Abstract
RNA transcripts encoding the 2C-subtype of serotonin (5HT2C) receptor undergo up to five adenosine-to-inosine editing events to encode twenty-four protein isoforms. To examine the effects of altered 5HT2C editing in vivo, we generated mutant mice solely expressing the fully-edited (VGV) isoform of the receptor. Mutant animals present phenotypic characteristics of Prader-Willi Syndrome (PWS) including a failure to thrive, decreased somatic growth, neonatal muscular hypotonia, and reduced food consumption followed by post-weaning hyperphagia. Though previous studies have identified alterations in both 5HT2C receptor expression and 5HT2C-mediated behaviors in both PWS patients and mouse models of this disorder, to our knowledge the 5HT2C gene is the first locus outside the PWS imprinted region in which mutations can phenocopy numerous aspects of this syndrome. These results not only strengthen the link between the molecular etiology of PWS and altered 5HT2C expression, but also demonstrate the importance of normal patterns of 5HT2C RNA editing in vivo.
Keywords: RNA Editing, serotonin receptor, Prader-Willi Syndrome, feeding behavior, metabolism, hyperphagia, failure-to-thrive, hypotonia
Introduction
The conversion of adenosine to inosine (A-to-I) by RNA editing is a widespread RNA processing event by which genomically encoded sequences are altered through the site-specific deamination of adenosine residue(s) in precursor and mature mRNA transcripts (Gott and Emeson, 2000). The majority of well characterized A-to-I editing events involve non-synonymous codon changes in RNAs encoding proteins involved in nervous system function including ligand- and voltage-gated ion channels, a G-protein coupled receptor and components of the synaptic release machinery (Burns et al., 1997; Gott and Emeson, 2000; Hoopengardner et al., 2003). Transcripts encoding the 5HT2C receptor can be modified by five A-to-I editing events (sites A-E) to generate as many as 24 protein isoforms that differ by up to three amino acids within the predicted second intracellular loop of the receptor, a region involved in receptor:G-protein coupling (Burns et al., 1997; Pin et al., 1994). Though initial sequence analyses of cDNAs isolated from dissected rat and human brains predicted the region-specific expression of as many as 12 major 5HT2C receptor isoforms encoded by eighteen distinct RNA species (Burns et al., 1997; Niswender et al., 1999), more recent studies identified the expression of 26 of the 32 possible mRNA isoforms and determined that only 4-6 of these mRNAs represent more than 5% of total 5HT2C transcripts in rats and humans, respectively (Dracheva et al., 2009). Alterations in 5HT2C receptor editing have been observed in suicide victims with a history of major depression, schizophrenia, or bipolar disorder (Dracheva et al., 2008; Gurevich et al., 2002; Iwamoto and Kato, 2003; Niswender et al., 2001) and in response to antidepressant and antipsychotic treatment (Englander et al., 2005; Gurevich et al., 2002; Sodhi et al., 2005). The fully-edited (VGV) isoform of the human 5HT2C receptor, encoding valine, glycine and valine at amino acid positions 156, 158 and 160, respectively, exhibits reduced constitutive activity and decreased G-protein coupling efficacy when compared to the genomically-encoded (INI) isoform in heterologous expression systems (Berg et al., 2001; Burns et al., 1997; Fitzgerald et al., 1999; Niswender et al., 1999; Wang et al., 2000), yet the physiologic relevance of 5HT2C RNA editing in nervous system function remains unclear.
Prader-Willi Syndrome is a maternally-imprinted human disorder resulting from a loss of paternal gene expression on chromosome 15q11-13 that is characterized by a complex phenotype including cognitive deficits, infantile hypotonia and failure to thrive, short stature, hypogonadism and hyperphagia which can lead to morbid obesity (Goldstone, 2004; Nicholls and Knepper, 2001). Multiple mouse models with deficiencies of one or more PWS candidate genes have partially correlated individual genes with aspects of the PWS phenotype (Bischof et al., 2007; Cattanach et al., 1992; Ding et al., 2008; Gabriel et al., 1999; Gerard et al., 1999; Muscatelli et al., 2000; Skryabin et al., 2007; Tsai et al., 1999b; Yang et al., 1998). Among these imprinted candidate genes are the brain-specific small nucleolar RNAs (snoRNAs), HBII-13, HBII-85 and HBII-52 (Cavaille et al., 2000). HBII-52 (SNORD115), and its mouse orthologue (MBII-52; Snord115), are members of the box C/D family of snoRNAs that are responsible for directing the 2′-O-methylation of specific ribose moieties in pre-ribosomal RNA transcripts and U small nuclear RNAs (Kiss, 2002). HBII-52/MBII-52 is complementary to an 18 nucleotide segment of 5HT2C mRNA containing three of five editing sites (E, C and D), predicting 2′-O-methylation of the ribose for the adenosine nucleoside at the C-site (Cavaille et al., 2000), an editing position that can significantly affect the function of encoded 5HT2C protein isoforms (Burns et al., 1997; Niswender et al., 1999). HBII-52/MBII-52 also has been observed to affect 5HT2C RNA editing/splicing patterns using tissue culture model systems (Kishore and Stamm, 2006; Vitali et al., 2005). Analyses of 5HT2C transcripts have indicated that site-specific editing is increased in brain samples from both PWS patients (Kishore and Stamm, 2006) and a mouse model of PWS (PWS-ICdel) that further exhibits deficits in specific 5HT2C-mediated behaviors (Doe et al., 2009). Though these studies have correlated changes in both 5HT2C receptor expression and function in the absence of 15q11-13 gene expression, here we show that increased 5HT2C RNA editing in mutant mice recapitulates many aspects of this disorder, suggesting an important role for altered 5HT2C function in the etiology of Prader-Willi Syndrome.
Materials and Methods
Generation of 5HT2C-VGV mice
A genomic fragment containing a portion of the mouse 5HT2C gene was isolated from a 129S6 BAC library (Genome Systems, St. Louis, MO) and a 7.4-kb Avr II restriction fragment containing exon 5 and a portion of the flanking introns was subcloned into a modified pBKSII− vector (Stratagene, La Jolla, CA). The five edited adenosine residues within exon 5 were mutated to guanosine moieties using overlap-extension PCR (Ho et al., 1989). A DNA fragment encoding loxP-flanked neomycin phosphotransferase, under control of the phosphoglycerate kinase (PGK) promoter, was inserted into an endogenous Kpn I site located 184 bases downstream of the 5′-splice site for exon 5. A selection cassette encoding PGK-driven herpes simplex virus thymidine kinase was inserted outside the region of homology as a negative selectable marker. The Not I-linearized targeting vector was introduced into TL-1 embryonic stem cells by electroporation; cells were selected with Geneticin® (G418) and ganciclovir and antibiotic resistant clones were screened for homologous recombination by PCR using the Expand Long Template PCR kit (Roche, USA) using primers located outside the region of homology (sense, 5′-CTGAGTGCATTGGAAAAGAGATCC-3′; antisense, 5′-CCATATATCAGGATGCAGTCTTGTCA-3′). Heterozygous mice obtained from chimeras were mated to mice expressing Cre recombinase under the control of the mouse protamine 1 promoter (The Jackson Laboratory, Bar Harbor, ME) and mutant male offspring were screened for the elimination of the loxP-flanked neomycin resistance cassette. Mutations were verified by DNA sequence analysis and all mice were maintained on the 129S6 strain (Taconic, USA). PCR-based genotyping was employed using primers upstream of the editing sites (5′-AATATCAATAGGTAATTATACC-3′) and downstream of the remaining loxP site within intron 5 (5′-GGGCAAATATTCTGAAAAGATGTT-3′), resulting in the production of 371 and 468 bp amplicons for the wild-type and mutant alleles, respectively. Mutant mice heterozygous for the 5HT2C-VGV allele were mated with wild-type 129S6 animals and subsequent offspring were assessed for the presence of the modified allele and loss of the Cre transgene by segregation.
RNA characterization
Total RNA from male hemizygous 5HT2C-VGV mice and wild-type adult littermates (14 weeks) was isolated from whole brain using Tri-Reagent (Molecular Research Center, Cincinnati, OH) or from dissected brain regions using the PerfectPure RNA tissue kit (5 Prime, Gaithersburg, MD) according to the manufacturer's instructions with all specimens processed in an identical manner. To verify that the introduced A-to-G mutations resulted in the sole production of 5HT2C mRNAs encoding the VGV isoform of the receptor, first-strand cDNA was synthesized using AMV reverse transcriptase (Promega, Madison, WI) and amplified using PCR with primers flanking the duplex region (sense primer 5′-AATATCAATAGGTAATTATACC-3′, antisense primer 5′-GGGCAAATATTCTGAAAAGATGTT-3′). Resultant PCR amplicons from both 5HT2C-VGV and wild-type mice were subjected to direct dideoxynucleotide sequence analysis and subsequently subcloned into pBKSII− (Stratagene, La Jolla, CA) where individual cDNA isolates from each genotype were sequenced.
Quantification of total and alternatively spliced 5HT2C mRNAs was performed using either a ribonuclease (RNase) protection assay as described (Emeson et al., 1989), a semi-quantitative RT-PCR strategy, or real-time RT-PCR. For RNase protection analyses, an antisense riboprobe directed against 5HT2C pre-mRNA, extending from nucleotide −276 to +316 (relative to the first nucleotide of exon 6), resulted in protected fragments of 602 and 316 nt for pre-mRNA and total 5HT2C transcripts, respectively. Three additional antisense riboprobes, specific for alternatively spliced 5HT2C RNA isoforms, were developed to unique regions of exon 5 (RNA 1, RNA 2) or the proximal region of intron 5 (RNA 3) and were contiguous with the first 182 nucleotides of exon 6. A cyclophilin antisense probe was also generated as an internal loading control as previously described (nucleotides +34-144, GenBank accession number M19533) (Singh et al., 2007). The relative expression of protected fragments was quantified using a Typhoon 9400 phosphorimager (GE Healthcare, Piscataway, NJ) with ImageQuant TL software.
Semi-quantitative analysis of 5HT2C mRNA isoform expression by end-point RT-PCR was performed using primers in exons 5 and 6 (sense, 5′-GATATTTGTGCCCCGTC-3′; antisense, 5′-ATCAAAGCTTGACGGCGTAGGACGTAG-3′) and amplified for 30 cycles. Resultant amplicons were resolved on a 2% agarose gel and ethidium bromide fluorescence was quantified by phosphorimager analysis and values were corrected for the length of each amplicon. Individual 5HT2C mRNA isoform expression was determined by summing values corresponding to the RNA1, RNA2, and RNA3 isoforms for each sample and dividing each isoform expression value by this total.
To determine the steady-state mRNA expression levels for 5HT2A, 5HT2B, 5HT2C, and 5HT7 receptors by quantitative RT-PCR, first-strand cDNA was synthesized using random hexamers and subjected to Taqman real-time PCR analysis (Applied Biosystems, Foster City, CA). All primers and probes used for the real-time PCR reactions were products of Assay-On-Demand from Applied Biosystems, (5HT2A, Assay ID Mm00555764_m1; 5HT2B, Assay ID Mm00434123_m1; 5HT2C, Assay ID Mm00434127_m1; 5HT7, Assay ID Mm00434133_m1). Eukaryotic 18S rRNA (product # 4319413E, Applied Biosystems) was included in each multiplex PCR reaction as an internal control. Real-time PCR reactions and subsequent analyses were performed with the ABI Prism 7900HT Sequence Detection System (SDS v2.3, Applied Biosystems). To quantify levels of pro-opiomelanocortin (POMC) and neuropeptide Y (NPY), hypothalamic tissue was dissected by the method of Glowinski and Iverson (Glowinski and Iversen, 1966); total RNA was isolated and subjected to real-time RT-PCR as described above (NPY, Assay ID Mm00445771_m1; POMC, Assay ID Mm00435874_m1). The amplification efficiencies for all reactions were calculated individually using Real-time PCR Miner software and used to determine the average Ct value for each sample (Zhao and Fernald, 2005). To achieve an accurate quantification of target gene expression, target Ct values were normalized to the corresponding 18S rRNA Ct value for each sample.
Protein expression
Male 5HT2C-VGV mice and wild-type littermates were sacrificed at either P2 or 14 weeks of age, whole brains were dissected and immediately frozen on dry ice, followed by manual homogenization in ice-cold sucrose buffer [0.32M sucrose; 10mM K+-HEPES, pH 7.4, complete EDTA-free protease inhibitor (Roche, Piscataway, NJ)]. After whole cells and nuclei were eliminated by centrifugation at 1000 × g for 10 minutes, microsomes were pelleted at 45000 × g for 15 minutes and resuspended in CHAPS lysis buffer (50mM HEPES, 150mM NaCl, 1mM EDTA, 10mM sodium pyrophosphate, 25mM DTT, 1.5% (w/v) CHAPS, complete EDTA-free protease inhibitor, pH 7.5) by triturating 10x through a 21-gauge needle. Insoluble particles were pelleted at 40000 × g for 20 minutes and the resulting supernatant was incubated overnight with a lectin from Triticum vulgaris conjugated to agarose beads (Sigma-Aldrich, St. Louis, MO) at 4°C. Beads were pelleted at 4000 × g for 1 minute, rinsed with wash buffer (50mM HEPES, 150mM NaCl, 0.1% Triton X-100, 10% glycerol, pH 7.5), and pelleted again. Protein was eluted in 2x SDS sample buffer (125 mM Tris-HCl, pH 6.8, 4% SDS, 20% glycerol, 200 mM DTT, and 0.2% bromophenol blue) at 65°C for 5 minutes before pelleting the beads and loading the supernatant on a 10% polyacrylamide gel. Following electrophoretic transfer to nitrocellulose, 5HT2C protein levels were determined by Western blotting analysis using an anti-5HT2C receptor monoclonal antibody (1:250 [v/v], Santa Cruz Biotechnology, Santa Cruz, CA; SR-2C (D-12)) and an anti-β-actin monoclonal antibody (1:10000 [v/v], Sigma-Aldrich, St. Louis, MO; A1978), respectively, followed by an HRP-labeled anti-mouse secondary antibody (1:1500 [v/v], Vector Labs, Burlingame, CA). The secondary antibody was detected with SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific, Rockford, IL).
To quantify receptor expression, saturation binding experiments were performed on dissected frontal cortex, striatum, and hippocampus from adult animals. Following anesthesia with isofluorane, mice were cervically dislocated, decapitated, and whole brains were removed from the cranium. A razor blade was used to coronally block the brain immediately anterior to the optic chiasm. The anterior portion of the striatum was identified in the front tissue block. Landmarks used included the white matter separating it from the overlying cortex and the lateral ventricle separating it from the medial wall of the telenchephalon. The caudate-putamen was removed and reserved, with the remainder of the tissue in this block considered frontal cortex. The two hemispheres of the remaining brain tissue were carefully separated from the dorsal surface using forceps to expose the hippocampus and the posterior portion of the striatum. Forceps were used to gently separate both the left and right entire hippocampal structures as a single unit from the rest of the brain, including CA1, CA2, CA3, and dentate gyrus. Following removal of whole hippocampus, the posterior portions of the striatum were visually identified by the striated nature of the tissue and nearby white matter tracts, carefully isolated with forceps, and added to the anterior striatum previously isolated to complete the striatal sample.
Dissected tissue was mechanically homogenized at 15000 rpm for 10 seconds (Tissue Tearor; Biospec Products, Bartlesville, OK) in 2 ml of standard binding buffer (SBB, 50 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 0.1 mM EDTA) and then centrifuged at 25000 × g for 10 minutes. The supernatant was removed and the pellet was resuspended in 1 ml of SBB and transferred to 1.7 mL eppendorf tubes. The tissue was then spun at 16000 × g in a microcentrifuge for 5 minutes at 4°C. The supernatant was removed and the tissue pellets were frozen at −80°C until used for binding experiments. Total binding was determined using N6-methyl-[3H]-mesulergine (8-point curves were generated using [3H]-mesulergine concentrations of 0.25, 0.5, 1.0, 1.5, 2.0, 4.0, 6.0, and 8.0 nM) with non-specific binding measured using those same concentrations of [3H]-mesulergine plus 8μM ritanserin; both sets of conditions included 100 nM spiperone to block 5HT2A receptor binding. Bradford protein assays were performed to quantify protein in each pellet (BioRad Laboratories, Hercules, CA). All incubations were performed in SBB for 1.5 hours at room temperature. Incubations were terminated by rapid filtration using Whatman GF/C glass fiber filters, washed 3 times with cold 50 mM Tris-HCl (pH 6.9 at room temperature, pH 7.4 at 4°C). Samples were measured by liquid scintillation spectrometry in a Perkin-Elmer Tri-Carb 2800TR counter. Data were analyzed using GraphPad Prism (Graphpad Software, San Diego, CA) using nonlinear regression to fit the experimental data to saturation binding equations (assuming ligand depletion). Bmax values from Graphpad were normalized to protein concentration as determined using the Bradford assay. Control studies with membranes from 5HT2C receptor-null mice (Tecott et al., 1995) demonstrated no appreciable binding (data not shown).
Behavioral analysis
Mice were maintained on a 12-h light, 12-h dark cycle in a humidity- and temperature-controlled room with ad libitum access to water and standard laboratory chow (rodent chow 5001; Ralston Purina Co., St. Louis, MO). All animal studies were approved by the Institutional Animal Care and Use Committee of Vanderbilt University or the University of Alabama at Birmingham. Growth rates of mutant mice and control littermates were monitored in separate cohorts either daily prior to weaning (P21) or weekly beginning at weaning. Cumulative food consumption was assessed for age-matched, adult 5HT2C-VGV male mice and wild-type littermates using an automated feeding apparatus that measured free-feeding behavior by allowing animals ad libitum access to food cups that were mounted on a balance and monitored every 30 seconds (Singh et al., 2007). All feeding studies were performed after animals have been acclimated to the apparatus for at least 72 hrs. Metabolic analyses were assessed for age-matched, adult 5HT2C-VGV male mice and wild-type littermates as previously described (Powell et al., 2002; Singh et al., 2007). Average daily locomotor activity was monitored by both wheel-running activity and infrared motion detection for 21 days, following a 7-day acclimation period and analyzed using ClockLab software (ActiMetrics Software, Wilmette, IL). Acute locomotor activity was measured using open field chambers (Med Associates, St. Albans, VT) and monitored for 60 minutes on 3 successive days as described (Crawley, 2008).
To measure pre-weaning food intake, dams were injected with 6.63mg/g 99.9% deuterium oxide (2H2O; Cambridge Isotope Laboratories, Andover, MA) and pups were injected with 3.33mg/g H 182O (Sigma-Aldrich, St. Louis, MO). A graduated glass tuberculin syringe (Becton-Dickson, Franklin Lakes, NJ) used to deliver the isotopic tracers was weighed before and after injection to obtain an accurate mass of the injected dose. Pups were decapitated and blood was harvested at 6, 36, and 72 hours post-injection, with tail-blood simultaneously collected from dams. Samples were centrifuged (14,000 × g) at 4°C for 15 minutes and plasma was analyzed by cavity ring-down infrared spectroscopy to determine the ratios of 2H2O/H2O and H2 18O/H2 16O (Metabolic Solutions, Nashua, NH) (Crosson et al., 2002). Estimates of water flux in the pups and milk transfer between the dam and pups were made using the methods of Lifson, et al. (Lee and Lifson, 1960; Lifson, 1966; McClintock and Lifson, 1957) where the tracer data were expressed as tracer to tracee ratios. A compartmental model was constructed which described the water dilution space and turnover in dams and pups based on the respective dose of 2H2O or H2 18O. The original single isotope method used in animal studies was modified by incorporation of H2 182O to account for independent water turnover in the pups due to metabolic production of water, which leads to about 8% bias, if uncorrected (Butte et al., 1988). SAAM II software (SAAM Institute, University of Washington, Seattle) was used for compartmental analysis (Barrett et al., 1998). Milk intake by each pup was estimated from the rate of transfer of 2H2O from the dam to the pup (Infante et al., 1985).
Grip force was measured as described (Meyer et al., 1979) using a force gauge (San Diego Instruments, San Diego, CA). Muscle endurance and motor coordination were assessed using an accelerating Rotarod (Ugo Basile, Comerio, Italy) paradigm. Briefly, mice were placed on a rotating 3.2 cm rod that accelerated from 5-40 revolution/minute over a 5 minute period; latency to fall off of the rod was measured using a stopwatch. Mice that fell from the rod within 15 seconds were retested; mice that grasped the rod and rotated with it for ≥2 revolutions were scored as having fallen.
Statistical analysis
Body weight data were analyzed by two-way analysis of variance (ANOVA). The survival curve was analyzed by the logrank test. Hypothalamic mRNA expression at the P21 timepoint was analyzed by randomized block analysis to eliminate variability in independently collected sample sets. All other data were analyzed by two-tailed Student's t test; for data sets with unequal variances, Welch's correction was applied. P values less than 0.05 were considered significant.
Results
Generation of mutant mice solely expressing the fully-edited (VGV) isoform of the 5HT2C receptor
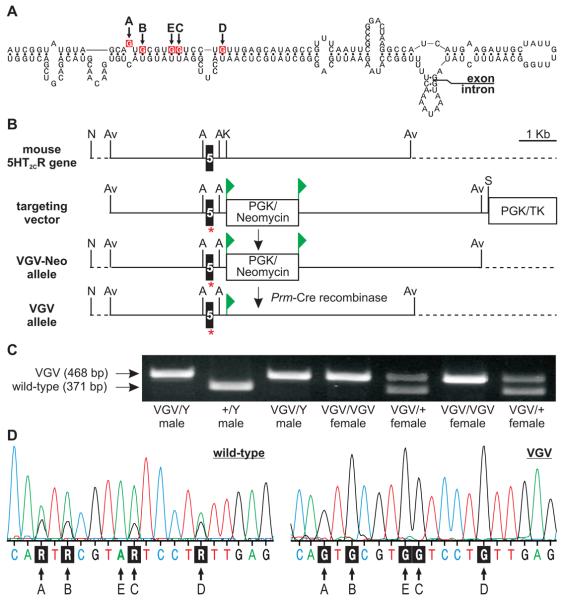
To investigate the importance of normal 5HT2C RNA editing patterns, we generated mutant mice solely expressing the fully-edited isoform of the receptor (5HT2C-VGV). We chose this receptor isoform based upon its decreased G-protein coupling efficacy and lack of constitutive activity compared to other 5HT2C isoforms (Berg et al., 2001; Burns et al., 1997; Fitzgerald et al., 1999; Niswender et al., 1999; Wang et al., 2000). Mice solely expressing 5HT2C-VGV receptors were generated by homologous recombination using a replacement-type targeting vector with the five edited adenosine residues of the 5HT2C receptor gene (htr2C) mutated to guanosine to mimic the base-pairing properties of inosine (Figs. 1A and B). Since the mouse 5HT2C gene is located on the X chromosome, obtaining all possible genotypes of mutant mice and wild-type littermates required two distinct breeding strategies (Table 1). The resultant offspring were genotyped at weaning (postnatal day 21; P21) and demonstrated a normal Mendelian distribution (Fig. 1C and Table 1), indicating that the 5HT2C-VGV mutation does not result in embryonic or early postnatal lethality. To confirm that mutant mice bearing the modified 5HT2C-VGV allele expressed transcripts solely encoding the VGV isoform of the receptor, whole-brain derived cDNA was generated by reverse-transcription polymerase chain reaction (RT-PCR) amplification of 5HT2C pre-mRNA and the resulting amplicon was sequenced directly. Complementary DNA from wild-type animals exhibited overlapping adenosine/guanosine peaks in electropherogram traces resulting from a mixture of edited 5HT2C transcripts, whereas cDNAs from mutant animals showed only VGV-encoding guanosine moieties at the five editing sites (Fig. 1D). Due to potential alterations in the structure of the extended RNA duplex required for site-selective A-to-I conversion (Burns et al., 1997) resulting from the introduced guanosine mutations at the five editing sites, further sequence analysis of 50 individual cDNA clones derived from 5HT2C pre-mRNAs indicated no aberrant editing throughout the predicted region of double-stranded RNA and sole expression of the VGV isoform in 5HT2C-VGV mice (data not shown).
Fig. 1.
Targeting strategy and genotype analysis for 5HT2C-VGV mice. (A) The predicted secondary structure for 5HT2C pre-mRNA near the distal end of exon 5 is presented. The positions of the five editing sites (A-E) are shown with site-specific adenosine to guanosine mutations indicated in red. (B) Schematic diagram and abbreviated restriction map of the mouse 5HT2C gene before and after targeted gene modification; the location of exon 5, the loxP sites (▶) flanking the PGK/neomycin resistance cassette, the negative selectable marker (PGK/TK) outside the region of homology, the approximate position of the introduced mutations (*) and sequences outside the region of homology (dotted line) are indicated; Av, Avr II; N, Not I; A, Acc I; K, Kpn I; S, Sfi I. (C) Genotype analysis of offspring from a hemizygous 5HT2C-VGV male (VGV/Y) x heterozygous mutant female (VGV/+) mating; migration positions of PCR amplicons corresponding to the wild-type (371 bp) or mutant (468 bp) 5HT2C alleles and animal genotypes are indicated. (D) Sequence electropherogram traces of 5HT2C receptor-derived RT-PCR products generated from wild-type and 5HT2C-VGV hemizygous male mice. The positions of the five editing sites and the corresponding nucleotide sequences are indicated; R, purine.
Table 1.
χ2-analysis of genotype distribution for wild-type and 5HT2C-VGV mutant mice.
| +/Y male × VGV/+ female | VGV/Y male × VGV/+ female | |||||
|---|---|---|---|---|---|---|
| Genotype | Expected | Observed | Genotype | Expected | Observed | |
| male | +/Y | 24.5 | 21 | +/Y | 34.75 | 36 |
| VGV/Y | 24.5 | 26 | VGV/Y | 34.75 | 31 | |
| female | +/+ | 24.5 | 22 | VGV/+ | 34.75 | 43 |
| VGV/+ | 24.5 | 29 | VGV/VGV | 34.75 | 29 | |
|
| ||||||
| Total: | 98 | 98 | 139 | 139 | ||
|
| ||||||
| p ≤ 0.65 | p ≤ 0.35 | |||||
Altered 5HT2C expression in VGV mutant mice
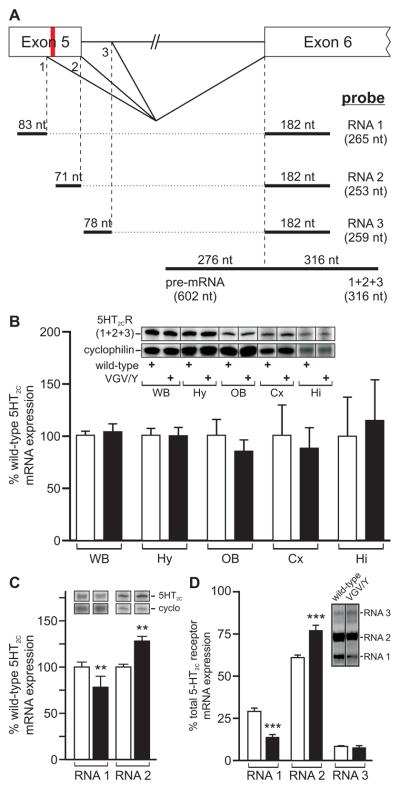
Co-expression of the HBII-52 snoRNA with 5HT2C minigenes has been shown to direct distal 5′-splice site selection in heterologous expression systems through blockade of a splicing silencer that overlaps with the edited region in exon 5 (Kishore and Stamm, 2006; Vitali et al., 2005). Editing of the mutant allele at the E, C, and D sites also weakens this splicing silencer to promote expression of RNA 2 (Fig. 2A) (Flomen et al., 2004), the 5HT2C mRNA isoform encoding the full-length receptor protein (Canton et al., 1996; Wang et al., 2000), even in the absence of HBII-52/MBII-52 (Kishore and Stamm, 2006). The other 5HT2C splice variants result from the use of a more proximal splice donor in exon 5 (RNA 1) or a distal 5′-splice site in intron 5 (RNA 3), each encoding truncated receptor proteins resulting from premature translation termination that are presumed to be nonfunctional (Fig. 2A) (Canton et al., 1996; Wang et al., 2000). To assess whether the 5HT2C-VGV allele produced alterations in 5HT2C mRNA expression, we employed a ribonuclease (RNase) protection strategy using an antisense riboprobe that could distinguish between unspliced premRNA and mature 5HT2C transcripts to quantify the level of 5HT2C mRNA expression in either whole brain or dissected brain regions (Fig. 2A). Results from this analysis revealed no detection of pre-mRNA expression nor any difference in total 5HT2C mRNA levels (RNA 1 + RNA 2 + RNA 3) for whole brain or dissected brain regions between wild-type and VGV/Y mutant animals (Fig. 2B), although a significant increase and decrease was observed in the levels of RNA 2 and RNA 1, respectively (Figs. 2C and D). Similar results for total 5HT2C mRNA expression were observed using a real-time RT-PCR-based strategy (data not shown). Since expression of RNA 3 was not detectable using this assay due to its low level of expression, we developed a more sensitive, semi-quantitative RT-PCR-based strategy to amplify the alternatively spliced region for all three 5HT2C mRNA isoforms with a single primer pair (Fig. 2D). These results revealed no alteration in RNA 3 expression and confirmed the observed changes in RNA 1 and RNA 2 in mutant mice (Fig. 2C), consistent with previous studies in which increased editing of 5HT2C pre-mRNAs promoted the production of RNA 2 at the expense of RNA 1 (Flomen et al., 2004).
Fig. 2.
Analysis of 5HT2C mRNA expression in wild-type and 5HT2C-VGV mice. (A) A schematic diagram of a portion of the 5HT2C gene is shown in which 5′-splice site selection can generate three alternatively spliced 5HT2C transcripts (RNA 1, RNA 2 and RNA 3). The relative positions of contiguous antisense riboprobes used for ribonuclease protection analysis (bold lines) to quantify either total 5HT2C mRNA (1+2+3) or the three individual 5HT2C splice variants is presented with the size of the expected protected fragment (nucleotides, nt) for each mRNA isoform; the location of the 5 editing sites is indicated in red. (B) Ribonuclease protection analysis of total 5HT2C mRNA expression (1+2+3) in whole brain and dissected brain regions for 5HT2C-VGV (■) male mice is presented as a percentage of mean wild-type (□) expression; mean ± SEM, n ≥ 4 animals/genotype for whole brain samples and n ≥ 6 animals/genotype for dissected brain regions. Inset, Representative ribonuclease protection analysis of total 5HT2C RNA expression in whole brain and dissected brain region samples from individual mice; WB, whole brain; Hy, hypothalamus; OB, olfactory bulb; Cx, frontal cortex; Hi, hippocampus. (C) Quantitative analysis of the relative expression levels for 5HT2C alternative splicing variants (RNA 1 and RNA 2) in whole brain samples from wild-type (□) and 5HT2C-VGV (■) male mice (mean ± SEM; n=7; **p ≤ 0.01). Inset, Representative ribonuclease protection analysis of alternatively spliced 5HT2C RNAs in whole brain samples from individual mice. (D) Semi-quantitative analysis of 5HT2C mRNA splicing patterns in wild-type and 5HT2C-VGV male mice. Quantification of each 5HT2C splice variant is presented as a percentage of total 5HT2C mRNA expression (n=7; mean ± SEM; ***p ≤ 0.001); inset, Representative RT-PCR amplification using common primers in exons 5 and 6 to detect three 5HT2C mRNA isoforms; the expected migration positions of amplicons corresponding to each 5HT2C splice variant are indicated.
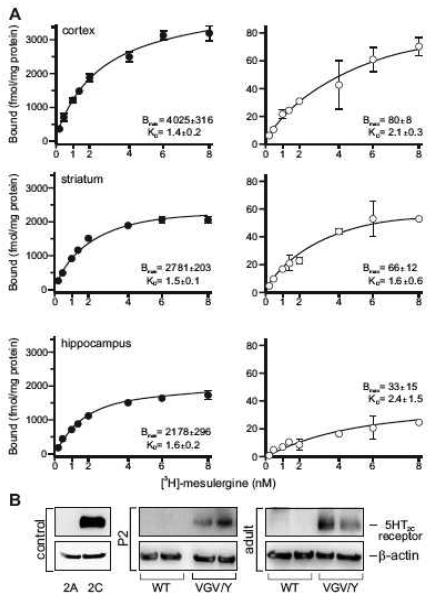
While no change in the steady-state level of total 5HT2C mRNA was observed in 5HT2C-VGV mice, mutant animals demonstrated a dramatic increase in receptor protein expression relative to wild-type littermates (40-to-70-fold) that varied between dissected brain regions (Fig. 3). Saturation binding with total membrane fractions isolated from frontal cortex, hippocampus and striatum revealed an increase in maximal binding (Bmax) with no change in affinity (Kd) for [3H]-mesulergine, a selective 5HT2A/2C antagonist (Fig. 3A). Since 5HT2A receptor binding was blocked by the addition of spiperone (100 nM) to all samples, these results indicate a specific increase in 5HT2C receptor levels. To determine if a compensatory increase in the expression of other serotonin receptors that can bind [3H]-mesulergine is responsible for the observed increase in binding, real-time RT-PCR analyses were performed to quantify the expression of 5HT2A, 5HT2B and 5HT7 receptors in whole brain RNA samples. No significant differences in the steady-state levels of 5HT2A and 5HT7 receptor transcripts were observed between wild-type and 5HT2C-VGV mutant animals, while the expression of 5HT2B mRNA was not detectable in whole brain RNA from either genotype (data not shown).
Fig. 3.
Analysis of 5HT2C protein expression in wild-type and 5HT2C-VGV mice. (A) Saturation binding isotherms demonstrating 5HT2C receptor-specific binding for total membranes prepared from dissected cortex, striatum and hippocampus in wild-type (□) and 5HT2C-VGV (●) male mice (wild-type, n=4 groups of five pooled mice per brain region; 5HT2C-VGV, n=7 brain regions from individual animals; mean ± SEM). (B) Western blotti ng analysis of 5HT2C receptor and β-actin expression in whole brain samples isolated from individual wild-type and 5HT2C-VGV male mice at postnatal day 2 (P2) or in adulthood (14-16 weeks); n ≥ 8 for mice of each genotype, two representative samples are shown for each age and genotype. Representative Western blotting controls from NIH-3T3 cells stably expressing either 5HT2A or 5HT2C receptors are presented.
We confirmed that the observed differences in 5HT2C protein level did not result from in vivo differences in pharmacologic properties using a Western blotting strategy from whole brain samples isolated from neonatal (P2) and adult animals (Fig. 3B). Control studies with transiently transfected mouse fibroblasts (NIH-3T3) expressing either 5HT2A or 5HT2C receptors verified antibody specificity (Fig. 3B). While 5HT2C receptor expression was detected readily in protein samples isolated from the whole brain of individual 5HT2C-VGV mice at both developmental time points, it was indistinguishable from background in wild-type animals, owing to the low levels of receptor expression observed in many brain regions (Fig. 3A, right) (Mengod et al., 1990). These results confirm that the dramatic changes in 5HT2C protein expression observed using radio-ligand binding are due to increases in receptor protein expression rather than isoform-specific differences in ligand affinity.
Failure to thrive in 5HT 2C-VGV mutant mice
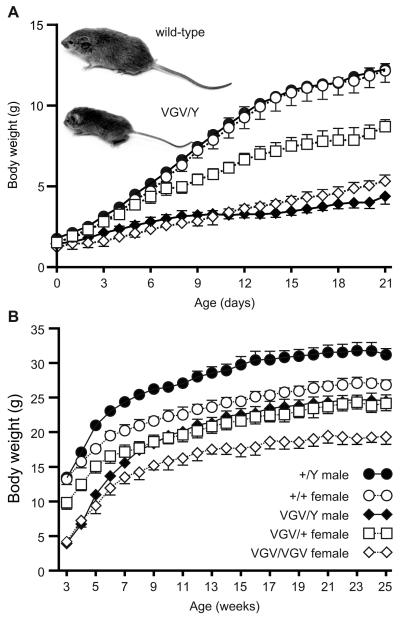
Initial characterization of 5HT2C-VGV mutant animals revealed a 70% decrease in body mass for hemizygous mutant males (VGV/Y) at P21 despite no significant differences in body mass at birth (Fig. 4A), suggesting a postnatal failure to thrive. While a similar reduction in body mass also was observed in homozygous (VGV/VGV) mutant females, heterozygous mutant females (VGV/+) demonstrated an intermediate growth phenotype indicative of haploinsufficiency. Mutant animals appeared hypotonic with reduced milk content in their stomach, subsequently showing clinical signs of dehydration that included piloerection, a mottled appearance and hunched posture (data not shown). Mutant mice also demonstrated a post-weaning mortality with 10-20% of mutant offspring dying between P21 and P24 that could be minimized with nutritional supplementation by placing a wet food paste in the bottom of the cage. In addition to reduced body mass, mutant mice displayed decreased linear growth (Fig. 4A, inset), as the body length (nose-to-rump) of VGV/Y males and control littermates (P21) was 60.4 ± 1.3 mm and 87.6 ± 1.5 mm, respectively (n ≥ 5; p ≤ 0.0001). Despite their decreased growth rate prior to P21, all mutant mice bearing the 5HT2C-VGV allele quickly established a normal growth pattern following weaning, gaining mass at a rate roughly parallel to that of their wild-type littermates (Fig. 4B). A gene-dosage effect remained evident in female mice, as homozygous mutants weighed significantly less than heterozygous animals and both groups weighed less than wild-type female littermates (p ≤ 0.0001). This unusual pattern of growth, switching from an initial failure to thrive to a post-weaning weight gain that parallels wild-type littermates, is similar to that previously observed for a mouse model of PWS (PWS-ICdel) in which a 42 kilobase (kb) region was deleted from the distal portion of the putative mouse imprinting center on chromosome 7 (Chamberlain et al., 2004; Yang et al., 1998).
Fig. 4.
Body weight analysis of wild-type and 5HT2C-VGV mice. (A) Pre- and (B) post-weaning growth curves are shown for wild-type and 5HT2C-VGV mutant mice (mean ± SEM; n=6 for each genotype and gender; p ≤ 0.0001 using two-way ANOVA). Inset, A representative photograph of wild-type and mutant (VGV/Y) littermates at postnatal day 21 is presented.
Altered feeding behavior and increased metabolism in 5HT2C-VGV mice
Infants with PWS initially present with feeding difficulties and a failure to thrive, often requiring gavage or other medical intervention to maintain proper nutrition (Goldstone, 2004). To determine if the observed failure to thrive in 5HT2C-VGV mice resulted from similar feeding deficits, we measured milk consumption in mice from postnatal day 9-12. Offspring derived from a wild-type male x heterozygous mutant female breeding strategy were genotyped at P8 and the individual litters were culled to 3 pups at P9, with one pup of each possible genotype represented in each litter (wild-type, VGV/+, and VGV/Y). Pups and dams were injected with separate, stable heavy water isotopes (H2 18O and 2H20, respectively) on P9 and the accumulation and clearance of the isotopes was measured at regular intervals over a 72 hour period. 5HT2C-VGV mice ingested significantly less milk than wild-type littermates (0.580 ± 0.097 and 1.345 ± 0.301 grams milk/day, respectively; p < 0.05), with heterozygous mutant animals again exhibiting an intermediate phenotype (1.092 ± 0.212 grams milk/day), suggesting that decreased pre-weaning milk consumption contributes to the reduced growth rate of mutant mice.
The 5HT2C receptor has been implicated directly in the anorectic modulation of food intake using both transgenic and pharmacologic approaches (Heisler et al., 2002; Hewitt et al., 2002; Nonogaki et al., 1998; Tecott et al., 1995). Despite the 22% reduction in the mean body mass for adult 5HT2C-VGV mice compared to their wild-type littermates (Fig. 4B), age-matched adult mutant and wild-type animals consume roughly equivalent amounts of food (data not shown). Since adult animals of even slightly different weights would be expected to demonstrate differences in food consumption due to the dominance of anabolic pathways to increase feeding to maintain adipose mass (Augustine and Rossi, 1999), food intake was corrected for body mass to reveal that 5HT2C-VGV mice were functionally hyperphagic with an 18% increase in daily food consumption (Fig. 5A).
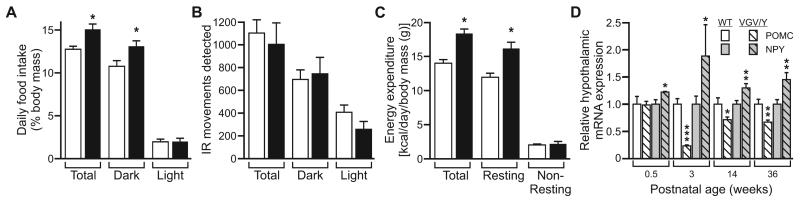
Fig. 5.
Feeding behavior and metabolic analysis of wild-type and 5HT2C-VGV mice. (A) Food intake for 15-week old male wild-type (□) and 5HT2C-VGV (■) mice per 24-hours (hr), 12-hr light, or 12-hr dark period, corrected for total body mass (mean ± SEM, n=6, *p ≤ 0.05). (B) Average daily activity detected by infrared sensors for wild-type (□) and 5HT2C-VGV adult (■) mice maintained in a 12hr-12hr light/dark environment (mean ± SEM; n=6). (C) Indirect calorimetric analysis of energy expenditure for wild-type (□) and 5HT2C-VGV (■) mice per 24-hr, while resting, or while active, corrected for total body mass (mean ± SEM, n=6, *p ≤ 0.05). (D) Quantitative analysis of hypothalamic POMC and NPY mRNA expression at 0.5 weeks (n=4), 3 weeks (n=8), 14 weeks (n=12) and 36 weeks (n=11) in wild-type and 5HT2C-VGV male mice (mean ± SEM, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001).
Individuals with PWS classically present with hyperphagia and impaired satiety, often leading to severe obesity in affected patients as young as two years of age (Goldstone, 2004). Despite their hyperphagia, 5HT2C-VGV mice do not become obese and maintain the 7-10 gram reduction in body mass compared to wild-type littermates initially observed at weaning (Fig. 4B). This absence of obesity was not associated with a corresponding increase in locomotor activity, as no significant differences were observed between 5HT2C-VGV and wild-type animals when assessed by home cage analysis of infrared motion detection (Fig. 5B) or wheel running activity (data not shown). However, indirect calorimetry revealed that 5HT2C-VGV mice expend more total and resting energy than wild-type littermates per gram body mass (Fig. 5C), suggesting that an elevated metabolic rate in mutant animals may compensate for hyperphagia to prevent the subsequent onset of obesity.
Perceived energy deficits increase expression of the orexigenic neuropeptide Y (NPY) (Mizuno et al., 1999) while simultaneously decreasing the expression of anorectic peptides derived from the pro-opiomelanocortin (POMC) prohormone (Schwartz et al., 1997) to increase food consumption. Quantitative analysis of hypothalamic RNA expression in mutant animals revealed a 22-89% increase in NPY mRNA relative to wild-type mice at all time points examined, whereas steady-state POMC mRNA levels were significantly decreased 76% at weaning (P21), 29% at 14 weeks and 33% at 36 weeks of age, but were unaltered at P4 (Fig. 5D). These changes in hypothalamic neuropeptide mRNA expression are consistent with the observed post-weaning increase in food consumption in 5HT2C-VGV animals (Ellacott and Cone, 2004).
5HT2C-VGV mice display additional PWS characteristics observed in humans or mouse models
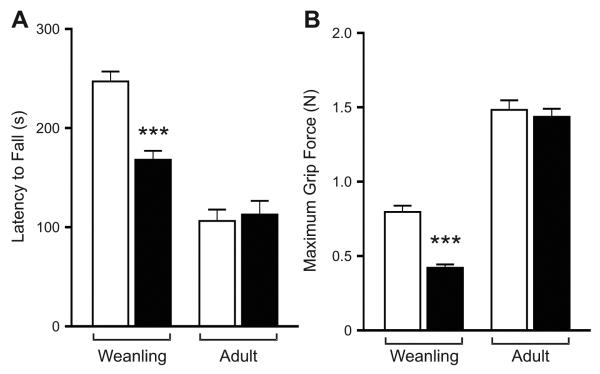
Infants affected with PWS exhibit other nonspecific symptoms including hypotonia and growth retardation (Goldstone, 2004), with similar phenotypes also observed in several mouse models of PWS (Ding et al., 2008; Gabriel et al., 1999; Skryabin et al., 2007; Tsai et al., 1999b; Yang et al., 1998). To examine potential changes in muscle tone, we initially employed an accelerating Rotarod® paradigm. Recently weaned wild-type animals outperformed their mutant littermates on this task, with no difference observed in adults (Fig. 6A). Since decreased performance using the Rotarod® could reflect alterations in either muscle tone or motor coordination, we also measured the grip strength of wild-type and hemizygous male 5HT2C-VGV mice (Fig. 6B). Though no differences were observed in adult mice, weanling mutant animals (P21-P28) exhibited a ~50% reduction in grip force, suggesting that an early hypotonia in 5HT2C-VGV mice improves with age similar to patients affected with PWS (Goldstone, 2004).
Fig. 6.
Analysis of muscle strength in wild-type and 5HT2C-VGV mice. (A) Motor coordination and endurance of post-weaning (P22-P28, n=10) and adult (25 weeks, n=12) wild-type (□) and 5HT2C-VGV mutant (■) male mice (mean ± SEM; *p ≤0.05, **p ≤ 0.01, ***p ≤ 0.001). (B) Average grip strength of recently weaned (P21-P28; n=5) or adult (n=7) wild-type (□) and mutant (■) mice (mean ± SEM; ***p ≤ 0.001).
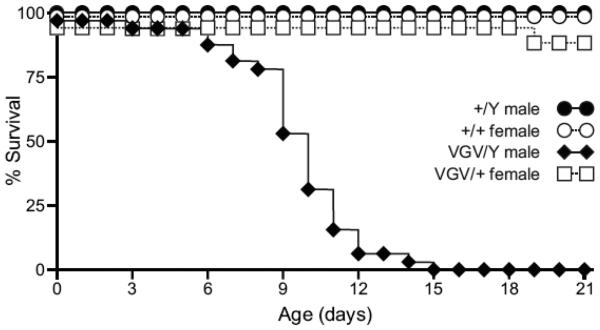
Several previously described murine models of PWS report feeding difficulties, failure to thrive and/or growth deficiency, yet the utility of these models has been hampered by substantial neonatal lethality (Cattanach et al., 1992; Gerard et al., 1999; Tsai et al., 1999b; Yang et al., 1998). More recent studies have identified permissive (FVB/NJ, C3H/HeJ, 129S1/Sv and BALB/cJ) and non-permissive (C57BL/6J and DBA/2J) mouse background strains that either allow survival or result in neonatal mortality, respectively, when bearing deletions within the PWS critical region on chromosome 7C (Chamberlain et al., 2004; Gerard et al., 1999). To compare background-dependent phenotypic differences between 5HT2C-VGV mutant mice and other PWS models, 129S6-derived heterozygous 5HT2C-VGV dams were mated with wild-type C57BL/6J males. Offspring were genotyped at weaning or at the time of death to reveal a completely penetrant neonatal lethality for hemizygous mutant males, with 75% dying between P8-P12 and none surviving beyond P15 (Fig. 7; p≤0.0001). These results are similar to the early neonatal lethality observed for other mouse models of PWS on the C57BL/6 background (Chamberlain et al., 2004; Gerard et al., 1999) and further indicate that while a single mutant 5HT2C allele can cause alterations in growth and receptor expression in both hemizygous males and heterozygous females (Fig. 4) (U. B. Olaghere da Silva, 2009), the presence of a single wild-type allele is sufficient for survival on a non-permissive background strain (Fig. 7).
Fig. 7.
Strain-specific lethality in 5HT2C-VGV mice. Cumulative survival rate for offspring resulting from the mating of heterozygous mutant 129S6 females (VGV/+) to wild-type C57BL/6 males (n=28, +/Y; n=32, VGV/Y; n=33, +/+; n=17, VGV/+).
Discussion
The editing of 5HT2C transcripts modulates multiple aspects of 5HT2C receptor signaling and expression in heterologous systems (Berg et al., 2001; Burns et al., 1997; Fitzgerald et al., 1999; Flomen et al., 2004; Marion et al., 2004; Niswender et al., 1999; Price et al., 2001; Wang et al., 2000), yet the physiologic importance for the existence of multiple 5HT2C isoforms has not been fully explored. Recent studies by Kawahara et al. have characterized mutant mice comparable to those described in the present study in which 5HT2C expression was limited to the fully-edited (VGV) isoform. These animals exhibited several similar phenotypic alterations, including reduced pre-weaning growth, post-weaning hyperphagia and an increased metabolic rate resulting from excessive activation of the sympathetic nervous system (Kawahara et al., 2008), yet several differences between these mutant lines also were readily apparent. Mice solely expressing the VGV isoform were previously reported to exhibit reduced locomotor activity in an open field chamber (Kawahara et al., 2008). While similar results were observed with 5HT2C-VGV mice using such a behavioral paradigm (data not shown), these animals exhibited normal activity in their home cage whether quantified by infrared motion detection (Fig. 5B) or wheel running (data not shown). This suggests that mice solely expressing the fully-edited 5HT2C isoform do not exhibit baseline hypoactivity, but rather decreased activity in a novel environment. Unlike the dramatic increase in 5HT2C receptor expression observed in 5HT2C-VGV animals using both radioligand binding (Fig. 3A) and Western blotting (Fig. 3B) analyses, the mutant animals described by Kawahara et al. did not exhibit any increase in 5HT2C receptor protein when analyzed by Western blotting (Kawahara et al., 2008). This result is particularly surprising given that 5HT2C-selective autoradiographic analyses by Kawahara et al. revealed an increase in receptor expression that was inconsistent with the Western blotting studies for these animals (Kawahara et al., 2008).
The 40-to-70-fold increase in total 5HT2C receptor protein observed in 5HT2C-VGV mutant animals (Fig. 3) was unexpected, given the minor nature of the mutations introduced into exon 5 of the endogenous htr2C locus (Figs. 1A and B) and the fact that there were no observed alterations in the steady-state levels of 5HT2C transcripts (Fig. 6B). Although a 27% increase in the production of RNA 2 was observed in mutant mice (Figs. 2C and D), the dramatic increase in functional 5HT2C protein levels cannot be explained readily by this small alteration in 5HT2C splice-site selection. Increased receptor protein could represent an adaptive mechanism to compensate for the diminished coupling efficacy and constitutive activity of the 5HT2C-VGV isoform that was observed initially in transfected cell lines (Burns et al., 1997; Fitzgerald et al., 1999; Niswender et al., 1999; Wang et al., 2000). The absence of a change in total 5HT2C transcripts for mutant animals indicates the existence of previously unrecognized post-transcriptional mechanism(s) to regulate 5HT2C receptor density that could be mediated by potential differences in translation efficiency and/or stability for distinct edited receptor isoforms. Recent studies by Marion et al. have provided a potential mechanism for isoform-specific differences in protein stability based upon a constitutive activity-dependent alteration in the subcellular compartmentalization of the receptor, resulting from an inability of highly edited 5HT2C receptor isoforms to effectively interact with β-arrestin2 (Marion et al., 2004).
While the hyperphagia observed in 5HT2C-VGV mutant mice is similar to that exhibited by 5HT2C receptor-null animals, null mutants develop adult-onset obesity with no change in metabolic rate (Nonogaki et al., 2003; Nonogaki et al., 1998); however, 5HT2C-VGV mutant mice display an increase in their resting energy expenditure in the absence of hyperactivity, which likely compensates for the hyperphagia observed in these mutants to prevent the onset of obesity. The orexigenic changes in hypothalamic POMC and NPY mRNA expression in 5HT2C-VGV mutant animals are consistent with the observed increase in relative food consumption (Fig. 5A). These data support previous evidence for a functional role for 5HT2C receptors in the modulation of food intake via a POMC-mediated mechanism, including the expression of 5HT2C transcripts in ~80% of POMC neurons within the arcuate nucleus of the hypothalamus (Heisler et al., 2002) and recent studies demonstrating that the selective ablation of 5HT2C in these neurons is sufficient to eliminate the effects of serotonergic compounds on food intake and result in hyperphagia-mediated obesity (Xu et al., 2008).
Although multiple mouse models lacking expression of one or more PWS candidate genes have implicated several loci with elements of the PWS phenotype, these models vary widely in their capacity to recapitulate the human disorder (Table 2). Mouse models lacking expression of individual genes located within the imprinted region on chromosome 7C display either no obvious PWS characteristics (Jong et al., 1999; Tsai et al., 1999a; Tsai et al., 1999b; Yang et al., 1998), neonatal lethality from respiratory distress (Gerard et al., 1999), or body mass changes that never differ from wild-type animals by more than 10% (Bischof et al., 2007). However, several functional deletions encompassing larger portions of the imprinted region result in neonatal lethality preceded by a failure to thrive (Cattanach et al., 1992; Chamberlain et al., 2004; Gerard et al., 1999; Tsai et al., 1999b; Yang et al., 1998). It is important to note that none of the mouse models of PWS reported to date demonstrate either the infertility or the morbid obesity characteristic of the human syndrome, suggesting that mice may not be capable of manifesting the full spectrum of phenotypic changes associated with this genetic disorder. Reasons for the limited PWS phenotype(s) observed in mice may include species-specific differences in gene structure, function, cell-expression patterns, neuroendocrine and metabolic pathways and/or modifier genes (Goldstone, 2004).
Table 2.
Summary of mouse models of Prader-Willi Syndrome
| Functional Chr. 7C Deletion |
Mouse Model | Growth Retardation |
Hypotonia | Obesity | Food Intake | C57BL/6 Lethality |
Survive on Other Strains |
References |
|---|---|---|---|---|---|---|---|---|
| Entire PWS Region |
Maternal Duplication | yes | n.d. | n.d. | n.d. | 100% | n.d. | [1] |
| Entire PWS Region |
PWS-ICdel | ↓~45% | n.d. | no | n.d. | 100% | yes | [2,3] |
| Entire PWS Region |
TgPWS | ↓~50% | yes | n.d. | n.d. | 100% | no | [4,5] |
| Entire PWS Region |
ΔSnrpn-Ube3a | ↓~55% | yes | no | n.d. | 78% | n.d. | [6] |
| Individual Gene |
ΔNecdin | no | n.d. | no | n.d. | no | yes | [7] |
| Individual Gene |
ΔNecdin | no | yes | no | n.d. | 95% | yes | [8] |
| Individual Gene |
ΔNecdin | no | n.d. | no | n.d. | ~65% | yes | [9] |
| Individual Gene |
ΔMagel2 | ↓~5% | n.d. | ↑~5-10% | ↓~10% | prenatal* | n.d. | [10] |
| snoRNA | Snord116del | ↓~40% | no | no | ↑13-31%** | no | n.d. | [11] |
| snoRNA | ΔPWScr | ↓~25% | n.d. | no | n.d. | 15% | yes | [12] |
| N/A | 5HT2C-VGV | ↓~70% | yes | no | ↑14-18% | 100% | yes | N/A |
[1] Cattanach, 1992; [2] Yang, 1998; [3] Chamberlain, 2004; [4] Gabriel, 1999; [5] Stefan, 2005; [6] Tsai, 1999b; [7] Tsai, 1999a; [8] Gerard, 1999; [9] Muscatelli, 2000; [10] Bischof, 2007; [11] Ding, 2008; [12] Skryabin, 2007
n.d., not determined; N/A, not applicable
incomplete penetrance
food intake normal at 7 weeks, hyperphagia after 3 months
Mutant mice solely expressing the fully-edited isoform of the 5HT2C receptor display several phenotypic characteristics of PWS, including a failure to thrive, decreased somatic growth and neonatal muscular hypotonia, followed by post-weaning hyperphagia, in addition to other similarities with mouse models of PWS where deletions within the imprinting center on chromosome 7C result in strain-specific neonatal lethality. These observations are consistent with recent analyses indicating that 5HT2C RNA editing is increased in autopsy samples from PWS patients (Kishore and Stamm, 2006) and a mouse model (PWS-ICdel) that also demonstrates alterations in 5HT2C-related behaviors (Doe et al., 2009). Previous studies have shown that the maternally imprinted snoRNA, HBII-52, can alter both the splicing and editing of 5HT2C transcripts (Kishore and Stamm, 2006), providing a provoking and straightforward mechanism by which to link the 5HT2C-VGV mutation with the 15q11-13 locus and multiple aspects of this disorder. However, several clinical cases have been identified recently that either suggest deletion of this snoRNA is not sufficient to cause a PWS-like phenotype (Runte et al., 2005) or that a specific paternal deletion of the HBII-85 snoRNA cluster can result in PWS (de Smith et al., 2009; Gallagher et al., 2002; Sahoo et al., 2008). Due to the nature of these human clinical studies however, it was impossible to assess potential changes in the brain-specific expression of other snoRNAs in these patients, negating the ability of such studies to conclusively eliminate a role for HBII-52. The generation of mutant mice bearing similar deletions have produced animals with multiple PWS-like phenotypes (Table 2) and have shown the expression of MBII-52 snoRNAs in the brains of these mutant animals, suggesting that the absence of MBII-52 alone is not sufficient to cause this syndrome (Ding et al., 2008; Ding et al., 2005; Skryabin et al., 2007). While most recent clinical and mouse studies have focused upon the HBII-85 snoRNA cluster as playing a causal role in PWS (de Smith et al., 2009; Ding et al., 2008; Gallagher et al., 2002; Runte et al., 2005; Skryabin et al., 2007), the targets of these imprinted snoRNAs remain largely unknown, raising the possibility that alternative mechanisms exist by which genes located within the 15q11-13 locus can alter 5HT2C receptor function independent of HBII-52 expression.
The present studies have demonstrated that alterations in the normal pattern(s) of editing can result in significant changes in growth, metabolism and feeding behavior and also have identified an unknown post-transcriptional mechanism to modulate 5HT2C receptor density. Given the ability of the 5HT2C-VGV mutation to phenocopy the failure-to-thrive, reduced early-postnatal growth, hypotonia, adult hyperphagia, and strain-specific lethality observed in previously-described mouse models of PWS (Table 2), these results strongly suggest a link between altered 5HT2C receptor function and the development of major criteria necessary for the diagnosis of this disorder (Gunay-Aygun et al., 2001). Though this model may implicate increased 5HT2C RNA editing in the molecular etiology of PWS, it does not suggest that the full spectrum of altered phenotypes observed in PWS patients result solely from altered 5HT2C receptor function. When combined with previously identified alterations in 5HT2C-mediated behaviors in PWS-ICdel mice (Doe et al., 2009), observed increases in the editing of 5HT2C transcripts in both mouse model of PWS and human patients (Doe et al., 2009; Kishore and Stamm, 2006), and the known involvement of the 5HT2C receptor in controlling energy homeostasis and satiety (Hewitt et al., 2002; Somerville et al., 2007; Xu et al., 2008), the presence of PWS-like phenotypes in mice with altered 5HT2C editing identifies the htr2C gene as the first locus outside the 15q11-13 imprinted region in which mutations can recapitulate multiple aspects of this human genetic disorder.
Acknowledgements
We thank Dr. Douglas McMahon and Christopher Ciarleglio for assistance with locomotor activity, Ryan Strachan, Michael Hughes, James Gilbert, Li Peng and Apoorwa Thati for technical assistance, and Drs. Tim Nagy and Maria Johnson of the UAB Small Animal Phenotyping Core for metabolic measurements. We also thank Drs. Elisabeth Dykens, Pat Levitt, and Larry Zwiebel for critical reading of the manuscript. This work was supported by grants from the National Institutes of Health (NS35891 to R.B.E.; MH61887 and MH82441 to B.L.R.), the Vanderbilt Silvio O. Conte Center for Neuroscience Research (MH78028), the Vanderbilt Digestive Disease Research Center (DK05840) and the Vanderbilt Kennedy Center (HD15052).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Augustine KA, Rossi RM. Rodent mutant models of obesity and their correlations to human obesity. Anat Rec. 1999;257:64–72. doi: 10.1002/(SICI)1097-0185(19990415)257:2<64::AID-AR7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Barrett PH, et al. SAAM II: Simulation, Analysis, and Modeling Software for tracer and pharmacokinetic studies. Metabolism. 1998;47:484–92. doi: 10.1016/s0026-0495(98)90064-6. [DOI] [PubMed] [Google Scholar]
- Berg KA, et al. RNA-editing of the 5-HT(2C) receptor alters agonist-receptor-effector coupling specificity. Br J Pharmacol. 2001;134:386–92. doi: 10.1038/sj.bjp.0704255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof JM, et al. Inactivation of the mouse Magel2 gene results in growth abnormalities similar to Prader-Willi syndrome. Hum Mol Genet. 2007;16:2713–9. doi: 10.1093/hmg/ddm225. [DOI] [PubMed] [Google Scholar]
- Burns CM, et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–8. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- Butte NF, et al. Human-milk intake measured by administration of deuterium oxide to the mother: a comparison with the test-weighing technique. Am J Clin Nutr. 1988;47:815–21. doi: 10.1093/ajcn/47.5.815. [DOI] [PubMed] [Google Scholar]
- Canton H, et al. Identification, molecular cloning, and distribution of a short variant of the 5-hydroxytryptamine2C receptor produced by alternative splicing. Mol Pharmacol. 1996;50:799–807. [PubMed] [Google Scholar]
- Cattanach BM, et al. A candidate mouse model for Prader-Willi syndrome which shows an absence of Snrpn expression. Nat Genet. 1992;2:270–4. doi: 10.1038/ng1292-270. [DOI] [PubMed] [Google Scholar]
- Cavaille J, et al. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci U S A. 2000;97:14311–6. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain SJ, et al. Evidence for genetic modifiers of postnatal lethality in PWS-IC deletion mice. Hum Mol Genet. 2004;13:2971–7. doi: 10.1093/hmg/ddh314. [DOI] [PubMed] [Google Scholar]
- Crawley JN. Behavioral phenotyping strategies for mutant mice. Neuron. 2008;57:809–18. doi: 10.1016/j.neuron.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Crosson ER, et al. Stable isotope ratios using cavity ring-down spectroscopy: determination of 13C/12C for carbon dioxide in human breath. Anal Chem. 2002;74:2003–7. doi: 10.1021/ac025511d. [DOI] [PubMed] [Google Scholar]
- de Smith AJ, et al. A Deletion of the HBII-85 Class of Small Nucleolar RNAs (snoRNAs) is Associated with Hyperphagia, Obesity and Hypogonadism. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, et al. SnoRNA Snord116 (Pwcr1/MBII-85) deletion causes growth deficiency and hyperphagia in mice. PLoS ONE. 2008;3:e1709. doi: 10.1371/journal.pone.0001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F, et al. Lack of Pwcr1/MBII-85 snoRNA is critical for neonatal lethality in Prader-Willi syndrome mouse models. Mamm Genome. 2005;16:424–31. doi: 10.1007/s00335-005-2460-2. [DOI] [PubMed] [Google Scholar]
- Doe CM, et al. Loss of the imprinted snoRNA mbii-52 leads to increased 5htr2c pre-RNA editing and altered 5HT2CR-mediated behaviour. Hum Mol Genet. 2009;18:2140–8. doi: 10.1093/hmg/ddp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dracheva S, et al. Editing of serotonin 2C receptor mRNA in the prefrontal cortex characterizes high-novelty locomotor response behavioral trait. Neuropsychopharmacology. 2009;34:2237–51. doi: 10.1038/npp.2009.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dracheva S, et al. Increased serotonin 2C receptor mRNA editing: a possible risk factor for suicide. Mol Psychiatry. 2008;13:1001–10. doi: 10.1038/sj.mp.4002081. [DOI] [PubMed] [Google Scholar]
- Ellacott KL, Cone RD. The central melanocortin system and the integration of short- and long-term regulators of energy homeostasis. Recent Prog Horm Res. 2004;59:395–408. doi: 10.1210/rp.59.1.395. [DOI] [PubMed] [Google Scholar]
- Emeson RB, et al. Alternative production of calcitonin and CGRP mRNA is regulated at the calcitonin-specific splice acceptor. Nature. 1989;341:76–80. doi: 10.1038/341076a0. [DOI] [PubMed] [Google Scholar]
- Englander MT, et al. How stress and fluoxetine modulate serotonin 2C receptor pre-mRNA editing. J Neurosci. 2005;25:648–51. doi: 10.1523/JNEUROSCI.3895-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald LW, et al. Messenger RNA editing of the human serotonin 5-HT2C receptor. Neuropsychopharmacology. 1999;21:82S–90S. doi: 10.1016/S0893-133X(99)00004-4. [DOI] [PubMed] [Google Scholar]
- Flomen R, et al. Evidence that RNA editing modulates splice site selection in the 5-HT2C receptor gene. Nucleic Acids Res. 2004;32:2113–22. doi: 10.1093/nar/gkh536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel JM, et al. A transgene insertion creating a heritable chromosome deletion mouse model of Prader-Willi and angelman syndromes. Proc Natl Acad Sci U S A. 1999;96:9258–63. doi: 10.1073/pnas.96.16.9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher RC, et al. Evidence for the role of PWCR1/HBII-85 C/D box small nucleolar RNAs in Prader-Willi syndrome. Am J Hum Genet. 2002;71:669–78. doi: 10.1086/342408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard M, et al. Disruption of the mouse necdin gene results in early post-natal lethality. Nat Genet. 1999;23:199–202. doi: 10.1038/13828. [DOI] [PubMed] [Google Scholar]
- Glowinski J, Iversen LL. Regional studies of catecholamines in the rat brain. I. The disposition of [3H]norepinephrine, [3H]dopamine and [3H]dopa in various regions of the brain. J Neurochem. 1966;13:655–69. doi: 10.1111/j.1471-4159.1966.tb09873.x. [DOI] [PubMed] [Google Scholar]
- Goldstone AP. Prader-Willi syndrome: advances in genetics, pathophysiology and treatment. Trends Endocrinol Metab. 2004;15:12–20. doi: 10.1016/j.tem.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu Rev Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- Gunay-Aygun M, et al. The changing purpose of Prader-Willi syndrome clinical diagnostic criteria and proposed revised criteria. Pediatrics. 2001;108:E92. doi: 10.1542/peds.108.5.e92. [DOI] [PubMed] [Google Scholar]
- Gurevich I, et al. Modulation of serotonin 2C receptor editing by sustained changes in serotonergic neurotransmission. J Neurosci. 2002;22:10529–32. doi: 10.1523/JNEUROSCI.22-24-10529.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler LK, et al. Activation of central melanocortin pathways by fenfluramine. Science. 2002;297:609–11. doi: 10.1126/science.1072327. [DOI] [PubMed] [Google Scholar]
- Hewitt KN, et al. Serotonin 2C receptor agonists and the behavioural satiety sequence in mice. Pharmacol Biochem Behav. 2002;71:691–700. doi: 10.1016/s0091-3057(01)00709-2. [DOI] [PubMed] [Google Scholar]
- Ho SN, et al. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–9. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Hoopengardner B, et al. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–6. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- Infante C, et al. Isotope dilution measurement of breast-milk production in Chilean urban mothers. Hum Nutr Clin Nutr. 1985;39:379–86. [PubMed] [Google Scholar]
- Iwamoto K, Kato T. RNA editing of serotonin 2C receptor in human postmortem brains of major mental disorders. Neurosci Lett. 2003;346:169–72. doi: 10.1016/s0304-3940(03)00608-6. [DOI] [PubMed] [Google Scholar]
- Jong MT, et al. Imprinting of a RING zinc-finger encoding gene in the mouse chromosome region homologous to the Prader-Willi syndrome genetic region. Hum Mol Genet. 1999;8:795–803. doi: 10.1093/hmg/8.5.795. [DOI] [PubMed] [Google Scholar]
- Kawahara Y, et al. Dysregulated editing of serotonin 2C receptor mRNAs results in energy dissipation and loss of fat mass. J Neurosci. 2008;28:12834–44. doi: 10.1523/JNEUROSCI.3896-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–2. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–8. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- Lee JS, Lifson N. Measurement of total energy and material balance in rats by means of doubly labeled water. Am J Physiol. 1960;199:238–42. doi: 10.1152/ajplegacy.1960.199.2.238. [DOI] [PubMed] [Google Scholar]
- Lifson N. Theory of use of the turnover rates of body water for measuring energy and material balance. J Theor Biol. 1966;12:46–74. doi: 10.1016/0022-5193(66)90185-8. [DOI] [PubMed] [Google Scholar]
- Marion S, et al. RNA editing induces variation in desensitization and trafficking of 5-hydroxytryptamine 2c receptor isoforms. J Biol Chem. 2004;279:2945–54. doi: 10.1074/jbc.M308742200. [DOI] [PubMed] [Google Scholar]
- McClintock R, Lifson N. Applicability of the D2018 method to the measurement of the total carbon dioxide output of obese mice. J Biol Chem. 1957;226:153–6. [PubMed] [Google Scholar]
- Mengod G, et al. The distribution and cellular localization of the serotonin 1C receptor mRNA in the rodent brain examined by in situ hybridization histochemistry. Comparison with receptor binding distribution. Neuroscience. 1990;35:577–91. doi: 10.1016/0306-4522(90)90330-7. [DOI] [PubMed] [Google Scholar]
- Meyer OA, et al. A method for the routine assessment of fore- and hindlimb grip strength of rats and mice. Neurobehav Toxicol. 1979;1:233–6. [PubMed] [Google Scholar]
- Mizuno TM, et al. Fasting regulates hypothalamic neuropeptide Y, agouti-related peptide, and proopiomelanocortin in diabetic mice independent of changes in leptin or insulin. Endocrinology. 1999;140:4551–7. doi: 10.1210/endo.140.10.6966. [DOI] [PubMed] [Google Scholar]
- Muscatelli F, et al. Disruption of the mouse Necdin gene results in hypothalamic and behavioral alterations reminiscent of the human Prader-Willi syndrome. Hum Mol Genet. 2000;9:3101–10. doi: 10.1093/hmg/9.20.3101. [DOI] [PubMed] [Google Scholar]
- Nicholls RD, Knepper JL. Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu Rev Genomics Hum Genet. 2001;2:153–75. doi: 10.1146/annurev.genom.2.1.153. [DOI] [PubMed] [Google Scholar]
- Niswender CM, et al. RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. J Biol Chem. 1999;274:9472–8. doi: 10.1074/jbc.274.14.9472. [DOI] [PubMed] [Google Scholar]
- Niswender CM, et al. RNA editing of the human serotonin 5-HT2C receptor. alterations in suicide and implications for serotonergic pharmacotherapy. Neuropsychopharmacology. 2001;24:478–91. doi: 10.1016/S0893-133X(00)00223-2. [DOI] [PubMed] [Google Scholar]
- Nonogaki K, et al. Hyperactivity and reduced energy cost of physical activity in serotonin 5-HT(2C) receptor mutant mice. Diabetes. 2003;52:315–20. doi: 10.2337/diabetes.52.2.315. [DOI] [PubMed] [Google Scholar]
- Nonogaki K, et al. Leptin-independent hyperphagia and type 2 diabetes in mice with a mutated serotonin 5-HT2C receptor gene. Nat Med. 1998;4:1152–6. doi: 10.1038/2647. [DOI] [PubMed] [Google Scholar]
- Pin JP, et al. Domains involved in the specificity of G protein activation in phospholipase C-coupled metabotropic glutamate receptors. EMBO J. 1994;13:342–8. doi: 10.1002/j.1460-2075.1994.tb06267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell CS, et al. Effects of energy expenditure and Ucp1 on photoperiod-induced weight gain in collard lemmings. Obes Res. 2002;10:541–50. doi: 10.1038/oby.2002.73. [DOI] [PubMed] [Google Scholar]
- Price RD, et al. RNA editing of the human serotonin 5-HT2C receptor alters receptor-mediated activation of G13 protein. J Biol Chem. 2001;276:44663–8. doi: 10.1074/jbc.M106745200. [DOI] [PubMed] [Google Scholar]
- Runte M, et al. Exclusion of the C/D box snoRNA gene cluster HBII-52 from a major role in Prader-Willi syndrome. Hum Genet. 2005;116:228–30. doi: 10.1007/s00439-004-1219-2. [DOI] [PubMed] [Google Scholar]
- Sahoo T, et al. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet. 2008;40:719–21. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MW, et al. Leptin increases hypothalamic pro-opiomelanocortin mRNA expression in the rostral arcuate nucleus. Diabetes. 1997;46:2119–23. doi: 10.2337/diab.46.12.2119. [DOI] [PubMed] [Google Scholar]
- Singh M, et al. Hyperphagia-mediated obesity in transgenic mice misexpressing the RNA-editing enzyme ADAR2. J Biol Chem. 2007;282:22448–59. doi: 10.1074/jbc.M700265200. [DOI] [PubMed] [Google Scholar]
- Skryabin BV, et al. Deletion of the MBII-85 snoRNA gene cluster in mice results in postnatal growth retardation. PLoS Genet. 2007;3:e235. doi: 10.1371/journal.pgen.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi MS, et al. A rapid new assay to detect RNA editing reveals antipsychotic-induced changes in serotonin-2C transcripts. Mol Pharmacol. 2005;68:711–9. doi: 10.1124/mol.105.014134. [DOI] [PubMed] [Google Scholar]
- Somerville EM, et al. 5-HT(2C) receptor activation inhibits appetitive and consummatory components of feeding and increases brain c-fos immunoreactivity in mice. Eur J Neurosci. 2007;25:3115–24. doi: 10.1111/j.1460-9568.2007.05567.x. [DOI] [PubMed] [Google Scholar]
- Tecott LH, et al. Eating disorder and epilepsy in mice lacking 5-HT2c serotonin receptors. Nature. 1995;374:542–6. doi: 10.1038/374542a0. [DOI] [PubMed] [Google Scholar]
- Tsai TF, et al. Necdin-deficient mice do not show lethality or the obesity and infertility of Prader-Willi syndrome. Nat Genet. 1999a;22:15–6. doi: 10.1038/8722. [DOI] [PubMed] [Google Scholar]
- Tsai TF, et al. Paternal deletion from Snrpn to Ube3a in the mouse causes hypotonia, growth retardation and partial lethality and provides evidence for a gene contributing to Prader-Willi syndrome. Hum Mol Genet. 1999b;8:1357–64. doi: 10.1093/hmg/8.8.1357. [DOI] [PubMed] [Google Scholar]
- Olaghere da Silva UB, C. CE, Morabito MV, Jacobs MM, Emeson RB, Li D, Airey DC, Sanders-Bush E. Impact of RNA editing on functions of the serotonin 2C receptor in vivo. 2009 doi: 10.3389/neuro.23.001.2010. in submission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali P, et al. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J Cell Biol. 2005;169:745–53. doi: 10.1083/jcb.200411129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, et al. Altered G protein-coupling functions of RNA editing isoform and splicing variant serotonin2C receptors. J Neurochem. 2000;74:1290–300. doi: 10.1046/j.1471-4159.2000.741290.x. [DOI] [PubMed] [Google Scholar]
- Xu Y, et al. 5-HT2CRs expressed by pro-opiomelanocortin neurons regulate energy homeostasis. Neuron. 2008;60:582–9. doi: 10.1016/j.neuron.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, et al. A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nat Genet. 1998;19:25–31. doi: 10.1038/ng0598-25. [DOI] [PubMed] [Google Scholar]
- Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. 2005;12:1047–64. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]