Abstract
The epidermal growth factor (EGF) receptor is a receptor tyrosine kinase involved in the control of cell proliferation. EGF receptor dimerization is a pre-requisite for kinase activation, and is the basis for exploring the feasibility of inhibiting EGF receptor activation by blocking dimer formation with small molecules. A compound, NSC56452, initially identified by virtual screening, was shown experimentally to inhibit EGF receptor kinase activation in a dose-dependent manner. This compound blocked EGF-stimulated dimer formation as measured by chemical cross-linking and luciferase fragment complementation. The compound was further shown to inhibit the growth of HeLa cells. This first-generation lead compound represents the first small-molecule inhibitor of EGF receptor activation that is not directed against the intracellular kinase domain.
Introduction
The EGF receptor is a transmembrane receptor tyrosine kinase that belongs to the ErbB super family. It plays a critical role in the regulation of signaling cascades that ultimately induce cell proliferation, survival, and migration (Yarden and Schlessinger, 1987). The receptor is composed of an extracellular ligand-binding domain, a single transmembrane helix, an intracellular tyrosine kinase domain, and an ~200 amino acid C-terminal tail (Ullrich et al., 1984). Thought to exist as monomer in cell membranes, the receptor is known to dimerize upon binding EGF (Yarden and Schlessinger, 1987). Dimerization leads to stimulation of the receptor’s tyrosine kinase activity (Zhang et al., 2006), resulting in transphosphorylation of specific tyrosine residues on the C-terminal tail of the receptor (Yarden and Schlessinger, 1987). These phosphotyrosines serve as binding sites for a variety of proteins that mediate activation of critical downstream signaling pathways (Yarden and Sliwkowski, 2001).
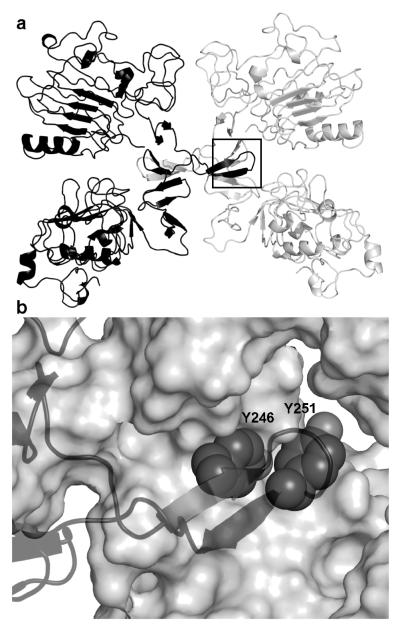
Dimerization is a prerequisite for EGF receptor activation (Yarden and Sliwkowski, 2001), and is driven by interactions between the extracellular domains of the two monomeric partners (Dawson et al., 2005; Ferguson et al., 2003; Garrett et al., 2002; Ogiso et al., 2002). Recent crystal structures of the dimeric extracellular domain revealed that the dimerization interface was centered on a protruding loop, known as the dimerization arm (Figure 1). Consistent with the importance of this loop, mutation of either Tyr-246 or Tyr-251 within this arm abolishes EGF receptor homodimer formation (Dawson et al., 2005; Walker et al., 2004).
Figure 1.
The target site at the EGF receptor dimerization interface. A) Crystal structures of the extracellular domain of the EGF receptor homodimer (PDB:1MOX) and the dimerization arm (box). B) Critical residues Y246 and Y251 pack into adjacent pockets at the dimer interface.
We reasoned that the critical requirement for ectodomain dimerization could be exploited to inhibit receptor activation, and set out to test the feasibility of targeting the dimerization process with small molecules. In the present work, we report the identification of a small-molecule lead compound capable of inhibiting the activation of the EGF receptor by blocking dimer formation. This inhibitor was initially identified by applying a consensus virtual high-throughput screening (vHTS) protocol to screen the National Cancer Institute Diversity (NCI-Diversity) library (http://dtp.nci.nih.gov/branches/dscb/diversity_explanation.html) for compounds with the potential to bind to the same pocket that Tyr-246 and Tyr-251 of the dimerization arm recognize. Subsequent biochemical assays confirmed that this compound selectively impaired EGF receptor dimerization and inhibited cell proliferation. This compound represents the first member of a new class of small-molecule inhibitors of EGF receptor activation and signal transduction.
Results
Assessment of the virtual high-throughput screening protocol
The vHTS employed in these experiments used AutoDock 4.0 (Huey et al., 2007; Morris et al., 1998) to dock approximately 2000 compounds present in the NCI Diversity database to a 25 Å3 docking box centered on the Tyr-246/Tyr-251 recognition pocket of the dimerization arm of the EGF receptor. For each compound, 100 docking poses were generated using AutoDock, and each pose was combined with the original protein structure without re-minimization to form a docked protein-ligand complex. These complexes were subsequently re-scored based on the optimality of the complexes and independently re-ranked accordingly by 8 additional scoring functions. These generated rankings were then summed together in an equal-weight consensus schema to yield a final ranking for each compound pose. The final ranking of each compound represents its best-ranked compound pose calculated by this consensus scheme. Details of the protocol are presented in the method section.
The performance of our consensus vHTS protocol was subsequently assessed by evaluating the enrichment power. The enrichment of a vHTS protocol is typically measured by its ability to recover true positives as early as possible in a ranked compound library. Protocol evaluation thus depends on the availability of existing reference active compounds. In the current case, because there were no existing dimerization inhibitors, it was not possible to evaluate the enrichment power of our vHTS protocol for the EGF receptor system a priori. As a result, robustness, measured as the average enrichment across different protein targets, became a critical criterion for evaluating the protocol performance. Our protocol was applied to four different protein targets: plasmepsin II (PMII), human cyclin-dependent kinase 2 (Cdk2), estrogen receptor (ER), and yeast heat shock protein (Hsp90). Structurally diverse compounds (positives) bound to these protein targets were extracted from co-crystal structures in the Protein Data Bank, and were mixed with 1926 decoy compounds (negatives) to construct a testing library. The ability of the vHTS protocol to recover positives was evaluated using enrichment-curve analysis (Chen et al., 2006).
The protocol recovered at least one true positive within the top 1% of the ranked library for PMII, Cdk2, and ER, and within the top 15% of the library for Hsp90 (Figure 2, Table 1). On average, this protocol is expected to recover at least one true ligand within the top 3.5%, and nearly 2/3 of all ligands within the top 15% of the representative libraries.
Figure 2.
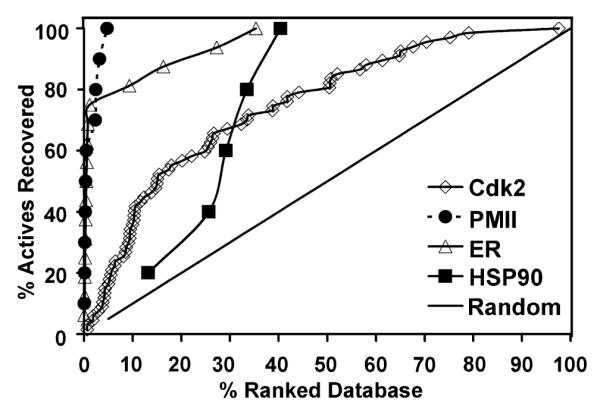
Evaluation of the vHTS protocol against four test cases shown in an enrichment curve analysis. In each case, multiple known ligands were mixed with ~2000 random compounds to form the screening library. The black diagonal line represents the random distribution of active molecules.
Table 1.
Efficacy and robustness of the vHTS protocol.
| Targets | Coverage1%a | Coverage15% | Coverage30% | Coverage50% | Best b |
|---|---|---|---|---|---|
| Cdk2 | 3% | 49% | 67% | 79% | 0.05% |
| PMII | 60% | 100% | 100% | 100% | 0.65% |
| ER | 69% | 81% | 94% | 100% | 0.05% |
| HSP90 | 0% | 20% | 60% | 100% | 13.21% |
| Avg | 33% | 63% | 80% | 100% | 3.5% |
Coveragefraction = Number of known actives recovered within the given fraction of the database / Total number of actives present in the database × 100%
Best = ranking of the best predicted active / database size × 100%
Inhibition of EGF receptor activation as the first-pass screen
We applied the vHTS protocol to the EGF receptor and obtained samples of the 80 top-ranked compounds (top 4%) along with 40 randomly chosen compounds from NCI for testing. Of the 80 compounds, 4 were not soluble in water or DMSO, and, therefore, not pursued further. The remaining 76 compounds were tested for their ability to inhibit EGF-stimulated receptor autophosphorylation in cells at a concentration of 100 μM.
Of the 76 compounds tested, 20 produced a significant (>60%) decrease in activation as measured by the level of phosphorylation at Tyr-1173, the major site of autophosphorylation on the EGF receptor. By contrast, none of the 40 compounds randomly chosen from the same library inhibited receptor phosphorylation when assayed under the same conditions (results not shown). This highlights the enrichment and the utility of our vHTS protocol in the present application to target the EGF receptor system.
Subsequent dose-response experiments following the initial screen at 100 μM confirmed the inhibitory effects of these compounds. Figure 3 presents the structure and the characterization of NSC56452, a compound that turned out to be the featured inhibitor in this work. In the phosphorylation assay, NSC56452 exhibited an estimated IC50 value of 0.4 μM, the most potent compound of all 20 first-round candidates.
Figure 3.
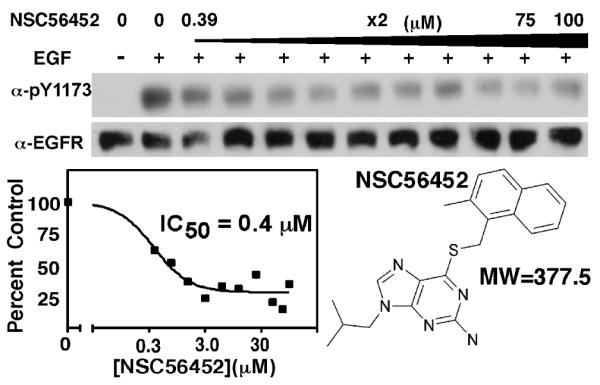
Inhibition of EGF receptor autophosphorylation by NSC56452. Cells were pre-incubated with NSC56452 or 1% DMSO for the control. Inhibition of EGF receptor autophosphorylation of the controls (lane 1 and 2) and increasing doses of NSC56452. Residual kinase activity was estimated by densitometry and plotted to obtain IC50 values. Structure and molecular weights of NSC56452 is also shown.
Specific inhibition of EGF receptor activation by lead compounds
To assess the specificity of inhibition, the 20 candidates were tested for their ability to inhibit two related receptor tyrosine kinases, the insulin receptor and the PDGF receptor. For the insulin receptor, insulin-stimulated tyrosine phosphorylation of IRS-1 in differentiated 3T3-L1 cells was assessed (Semenkovich et al., 1989). For the PDGF receptor, PDGF-stimulated receptor autophosphorylation in NIH3T3 cells was measured (Nakata et al., 2007). Neither 3T3-L1 cells nor NIH3T3 cells express the EGF receptor obviating potential problems associated with receptor crosstalk.
Of the 20 compounds that inhibited EGF receptor autophosphorylation, 2 inhibited insulin-stimulated IRS-1 phosphorylation and 4 others inhibited PDGF receptor autophosphorylation (Figure 4). An additional 3 compounds markedly enhanced PDGF receptor autophosphorylation. A total of 9 compounds were thus eliminated from further consideration due to lack of specificity for the EGF receptor.
Figure 4.
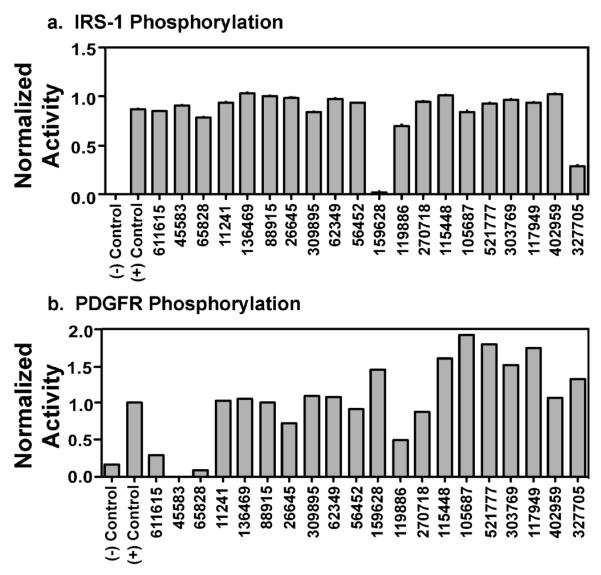
Specificity of the inhibitors for the EGF receptor. Cells expressing either the insulin receptor or the PDGF receptor were pre-incubated with 1% DMSO (controls) or 100 μM of each of the 20 candidate compounds. A) Insulin receptor kinase activity was assessed by measuring the phosphorylation of IRS-1 in response to 3 nM insulin for 1 minute. The data shown are representative of three separate experiments. B) PDGF receptor kinase activity was assessed by measuring autophosphorylation of the PDGF receptor in response to 2 nM PDGF for 3 minutes. The data shown are representative of two separate experiments.
Inhibition of EGF receptor dimerization by lead compounds
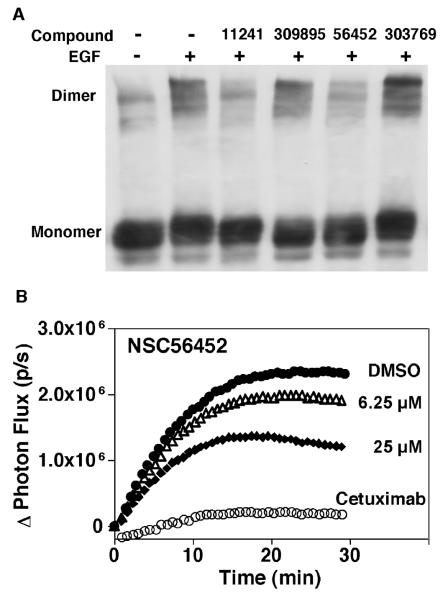
Since the lead inhibitors were initially chosen based on their potential to interfere with EGF receptor dimerization, we next determined whether the remaining 11 candidates inhibited EGF receptor autophosphorylation by directly blocking receptor dimerization. Two independent assays, chemical cross-linking and a novel live-cell reporting assay, were utilized for the purpose of obtaining confirmatory and complementary results. In the chemical cross-linking experiment, cells were preincubated with the inhibitors for 15 min at a final concentration of 100 μM. EGF at 25 nM was then added followed by 3 mM BS3, a membrane-impermeable chemical cross-linker. Figure 5A shows the effect of a subset of these inhibitors on the cross-linking of EGF receptor dimers.
Figure 5.
Testing for inhibition of EGF receptor dimerization by two methods. A) Chemical cross-linking assay. Cells were pre-incubated with 1% DMSO (lane 1 and 2) or 100 μM of different inhibitors (lane 3-6) prior to stimulation with 25 nM EGF (lane 2-6) for 5 minutes. All cells were then treated with 3 mM BS3. NSC11241 (lane 3) and NSC56452 (lane 5) significantly inhibit dimer formation, while Lane 4 and 6 show compounds that did not inhibit dimer formation. B) Luciferase fragment complementation. Cells stably expressing ΔC-EGFR-NLuc and ΔC-EGFR-CLuc were pre-treated with DMSO, the indicated concentrations of NSC56452 or 1 μg/ml cetuximab for 20 min in the presence of 0.6 mg/ml D-luciferin prior to the addition of 3 nM EGF. All assays were performed in quadruplicate. Data represent the change in photon flux between cells treated with or without EGF.
Among the 11 tested compounds, only NSC56452 and 1 other compound (NSC11241) significantly reduced the formation of high molecular weight oligomers while the remaining compounds, as represented by NSC309895 and NSC303769, failed to block oligomer formation. Because the cross-linker was used at a concentration 30-fold higher than that of the inhibitors (3 mM vs. 100 μM), it is unlikely that this inhibition was due to quenching of the cross-linking reaction by the compounds. Consistent with this conclusion, increasing the concentration of cross-linker BS3 to 5 mM yielded the same results (results not shown). It is possible, however, that false negatives may have been obtained if reaction of the test compound with cross-linker prevented that compound from binding to the EGF receptor.
The two hits from the chemical cross-linking assay, NSC56452 and NSC11241, were further characterized by the luciferase fragment complementation imaging assay recently developed in our lab (Yang et al., 2009). In this assay, an EGF receptor lacking the entire intracellular domain (referred to as ΔC-EGFR) is fused to either an N-terminal (NLuc) or a C-terminal (CLuc) fragment of firefly luciferase. Ligand-induced dimerization of the ΔC-EGFR brings the luciferase fragments into close proximity resulting in enzyme complementation and luciferase activity. The rate and extent of receptor dimerization could, therefore, be measured by following photon flux in the presence of luciferin. Figure 5B shows the dose-response of NSC56452 in comparison to DMSO and cetuximab treatments while figure S1 displays results from a wider range of doses.
As expected, EGF stimulated a rapid increase in light production in DMSO-treated control cells reflective of ligand-induced dimer formation. Cetuximab, an FDA-approved antibody-based drug that binds to the extracellular domain of the EGF receptor (Kirkpatrick et al., 2004; Li et al., 2005), dramatically decreased EGF-induced luciferase activity thus serving as a positive control for dimer inhibition. Consistent with the cross-linking experiment, NSC56452 exhibited a dose-dependent inhibition of the dimer-driven luciferase complementation (IC50 of ~7 μM) and maximally inhibited dimerization by 50% to 60%. The absence of the intracellular domain in these EGF receptor/luciferase constructs eliminated the possibility that inhibition by the compound involves interaction with the kinase domain, and suggested that NSC56452 acted by directly inhibiting interaction of the extracellular domains. Consistent with this interpretation, erlotinib, an intracellular tyrosine kinase inhibitors, had no effect on luciferase complementation when tested under the same conditions (Figure S2). The other compound, NSC11241, failed to inhibit in a dose-dependent manner (Figure S3) and was not pursued further. NSC56452 displayed no inhibitory effect on the complementation of the NLuc and CLuc fragments themselves since it did not inhibit an independent control system in which luciferase fragments were fused to FRB-NLuc and its binding partner, CLuc-FKBP (Luker et al., 2004) (results not shown).
Additional mechanistic characterization of NSC56452
The luciferase complementation data using the ΔC-EGFR constructs suggest that NSC56452 inhibit dimerization by binding to a site on the extracellular domain of the receptor. However, it was possible that the observed inhibition might be due to interference with the binding of EGF to the receptor at the ligand-binding site. To address this possibility, we measured the effect of NSC56452 on the binding of 125I-EGF to CHO cells expressing the EGF receptor. As shown in Figure S4, NSC56452 had little effect on the binding of EGF but inhibited kinase activity by ~75%. This suggests that this compound does not compete with EGF for binding to the EGF receptor and is unlikely to inhibit kinase activity through an effect on ligand binding.
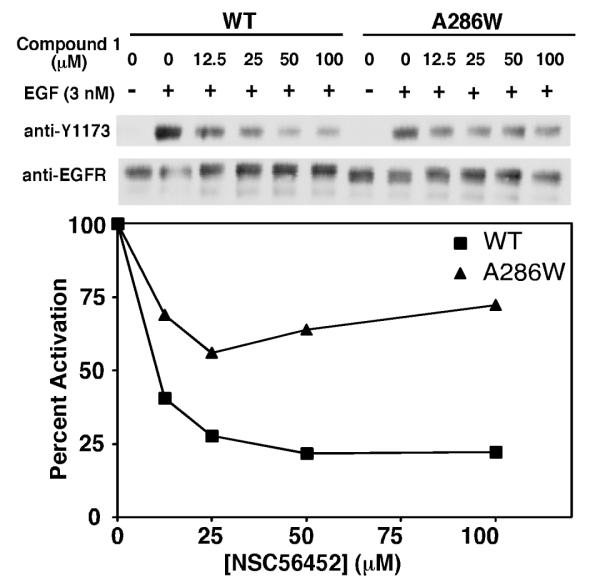
Based on the predicted docking site of NSC56452 (Figure S5), one of the compound’s ring systems overlaps with the pair of adjacent interaction sites for Tyr-246 and Tyr-251 of the partner dimerization arm, with a second ring system extending into a secondary pocket distal to the main dimerization interface (Figure S5, around residue Ala-286). To determine whether NSC56452 was likely to bind at the predicted site, we reasoned that a substitution of Ala-286 with a bulky Trp residue (A286W) at this distal secondary site would sterically hinder the binding of NSC56452 while preserving the natural process of inter-receptor dimerization.
The autophosphorylation of wild type and A286W-EGF receptors in the presence of NSC56452 is compared in Figure 6. Although the mutant showed a slight decrease in activation potency compared to wild type receptor, we were still able to measure the relative inhibition by the inhibitor. NSC56452 dose-dependently inhibited kinase activity of wild type EGF receptor by a maximum of 75%. By contrast, the inhibitory effect of NSC56452 was markedly weaker in cells expressing the A286W-EGF receptor, showing only about 25% maximal inhibition. These results are consistent with the hypothesis that the A286W mutation sterically hinders the binding of NSC56452, and thus, support the predicted docking site of the compound.
Figure 6.
Effect of the A286W mutation on the sensitivity to NSC56452 inhibition. Mixed CHO cell culture expressing either wild type or A286W-EGF receptors were pre-incubated with 1%DMSO (control) or NSC56452 at the indicated doses for 20 min at room temperature before stimulation with 3 nM EGF for 1 minute. Data shown are representative of two separate experiments.
Growth inhibition of HeLa cells by NSC56452
To assess the effect of NSC56452 in a longer term assay, we next tested its ability to inhibit the proliferation of HeLa cells which express endogenous EGF receptors (Figure 7). For comparison, the cells were also treated with the EGF receptor tyrosine kinase inhibitor erlotinib. By itself, NSC56452 induced significant inhibition of cell growth at 50 μM but had little effect at the lower dose of 12.5 μM. Interestingly, the growth inhibition at this sub-optimal dose of 12.5 μM was significantly enhanced when NSC56452 was given in combination with a dose of erlotinib that by itself failed to inhibit HeLa cell proliferation. The apparent synergistic effect of the two compounds is consistent with the hypothesis that NSC56452 inhibits EGF receptor activity through a mechanism different from that of the classical tyrosine kinase inhibitors.
Figure 7.
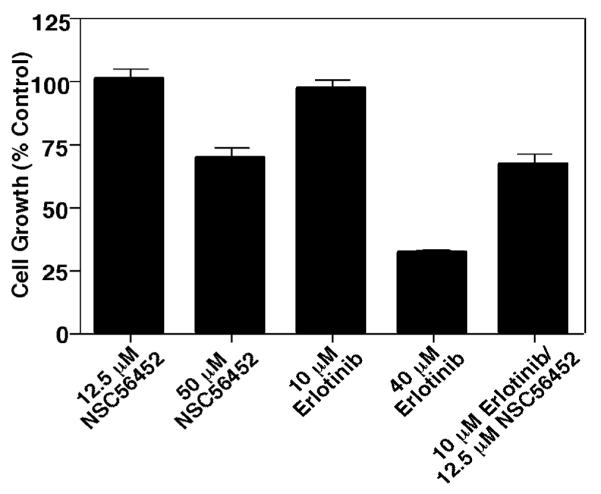
Inhibition of HeLa cell proliferation. Cells were grown in the absence or presence of erlotinib, NSC56452, or a combination of the two inhibitors at the indicated doses. Cell proliferation was measured by the cellTiter 96 Aqueous One Solution Cell Proliferation Assay after 48 hr incubation with the inhibitors. All experiments were performed in triplicate. All cultures contained 1% DMSO.
Discussion
Recent X-ray crystallographic structures of the EGF receptor have identified the mechanism through which the receptor dimerizes (Ferguson et al., 2003; Garrett et al., 2002; Ogiso et al., 2002). In these structures, residues 242-259 comprise a dimerization arm that mediates receptor-receptor interactions. Based on the sensitivity of receptor dimerization to mutations of critical residues on the arm (Dawson et al., 2005; Walker et al., 2004), we hypothesized that these interactions could be exploited to discover compounds capable of interfering with EGF receptor dimerization and activation.
In this work, we adopted a top-down approach that combines virtual high-throughput screening with biochemical assays to identify inhibitors of EGF receptor dimerization. By testing only 4% of the NCI-diversity library, we identified a lead compound, NSC56452, which selectively inhibits EGF-stimulated tyrosine kinase activity while having no effect against other related PDGF and insulin membrane receptor tyrosine kinases. Several lines of evidence suggest that NSC56452 works by inhibiting EGF receptor dimerization. In both the chemical cross-linking assay and the luciferase fragment complementation assay, NSC56452 significantly reduced the formation of EGF-induced high molecular-weight oligomers and luciferase complementation, respectively. It should be noted that since the luciferase fragment complementation assay utilized only the extracellular portion of the EGF receptor, the ability of NSC56452 to inhibit luciferase activity thus indicates that the effect is due to its interaction with the ectodomain of the EGF receptor. This is a significant contrast to classical tyrosine kinase inhibitors. Interestingly, NSC56452 acted synergistically with the EGF receptor-specific tyrosine kinase inhibitor, erlotinib, to inhibit HeLa cell proliferation which provided further support that the two inhibitors act via different mechanisms.
At the dimerization interface, NSC56452 is predicted to bind to two distinct pockets (see Supplemental Figure S4). The first is the pocket that recognizes Tyr-246/Tyr-251 which would explain NSC56452’s ability to inhibit receptor dimerization. The second pocket is distal to the main receptor dimerization interface and appears to serve as an auxiliary anchor point for inhibitor binding.
Based on the computational docking model of NSC56452 (Supplemental Figure S4), mutation of residue Ala-286 to a bulky Trp residue at the secondary binding pocket was predicted to impair the binding of the compound to the receptor. Indeed, EGF receptors bearing the A286W mutation were significantly less sensitive to inhibition by NSC56452 than wild type receptors. While a co-crystal structure is necessary to definitively identify the binding mode of NSC56452, the results of our mutational analysis are consistent with the computational model and the proposed inhibition mechanism.
We do note that NSC56452 exhibits different IC50 values for inhibiting receptor autophosphorylation (~0.4 μM) and receptor dimerization, asmeasured by the luciferase fragment complementation assay (~7 μM). The basis for this discrepancy is not known but may be due to the fact that the autophosphorylation studies utilized full length EGF receptor where the luciferase assay uses a receptor lacking the entire intracellular domain. As the intracellular domain is known to contribute to receptor-receptor interactions (cite Jura et al paper), the dimerization interface of the extracellular domains may be affected by this truncation. In addition, receptor autophosphorylation may occur within dimers as well as higher-order oligomers (Clayton et al., 2005), which would make this process more sensitive to changes in the upstream receptor dimerization status.
In addition to its utility as a research tool, NSC56452 may serve as a lead for the development of anticancer agents to complement existing therapeutics. In the recent release of the NCI Cancer Screening Data against 60 cancer cell lines (http://dtp.nci.nih.gov/compare-web-public_compare/login.do, accessed in April of 2009), NSC56452 was reported to inhibit the growth of three cancer cell lines, SK-MEL-5, UO-31, and COLO205 cells. SK-MEL-5 (Liu et al., 2007) and UO-31 (Bishop et al., 2002) cells both overexpress EGF receptors while COLO205 cells overexpress the highly homologous ErbB2 receptor (Vernimmen et al., 2003). The apparent sensitivity of these EGF receptor/ErbB-expressing cells to NSC56452 is intriguing and suggests that optimized versions of this compound may be useful chemotherapeutic agents.
Significance
The epidermal growth factor receptor is a membrane receptor tyrosine kinase whose over-activation has been implicated to cause many human cancers. Novel strategies to inhibit the activation of EGF receptors other than the conventional antibody-based and tyrosine kinase inhibitors are virtually non-existent but could provide additional benefits both in laboratory and clinical settings. In particular, ongoing researches have continuously highlighted resistance as an elevated challenge based on reports that tumors characterized by abnormal levels of EGFR homodimerization (Abd El-Rehim et al., 2004; Wong et al., 1992), EGFR-ErbB2 heterodimerization (Zhan et al., 2006), and mutations within the ATP-binding site of the kinase domain (Bell et al., 2005; Sharma et al., 2007) often display increased resistance to current treatments. In this setting, inhibitors that block signaling by multiple ErbB receptors could potentially represent more effective chemotherapeutic agents than those targeting one of the ErbB receptors. In an effort to “think outside of the kinase”, we focused on targeting the protein-protein interactions that drives the homodimerization of the EGF receptors. The top-down workflow of our screening approach combined virtual screening with a series of biochemical assays that included a novel real-time luciferase fragment imaging assay to tackle the long-standing challenge of measuring EGF receptor dimerization. NSC56452, as a proof-of-concept dimer-inhibiting compound, also represents the first application of the luciferase assay that demonstrates its compound-screening utility. Further optimization of both the lead compound and the assay can be extended to targeting the heterodimeric interactions among all ErbB receptors.
Methods
Virtual Screening
Autodock 4.0 (Huey et al., 2007; Morris et al., 1998) was used to screen the NCI-diversity database (1990 compounds). The database was initially downloaded from the Autodock website and processed by in-house scripts to fix incorrectly formatted structures, and to exclude structures that contained metals: iron, zinc, mercury and copper (final size = 1926 compounds). A cubic docking box of dimension 25 Å3 was centered at the Tyr-246/Tyr-Y251 recognition site on monomer A of the extracellular dimer crystal structure (PDB: 1MOX). Larmackian genetic algorithm with Solis and Wets local search was used to generate 100 docking poses per compound. All poses were subsequently scored using: HP, HM, HS (implemented in X-score 1.2.1 (Wang et al., 2002)), D-score, PMF, G-score, Chem-score (implemented in Sybyl 7.3 CSCORE module), and Dfire (Zhang et al., 2005). A consensus score for each pose was calculated by summing the rankings given by each of these 8 scoring functions. Three compounds that ranked high using the consensus scores were excluded because they displayed high rankings against other protein targets suggesting poor specificity.
EGF receptor autophosphorylation
CHO cells stably expressing wild type EGF receptor were grown to 80% confluency in 35 mm plates in Hams’ F-12 containing 10% FBS, penicillin/streptomycin, and 100 μg/ml hygromycin. Prior to use, the cells were incubated for 3 hours in F-12 medium containing 0.1% FBS. For the experiments, cultures were incubated with the test compounds at a final concentration of 100 μM in 1% DMSO for 30 min at 25° C in F-12 containing 1 mg/ml bovine serum albumin and 25mM Hepes, pH 7.2. Control cultures were incubated for the same length of time with 1% DMSO. EGF (Biomedical Technologies, Inc) was then added at a final concentration of 3 nM and the cultures incubated at 25° C for an additional 1 min. Subsequently, the monolayers were washed twice with ice-cold phosphate-buffered saline and scraped into RIPA buffer (10 mM Tris, pH 7.2, 150 mM NaCl, 0.1% sodium dodecyl sulfate, 1% Triton X-100, 17 mM deoxycholate, and 2.7 mM EDTA) containing 1 mM sodium orthovanadate, 20 mM p-nitrophenylphosphate, and protease inhibitors. Equal amounts of protein (BCA assay, Pierce) were separated by electrophoresis on a 9% SDS polyacrylamide gel, and transferred to PVDF or nitrocellulose (Millipore). Western blotting was performed using anti-pY1173 (Cell Signaling), or anti-EGF receptor antibodies (Cell Signaling and Santa Cruz). Time-course and dose-response experiments were done using the same procedure except that the dose or preincubation time with inhibitors was varied. A similar protocol was used for assessing insulin-stimulated phosphorylation of IRS-1 or PDGF-stimulated receptor autophosphorylation except that differentiated 3T3-L1 cells or NIH3T3 cells were used, respectively. In all cases, phosphorylation was quantified using ImageJ and normalized to that observed in control samples.
Phosphorylation using A286W mixed culture
The DNA primers for the A286W construct were 5′-CTCGTGCGTCCGATGGTGTGGCGCCGACAGC-3′ and 3′-GCTGTCGGCGCCACACCATCGGACGCACGAG-5′. FI CHO cells (Invitrogen) were stably co-transfected with pOG44 (Invitrogen) and the A286W-EGF receptor in pcDNA5/FRT (Invitrogen). Stable clones were selected in F-12 containing 600 μg/ml hygromycin (InvivoGen). The mixed culture was grown to confluency and maintained in F-12 containing FetalPlex (Gemini bioproducts), 1000 μg/ml penicillin/streptomycin, and 100 μg/ml hygromycin. The experimental phosphorylation protocols were identical as the one described above.
Chemical cross-linking of the EGF receptor
CHO cells stably expressing EGF receptor were preincubated with the test compounds for 15 min at a final concentration of 100 μM. EGF (25 nM) was then added for 3 min followed by the addition of BS3 (Pierce) at a final concentration of 3 mM for 30 min. The reaction mixture was buffered at pH 8. The cross-linking reactions were quenched by the addition of glycine to a final concentration of 1 M (pH 7.5). Cells were lysed as above, and equal amounts of protein were loaded onto a 4%-7.5% gradient SDS-polyacrylamide gel. After electrophoresis and transfer to PVDF, EGF receptor dimerization was measured by Western blotting using anti-EGF receptor antibodies.
Luciferase fragment complementation imaging
CHO-K1 Tet-On cells stably expressing ΔC-EGFR-NLuc and ΔC-EGFR-CLuc (Yang et al., 2009) were plated 48 hrs prior to imaging in DMEM containing 1 μg/ml doxycycline. On the day of imaging, cells were serum-starved for 4 hrs followed by treatment with vehicle, the indicated concentration of each compound, or 1 μg/ml cetuximab for 20 min in the presence of 0.6 mg/ml D-luciferin. 3 nM EGF was then added and the photon flux immediately measured using an IVIS imaging system. Data represent the change in photon flux between EGF-treated cells and control cells. For the control experiments using the FRB-NLuc and CLuc-FKBP system (Luker et al., 2004; Villalobos et al., 2008), CHO-K1 Tet-On cells were plated 48 hrs prior to use and transiently transfected with the cDNA encoding FRB-NLuc and CLuc-FKBP 24 later. On the day of assay, cells were pre-treated with vehicle or 80 nM rapamycin for 4 hrs. Media was removed and replaced with DMEM lacking phenol red containing 0.6 mg/ml D-luciferin and DMSO or 25 μM NSC56452. Photon flux was measured as above. To test the effect of tyrosine kinase inhibitor using this system, 5 μM erlotinib was used, an effective dose that completely inhibited EGF receptor kinase phosphorylation and MAP kinase on full-length receptors under the same conditions by Western blot.
125-I-EGF binding
125I-EGF binding was carried out by incubating the cells with 50 pM 125I-EGF for 24 hr at 4° C, following the previously described protocol (Macdonald and Pike, 2008), in the presence of 1% DMSO (control) or 100 μM NSC56452.
Cell Growth Assay
HeLa cells were grown in Dulbecco’s Modified Eagles’ Medium with 10% FBS. Cells were plated in triplicate in 96-well plates at 5000 cells per well and allowed to grow for 24 hours before the addition of DMSO (control), erlotinib (Genetech) or NSC56452. All cultures contained 1% DMSO in the final media. Cells were then incubated for 48 hours. The cell growth rate was then measured using the cellTiter 96 Aqueous One Solution Cell Proliferation Assay kit according to the manufacturer’s instructions (Promega). Readings were taken at 490 nm after 1 hour incubation with the MTS and PMS solution.
Identification and purity of NSC56452
NSC56452 was characterized by liquid chromatography coupled to mass spectrometry (figure S6). The compound samples were prepared at 1 mg/ml in 100% DMSO. a) Liquid chromatography of NSC56452. The LC column was a Gemini C18 110Å New column (50 mm × 2.0 mm, 5 μm particle size; Phenomenex Inc.) maintained at 20 °C. The mobile phase consisted of water and acetonitrile (95:5, v/v). The flow rate was 0.4 ml/min at a maximum pressure of 300 bar. The total run time was 12 min. The purity was calculated as 95% using MassLynx4.1 software (Waters Corp., Milford, MA). b) The liquid chromatograph was coupled to a mass spectrometer with a turbo electrospray ion source in negative ionization mode. Within the main LC peak at time 8.12 minute, NSC56452 was identified as having a molecular weight of 379.0, in agreement with the expected mass of 378.
Highlights.
- NSC56452 was identified as a novel EGF receptor inhibitor by virtual screening and subsequently confirmed by a series of biochemical assays.
- NSC56452 dose-dependently inhibits the ectodomain-mediated EGF receptor dimerization, which consequently inhibits receptor activation.
- NSC56452 inhibits HeLa growth by itself, and synergistically with erlotinib.
Supplementary Material
Acknowledgements
The authors acknowledge the Drug Synthesis and Chemistry Branch of NCI for supplying the compounds used in this work. This work was supported by NIH grants R01 GM064491 and RO1 GM082824 to LJP and R01 GM068460 to GRM. RYY is grateful for funding support from the Cancer Biology Pathway sponsored by the Siteman Cancer Center, a graduate fellowship from the Division of Biology and Biological Sciences, Washington University, St. Louis, MO, and a pre-doctoral fellowship in informatics by PhRMA foundation, Washington DC.
a Abbreviations
- Cdk2
human cyclin-dependent kinase 2
- CLuc
C-terminal luciferase fragment
- EGF
epidermal growth factor
- ER
estrogen receptor
- Hsp90
heat shock protein 90
- NLuc
N-terminal luciferase fragment
- PDGF
- PMII
plasmepsin II
- vHTS
virtual high-throughput screen
Footnotes
The authors declare no competing financial interests.
Supporting Information Available.
References
- Abd El-Rehim D, Pinder S, Paish C, Bell J, Rampaul R, Blamey R, Robertson J, Nicholson R, Ellis I. Expression and co-expression of the members of the epidermal growth factor receptor (EGFR) family in invasive breast carcinoma. Br J Cancer. 2004;91:1532–1542. doi: 10.1038/sj.bjc.6602184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell DW, Gore I, Okimoto RA, Godin-Heymann N, Sordella R, Mulloy R, Sharma SV, Brannigan BW, Mohapatra G, Settleman J, Haber DA. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet. 2005;37:1315–1316. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- Bishop PC, Myers T, Robey R, Fry DW, Liu ET, Blagosklonny MV, Bates SE. Differential sensitivity of cancer cells to inhibitors of the epidermal growth factor receptor family. Oncogene. 2002;21:119–127. doi: 10.1038/sj.onc.1205028. [DOI] [PubMed] [Google Scholar]
- Chen H, Lyne PD, Giordanetto F, Lovell T, Li J. On Evaluating Molecular-Docking Methods for Pose Prediction and Enrichment Factors. J Chem Inf Model. 2006;46:401–415. doi: 10.1021/ci0503255. [DOI] [PubMed] [Google Scholar]
- Clayton AHA, Walker F, Orchard SG, Henderson C, Fuchs D, Rothacker J, Nice EC, Burgess AW. Ligand-induced Dimer-Tetramer Transition during the Activation of the Cell Surface Epidermal Growth Factor Receptor-A Multidimensional Microscopy Analysis. J Biol Chem. 2005;280:30392–30399. doi: 10.1074/jbc.M504770200. [DOI] [PubMed] [Google Scholar]
- Dawson JP, Berger MB, Lin C-C, Schlessinger J, Lemmon MA, Ferguson KM. Epidermal growth factor receptor dimerization and activation require ligand-induced conformational changes in the dimer interface. Mol Cell Biol. 2005;25:7734–7742. doi: 10.1128/MCB.25.17.7734-7742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KM, Berger MB, Mendrola JM, Cho H-S, Leahy DJ, Lemmon MA. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- Garrett TPJ, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Zhu H-J, Walker F, Frenkel MJ, Hoyne PA. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor [alpha] Cell. 2002;110:763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. J Comput Chem. 2007;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick P, Graham J, Muhsin M. Cetuximab. Nature Reviews Drug Discovery. 2004;3:549–550. doi: 10.1038/nrd1445. [DOI] [PubMed] [Google Scholar]
- Li S, Schmitz KR, Jeffrey PD, Wiltzius JJW, Kussie P, Ferguson KM. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell. 2005;7:301–311. doi: 10.1016/j.ccr.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Liu W, Wu X, Zhang W, Montenegro RC, Fackenthal DL, Spitz JA, Huff LM, Innocenti F, Das S, Cook, Edwin H, Jr., et al. Relationship of EGFR Mutations, Expression, Amplification, and Polymorphisms to Epidermal Growth Factor Receptor Inhibitors in the NCI60 Cell Lines. Clin Cancer Res. 2007;13:6788–6795. doi: 10.1158/1078-0432.CCR-07-0547. [DOI] [PubMed] [Google Scholar]
- Luker KE, Smith MC, Luker GD, Gammon ST, Piwnica-Worms H, Piwnica-Worms D. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proceedings of the National Academy of Sciences. 2004;101:12288–12293. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald JL, Pike LJ. Heterogeneity in EGF-binding affinities arises from negative cooperativity in an aggregating system. Proc Natl Acad Sci U S A. 2008;105:112–117. doi: 10.1073/pnas.0707080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- Nakata S, Fujita N, Kitagawa Y, Okamoto R, Ogita H, Takai Y. Regulation of platelet-derived growth factor receptor activation by afadin through SHP-2: implications for cellular morphology. J Biol Chem. 2007;282:37815–37825. doi: 10.1074/jbc.M707461200. [DOI] [PubMed] [Google Scholar]
- Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim J-H, Saito K, Sakamoto A, Inoue M, Shirouzu M, Yokoyama S. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- Semenkovich C, Wims M, Noe L, Etienne J, Chan L. Insulin regulation of lipoprotein lipase activity in 3T3-L1 adipocytes is mediated at posttranscriptional and posttranslational levels. J Biol Chem. 1989;264:9030–9038. [PubMed] [Google Scholar]
- Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nature Reviews Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- Ullrich A, Coussens L, Hayflick JS, Dull TJ, Gray A, Tam AW, Lee J, Yarden Y, Libermann TA, Schlessinger J. Human epidermal growth factor receptor cDNA sequence and aberrant expression of the amplified gene in A431 epidermoid carcinoma cells. Nature. 1984;309:418–425. doi: 10.1038/309418a0. [DOI] [PubMed] [Google Scholar]
- Vernimmen D, Gueders M, Pisvin S, Delvenne P, Winkler R. Different mechanisms are implicated in ERBB2 gene overexpression in breast and in other cancers. Br J Cancer. 2003;89:899–906. doi: 10.1038/sj.bjc.6601200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalobos V, Naik S, Piwnica-Worms D. Detection of Protein-Protein Interactions in Live Cells and Animals with Split Firefly Luciferase Protein Fragment Complementation. In Genomics Protocols (Humana Press) 2008:339–352. doi: 10.1007/978-1-59745-188-8_23. [DOI] [PubMed] [Google Scholar]
- Walker F, Orchard SG, Jorissen RN, Hall NE, Zhang H-H, Hoyne PA, Adams TE, Johns TG, Ward C, Garrett TPJ, et al. CR1/CR2 interactions modulate the functions of the cell surface epidermal growth factor receptor. J Biol Chem. 2004;279:22387–22398. doi: 10.1074/jbc.M401244200. [DOI] [PubMed] [Google Scholar]
- Wang R, Lai L, Wang S. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J Comput Aided Mol Des. 2002;16:11–26. doi: 10.1023/a:1016357811882. [DOI] [PubMed] [Google Scholar]
- Wong AJ, Ruppert JM, Bigner SH, Grzeschik CH, Humphrey PA, Bigner DS, Vogelstein B. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proceedings of the National Academy of Sciences. 1992;89:2965–2969. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang KS, Ilagan MXG, Piwnica-Worms D, Pike LJ. Luciferase Fragment Complementation Imaging of Conformational Changes in the Epidermal Growth Factor Receptor. J Biol Chem. 2009;284:7474–7482. doi: 10.1074/jbc.M808041200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarden Y, Schlessinger J. Epidermal growth factor induces rapid, reversible aggregation of the purified epidermal growth factor receptor. Biochemistry. 1987;26:1443–1451. doi: 10.1021/bi00379a035. [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nature Reviews Molecular Cell Biology. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- Zhan L, Xiang B, Muthuswamy SK. Controlled Activation of ErbB1/ErbB2 Heterodimers Promote Invasion of Three-Dimensional Organized Epithelia in an ErbB1-Dependent Manner: Implications for Progression of ErbB2-Overexpressing Tumors. Cancer Res. 2006;66:5201–5208. doi: 10.1158/0008-5472.CAN-05-4081. [DOI] [PubMed] [Google Scholar]
- Zhang C, Liu S, Zhu Q, Zhou Y. A knowledge-based energy function for protein-ligand, protein-protein, and protein-DNA complexes. J Med Chem. 2005;48:2325–2335. doi: 10.1021/jm049314d. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.