was included as a positive control and indeed it was found that P-CREB bound to the c-Fos gene under our assay conditions as expected.
This study utilized GH3 cells because working with brain tissue is much more difficult [27]. But, it was also shown for the first time in this study that CREB and P-CREB extracted from the nuclei of rat pituitary neurons were able to specifically bind to the CRE site on the CART promoter in EMSA/SS analyses, in vitro (Figure 7). Previous studies showed that CREB and P-CREB from the rat NAc could bind to that CRE site [20] and the data presented in this study indicated that in the pituitary, CREB and P-CREB also existed in conformations and dimerization combinations with the ability to bind to the CART promoter CRE site. In conjunction with the ChIP data which showed that CREB and P-CREB were able to bind to the histone-bound CRE site in the GH3 cell nucleus, the fact that pituitary proteins also bound to that site makes it highly likely that CREB can directly bind to the CART gene promoter CRE site in brain in vivo, and regulate the CART gene in the nuclei of neurons while coiled around histones in the nuclear chromatin. Thus, these findings help extend the significance of our results in cell culture to mammalian tissues in vivo.
4. Experimental Procedure
Primer design and quantitative, real-time PCR
Quantitative, real-time PCR reactions were performed according to the manufacturer's directions with the Applied Biosystems Step-One Plus Real-time PCR system (Applied Biosystems, Carlsbad, CA). Primer Express Software v3.0 (Applied Biosystems) selected the PCR primer sequences (Table 1) used in real-time PCR amplifications of the rat CART gene promoter. Primers that amplified a CRE-containing region of the c-Fos promoter were adapted from a previous publication that performed chromatin immunoprecipitation (ChIP) with P-CREB antibodies [27]. In the case of CART primers, the sequences were BLASTED and compared to genome databases for potential non-specific interactions and found to have none. In each experiment, the relative quantities of DNA amplified by real-time PCR were determined by the StepOne Plus software and the ΔΔCt method [33].
DNA electrophoresis
The specificity of DNA enriched by antibody immunoprecipitations in ChIP assays and amplified by PCR were verified by their melting temperatures and amplicon sizes in 2% agarose gels, that were supplemented with ethidium bromide (Fisher Scientific, Pittsburgh, PA) electrophoresed at 100 volts at room temperature for 1 hour, and photographed using ImageOne software (Biorad, Hercules, CA). The two kilobase ladder (Fermentas, Burlington, Ontario, Canada), which was a conglomeration of different-sized DNA fragments ranging from 100 base pairs (bp) to 2,000 bp that were separated by gel electrophoresis, were used to determine the sizes of DNA amplicons.
Tissue culture and drug treatments
Rat pituitary-derived, GH3 cells (ATCC, Manassas, VA) were cultured on 100 cm2 dishes in Ham's F-12 media supplemented with 10% fetal bovine serum (FBS) and 5% penicillin/streptomycin (P/S) (Life technologies, Carlsbad, California). Dishes of cells to be immunoprecipitated with anti-CREB were harvested and paired with cells to be immunoprecipitated with IgG. Cells were grown to 80–85% confluency before a 20–24 hour period of serum-starvation. After serum-starvation, the media was replaced with 10ml of fresh Ham's F-12 media supplemented with 10% FBS and 5% P/S and either 10µl of 20mM 7β-Acetoxy-8,13-epoxy-1α,6β,9α-trihydroxylabd-14-en-11-one (forskolin, Sigma-Aldrich) dissolved in DMSO or 10µl of DMSO alone. Forskolin-treated cells were cross-linked in media supplemented with both 1% formaldehyde and 30nM okadaic acid (Sigma-Aldrich, St Louis, MO).
Chromatin Immunoprecipitation (ChIP) assays
ChIP assays were performed with a kit from Millipore (Billerica, MA) according to the manufacturer's instructions with the following modifications. Approximately 108 GH3 cells in 100 cm2 culture dishes were cross-linked in 1% formaldehyde solution (Fisher Scientific, Pittsburgh, PA) for 20 minutes at 37°C, washed with 1M glycine (Fisher Scientific), 1× phosphate-buffered saline solution (PBS) (Life Technologies) and harvested in 1-ml of 1× PBS + protease inhibitor cocktail + phenylmethylsulfonylfluoride (PMSF) (Sigma-Aldrich, St. Louis, MO). They were then pelleted by centrifugation at 125 × g for 8 minutes, 4°C and the pellet was resuspended in 200µl of fresh ChIP kit SDS lysis buffer + protease inhibitor cocktail + PMSF, rotated at 4°C for 30 minutes, and sonicated using the Sonic Dismembrator model 100 (Fisher Scientific) to shear the DNA to an average length of 300 to 500 base pairs (six, 8-s bursts at 50% maximum output power on ice). Samples were centrifuged at 14,000 × rpm for 20 minutes at 4°C and the supernatant diluted in a fresh 2.0 ml tube with ChIP kit dilution buffer supplemented with 75µl of 1:1 salmon sperm:protein A slurry and rocked at 4°C for 30 minutes. Afterwards, the samples were centrifuged at 1,000 × g for 1 minute, 4°C and the supernatant was transferred to a fresh 2.0 ml tube.
As recommended by Cell Signaling Technology (Boston, MA), CREB immunoprecipitations were carried out with 6 µg of CREB antibodies and P-CREB immunoprecipitations with 0.32µg of anti-P-CREB (all antibodies were from Cell Signaling Technolgy). Purified anti-rabbit IgG was used as a non-specific precipitator and control. After overnight immunoprecipitation on a rotator at 4°C, ChIP samples were then washed and eluted and DNA was un-crosslinked with NaCl and subsequently treated with proteinase K, Tris-Hcl and EDTA. DNA isolation occurred via phenol/chloroform/isoamyl alcohol extraction (Fisher Scientific). The final DNA pellets were resuspended in ultra-pure H2O (Life Technologies) and subjected to real-time PCR.
Animals
Male, Sprague-Dawley rats weighing 250–325g, aged 6–8 weeks were housed on a 7:00 to 19:00 light-dark cycle and fed and watered ad libitum. All animal care and experimentation were performed in accordance with the National Institutes of Health guide for the care and use of laboratory animals with IACUC approval. Rat pituitaries were collected after rats were anesthetized with isofluorane (Abbot, Chicago, IL), decapitated by guillotine, and their brains removed from the skull. The pituitaries were dissected from the base of the skull with pre-chilled forceps and immediately frozen at −80°C for use in EMSA/SS assays.
Electrophoretic mobility shift assay (EMSA)/antibody super shift (SS)
DNA–protein interactions were studied by EMSA/SS. Nuclear protein extracts were separated from cytoplasmic proteins in preparations as described by Xu and Cooper [34]. Total nuclear protein (15 µg) was determined by Bradford assays (Biorad) and incubated for 45 min at room temperature with 2 ng of 32P-5′ end-labeled oligonucleotide which was from the CART promoter containing the CRE cis-regulatory element (5'-CGG CGG GCA TTG ACG TCA AAC GGC AGC-3’), in binding buffer composed of 10 mM Tris–HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA, 5% glycerol, and 0.05 µg/µl Poly[d(I-C)] (Roche, Indianapolis, IN). Oligonucleotides were synthesized and HPLC purified by the Emory Core facility (Emory University, Atlanta, GA). After a 45 minute incubation with unlabeled oligonucleotide at room temperature, the 32P-labeled CART CRE oligonucleotide was added and incubation was continued for another 45 minutes.
In the case of antibody super shift assays, 2 µg of anti-CREB (Cell signaling technology, Boston, MA) or anti-P-CREB antibody (Santa Cruz Biotechnology, CA) were pre-incubated with the nuclear extract for 45 minutes at room temperature followed by incubation with the 32P-labeled CART CRE oligonucleotide for 45 minutes at room temperature.
The 32P-labeled CART CRE oligonucleotide + protein complex was separated by electrophoresis on a 6% non-denaturing (80:1) polyacrylamide gel (1× TBE, 2.5% glycerin). Gels were run at 200 V in the presence of 0.5× TBE buffer for 45 minutes at 4°C. Dried gels were exposed to Kodak BioMax MR Film (Eastman Kodak Company, Rochester, NY) and images were developed and analyzed.
Western blots
GH3 cells were harvested in fresh 1× PBS with protease inhibitor cocktail solution (Sigma-Aldrich), pelleted and resuspended in T-PER Tissue Protein Extraction Reagent (Thermo Scientific, Waltham, MA). Protein content was determined by Bradford assays (Biorad), and equal amounts of each sample were combined with 4× LDS sample buffer and 10× reducing buffer (Life technologies), heated to 70°C for 10 minutes, briefly centrifuged and loaded into 4–20% SDS-PAGE gels (Biorad, Hercules, CA). Electrophoresis occurred at 120 volts, room temperature for about two hours before overnight transfer at 25 volts, 4°C. After transfer, the membranes were blocked with 5% non-fat milk in 1× TBS-T (TBS + 0.1% Tween-20 [pH 7.6]) for 3 hours, and incubated with the primary antibody overnight. The immunoreactivities of CREB and P-CREB transcription factors (approximately 45 KDa) were visualized with the same antibodies used in ChIP assays (Cell Signaling Technology). Immunoreactive signals were detected by using horseradish peroxidase (HRP)-conjugated anti-rabbit IgG antibodies (Cell Signaling Technology) and an enhanced chemiluminescence kit (Amersham, Arlington Heights, IL).
Quantification of data and statistical analyses
All statistical analyses were calculated with Graphpad Prizm software (Graphpad Software, inc., La Jolla, CA). Fold-enrichment of DNA was determined by calculating the ratio of experimental values divided by control values (e.g. DNA enriched after anti-P-CREB IP divided by anti-rabbit IgG IP). All values were determined by the StepOne Plus Real-time PCR system (Applied Biosystems). Fold-enrichment values were averaged together from at least 3 ChIP assays/antibody/drug treatment and subjected to the student's one-sample t-test, which tested the hypothesis that the mean fold-enrichment was greater than 1.0. A fold-enrichment greater than 1.0 determined by p < 0.05 indicated that there was more DNA enriched in a ChIP assay (e.g. anti-P-CREB IP) compared to another ChIP assay (e.g. anti-rabbit IgG IP).
To be sure differences between samples (i.e. anti-P-CREB IP Vs anti-rabbit IgG IP) were not simply due to different amounts of DNA being subjected to the ChIP assay, the amount of DNA immunoprecipitated from a sample was normalized to the total amount of DNA. "Starting material" was defined as the amount of DNA in the same sample before the immunoprecipitation reaction. By normalizing the amount of DNA immunoprecipitated to the total amount of DNA, fold-enrichment values of ChIP products from different culture dishes controlled for possible differences in the total amount of DNA subjected to the ChIP protocol and comparisons of DNA enrichment could be made between samples. Depending on the experiments, the student's two-sample t-test or ANOVAs were used to determine significant differences.
Western blots for P-CREB were performed using pairs of treated vs. nontreated GH3 cells. Working with individual pairs and utilizing the ratios of each pair minimized inter-experimental variability. The ratios from individual time-points were determined by scanning Western blots into computer files and using Scion Image software (NIH, Bethesda, MD) to quantify the optical densities of immunoreactive bands after background subtraction.
Acknowledgements
The authors acknowledge the support of NIH grants RR00165, DA15162, DA015040, and DA021970, as well as helpful discussions with Dr Wei Chen.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature references
- 1.Douglass J, Daoud S. Characterization of the human cDNA and genomic DNA encoding CART: a cocaine- and amphetamine-regulated transcript. Gene. 1996;169(2):241–245. doi: 10.1016/0378-1119(96)88651-3. [DOI] [PubMed] [Google Scholar]
- 2.Douglass J, McKinzie AA, Couceyro P. PCR differential display identifies a rat brain mRNA that is transcriptionally regulated by cocaine and amphetamine. J Neurosci. 1995;15(3 Pt 2):2471–2481. doi: 10.1523/JNEUROSCI.15-03-02471.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunter RG, et al. The effects of cocaine on CART expression in the rat nucleus accumbens: a possible role for corticosterone. Eur J Pharmacol. 2005;517(1–2):45–50. doi: 10.1016/j.ejphar.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 4.Marie-Claire C, et al. Fos but not Cart (cocaine and amphetamine regulated transcript) is overexpressed by several drugs of abuse: a comparative study using real-time quantitative polymerase chain reaction in rat brain. Neurosci Lett. 2003;345(2):77–80. doi: 10.1016/s0304-3940(03)00307-0. [DOI] [PubMed] [Google Scholar]
- 5.Vrang N, Larsen PJ, Kristensen P. Cocaine-amphetamine regulated transcript (CART) expression is not regulated by amphetamine. Neuroreport. 2002;13(9):1215–1218. doi: 10.1097/00001756-200207020-00029. [DOI] [PubMed] [Google Scholar]
- 6.Jones DC, Kuhar MJ. Cocaine-amphetamine-regulated transcript expression in the rat nucleus accumbens is regulated by adenylyl cyclase and the cyclic adenosine 5'-monophosphate/protein kinase a second messenger system. J Pharmacol Exp Ther. 2006;317(1):454–461. doi: 10.1124/jpet.105.096123. [DOI] [PubMed] [Google Scholar]
- 7.Hubert GW, Kuhar MJ. Cocaine administration increases the fraction of CART cells in the rat nucleus accumbens that co-immunostain for c-Fos. Neuropeptides. 2008;42(3):339–343. doi: 10.1016/j.npep.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herdegen T, Waetzig V. AP-1 proteins in the adult brain: facts and fiction about effectors of neuroprotection and neurodegeneration. Oncogene. 2001;20(19):2424–2437. doi: 10.1038/sj.onc.1204387. [DOI] [PubMed] [Google Scholar]
- 9.Brenz Verca MS, et al. Cocaine-induced expression of the tetraspanin CD81 and its relation to hypothalamic function. Mol Cell Neurosci. 2001;17(2):303–316. doi: 10.1006/mcne.2000.0942. [DOI] [PubMed] [Google Scholar]
- 10.Fagergren P, Hurd YL. Mesolimbic gender differences in peptide CART mRNA expression: effects of cocaine. Neuroreport. 1999;10(16):3449–3452. doi: 10.1097/00001756-199911080-00034. [DOI] [PubMed] [Google Scholar]
- 11.Fagergren P, Hurd Y. CART mRNA expression in rat monkey and human brain: relevance to cocaine abuse. Physiol Behav. 2007;92(1–2):218–225. doi: 10.1016/j.physbeh.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 12.Albertson DN, et al. Gene expression profile of the nucleus accumbens of human cocaine abusers: evidence for dysregulation of myelin. J Neurochem. 2004;88(5):1211–1219. doi: 10.1046/j.1471-4159.2003.02247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S, Yoon HS, Kim JH. CART peptide 55–102 microinjected into the nucleus accumbens inhibits the expression of behavioral sensitization by amphetamine. Regul Pept. 2007;144(1–3):6–9. doi: 10.1016/j.regpep.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Jaworski JN, et al. Intra-accumbal injection of CART (cocaine-amphetamine regulated transcript) peptide reduces cocaine-induced locomotor activity. J Pharmacol Exp Ther. 2003;307(3):1038–1044. doi: 10.1124/jpet.103.052332. [DOI] [PubMed] [Google Scholar]
- 15.Jaworski JN, et al. Injection of CART (cocaine- and amphetamine-regulated transcript) peptide into the nucleus accumbens reduces cocaine self-administration in rats. Behav Brain Res. 2008;191(2):266–271. doi: 10.1016/j.bbr.2008.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JH, Creekmore E, Vezina P. Microinjection of CART peptide 55–102 into the nucleus accumbens blocks amphetamine-induced locomotion. Neuropeptides. 2003;37(6):369–373. doi: 10.1016/j.npep.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Rogge G, et al. CART peptides: regulators of body weight, reward and other functions. Nat Rev Neurosci. 2008;9(10):747–758. doi: 10.1038/nrn2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nestler EJ. Historical review: Molecular and cellular mechanisms of opiate and cocaine addiction. Trends Pharmacol Sci. 2004;25(4):210–218. doi: 10.1016/j.tips.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Nestler EJ. Molecular mechanisms of drug addiction. Neuropharmacology. 2004;47 Suppl 1:24–32. doi: 10.1016/j.neuropharm.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 20.Rogge GA, et al. Regulation of CART peptide expression by CREB in the rat nucleus accumbens in vivo. Brain Res. 2009;1251:42–52. doi: 10.1016/j.brainres.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett P, Davidson J, Morgan P. CART gene promoter transcription is regulated by a cyclic adenosine monophosphate response element. Obes Res. 2002;10(12):1291–1298. doi: 10.1038/oby.2002.175. [DOI] [PubMed] [Google Scholar]
- 22.Barrett P, et al. The differential regulation of CART gene expression in a pituitary cell line and primary cell cultures of ovine pars tuberalis cells. J Neuroendocrinol. 2001;13(4):347–352. doi: 10.1046/j.1365-2826.2001.00634.x. [DOI] [PubMed] [Google Scholar]
- 23.Dominguez G, Kuhar MJ. Transcriptional regulation of the CART promoter in CATH.a cells. Brain Res Mol Brain Res. 2004;126(1):22–29. doi: 10.1016/j.molbrainres.2004.02.027. [DOI] [PubMed] [Google Scholar]
- 24.Dominguez G, Lakatos A, Kuhar MJ. Characterization of the cocaine-and amphetamine-regulated transcript (CART) peptide gene promoter and its activation by a cyclic AMP-dependent signaling pathway in GH3 cells. J Neurochem. 2002;80(5):885–893. doi: 10.1046/j.0022-3042.2002.00775.x. [DOI] [PubMed] [Google Scholar]
- 25.Lakatos A, Dominguez G, Kuhar MJ. CART promoter CRE site binds phosphorylated CREB. Brain Res Mol Brain Res. 2002;104(1):81–85. doi: 10.1016/s0169-328x(02)00321-2. [DOI] [PubMed] [Google Scholar]
- 26.de Lartigue G, et al. Cocaine- and amphetamine-regulated transcript: stimulation of expression in rat vagal afferent neurons by cholecystokinin and suppression by ghrelin. J Neurosci. 2007;27(11):2876–2882. doi: 10.1523/JNEUROSCI.5508-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hao H, et al. A fast carrier chromatin immunoprecipitation method applicable to microdissected tissue samples. J Neurosci Methods. 2008;172(1):38–42. doi: 10.1016/j.jneumeth.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herdegen T, Leah JD. Inducible and constitutive transcription factors in the mammalian nervous system: control of gene expression by Jun, Fos and Krox, and CREB/ATF proteins. Brain Res Brain Res Rev. 1998;28(3):370–490. doi: 10.1016/s0165-0173(98)00018-6. [DOI] [PubMed] [Google Scholar]
- 29.McClung CA, Nestler EJ. Regulation of gene expression and cocaine reward by CREB and DeltaFosB. Nat Neurosci. 2003;6(11):1208–1215. doi: 10.1038/nn1143. [DOI] [PubMed] [Google Scholar]
- 30.Carlezon WA, Jr, Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci. 2005;28(8):436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35(4):605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 32.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2(8):599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Xu W, Cooper GM. Identification of a candidate c-mos repressor that restricts transcription of germ cell-specific genes. Mol Cell Biol. 1995;15(10):5369–5375. doi: 10.1128/mcb.15.10.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]