Abstract
Rapid development of anticancer therapies has occurred, but many challenges remain, including difficulties in early detection and the side effects from chemotherapy. To address these problems, aptamers, which are single-stranded DNA or RNA oligonucleotides with high selectivity, affinity and stability, have attracted considerable attention for biomedical applications. These oligonucleotides, which are selected by an in vitro process known as cell systematic evolution of ligands by exponential enrichment (cell-SELEX), have demonstrated the merits required to recognize disease cells. As such, they show great potential for applications in both clinical and laboratory settings. This review focuses on recently developed techniques utilizing aptamers in cancer research, including cancer cell detection, sorting and enrichment, as well as targeted drug delivery for cancer therapy.
Cancers originate from mutations of human genes. These mutations will result in molecular changes within the cell and then finally lead to changes in cell morphology and physiology. For decades, clinicians have primarily relied on the morphology of tumor cells to diagnose cancers, using techniques such as computed tomography and MRI [1]. But these methods are not capable of detection and diagnosis at a molecular level. In order to accurately predict cancer progression, the detection of the molecular level changes on the cell surface is required. In order to achieve this goal the technique of molecular probes has developed rapidly in recent years.
Among many new tools developed for cancer research, there is a new class of nucleic acid probes known as aptamers, which are single-stranded DNA or RNA oligonucleotides that can be selected to target a wide range of molecules or cells. Aptamers have many advantages over antibodies, particularly their selection, since cancer cells can be used as the target without knowing the number and arrangement of proteins on the cell surface.
These aptamers can distinguish normal cells from tumor cells by identifying molecular level differences and can even discriminate cancer cells by type, by stage of development or by patient profile. In this review, we will focus on the applications of aptamers selected from cell dystematic evolution of ligands by exponential enrichment (cell-SELEX), as a newly developed molecular tool for cancer studies.
Aptamers & cell-SELEX
■ Aptamers
Aptamers, a term derived from the Latin aptus, meaning ‘to fit’ [2], are single-stranded DNA or RNA oligonucleotides selected from an in vitro method known as SELEX, as originally developed by Gold, Szostak et al. in 1990 [2,3]. Aptamers have been selected for a wide range of target molecules, from simple organic and inorganic molecules to peptides, proteins and even living cells. In addition to specific recognition of their targets, manmade aptamers possess several advantages over naturally occurring antibodies [4-7]. These include the ease of synthesis, stability under room conditions, lack of immunogenicity, rapid tissue penetration, ease of immobilization and modification on the chemical devices and, most importantly, specific selectivity to their targets. According to all these advantages, aptamers have great potential as molecular probes for disease diagnosis and therapy, especially for cancer.
In recent years, many applications have been reported for the use of aptamers in bioanalysis and biomedicine [4]. Several kinds of aptamers have been selected against cancer-related proteins, such as PDGF, VEGF, HER3, NFκB, tenascin-C or prostate-specific membrane antigen (PSMA) [8-10]. The technique of aptamer selection against whole cancer cells was subsequently developed [11-17]. Compared with protein-based SELEX, this cell-SELEX can be carried out without prior knowledge of the number or types of proteins on the cell surface. Furthermore, selection can be performed against whole cells with many kinds of receptor proteins existing on their cellular surfaces, which makes it possible to select a panel of aptamer probes that can specifically recognize the biomarker on the cancer cells and then distinguish them from the normal ones, based on differences at the molecular level.
■ Cell-SELEX: cell-based selection of aptamers specific to cancer cells
To generate aptamers that can specifically target cancer cells, a library of ssDNA is used [17]. As shown in Figure 1, the library is first incubated with the target cells. After washing, the DNA sequences bound to the target cell surface are collected and then incubated with the negative control cells (usually the normal cells). All DNA sequences that show binding to the negative control cells are removed. This step is necessary because some proteins on cancer cell surfaces are also expressed by normal cells. To avoid recognition of normal cells, the aptamers binding to these nonspecific proteins must be removed. The remaining sequences are kept and amplified for the next round of selection. Generally, approximately 20 rounds of cell-SELEX are required to isolate aptamers with the highest selectivity affinity to the target cells.
Figure 1. Cell-based aptamer selection.
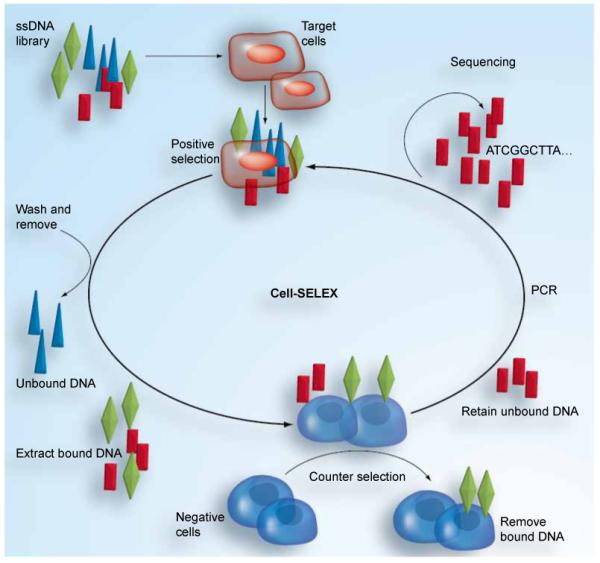
Cell-SELEX: Cell systematic evolution of ligands by exponential enrichment.
A panel of aptamer probes has been successfully selected from cell-SELEX for several types of cancer cells, including lymphocytic leukemia, myeloid leukemia, liver cancer, small-cell lung cancer and non-small-cell lung cancer [18-20]. These aptamers all showed high affinity and excellent specific selectivity against their targets. This result illustrates how aptamers can be selected without specific knowledge of the cell’s molecular signature or the number or type of proteins on the cell surface.
In order to test whether aptamers selected by cell-SELEX can specifically recognize cancer cells, four aptamers were selected for adenocarcinoma A549 and tested for their recognition of different cell lines, as shown in Table 1 [21]. The results indicate specific recognition of these aptamers to A549 cells. While all four aptamers can recognize the large carcinoma cell, only S11e also recognizes both squamous carcinoma cell lines. This result indicates that these aptamers can be used to distinguish different subtypes of non-small-cell lung carcinoma. In addition, these four aptamers show the differential binding to the different non-small-cell lung carcinoma patient samples, indicating that these aptamers can even distinguish molecular differences among patients with the same diagnosis.
Table 1. Recognition of different cancer cell lines by aptamers selected for adenocarcinoma.
| Cancer | Cell lines | S1 | S6 | S11e | S15 |
|---|---|---|---|---|---|
| Adenocarcinoma (NSCLC) | A549 | +++ | ++++ | ++++ | ++++ |
| Large cell carcinoma (NSCLC) | HLAMP | + | ++ | +++ | ++ |
| Large cell carcinoma (NSCLC) | NCI-H460 | + | ++ | +++ | ++ |
| Large cell carcinoma (NSCLC) | NCI-H299 | 0 | + | + | + |
| Squamous carcinoma (NSCLC) | NCI-H520 | 0 | 0 | + | 0 |
| Squamous carcinoma (NSCLC) | NCI-H157 | 0 | 0 | ++ | 0 |
| Small-cell lung cancer | NCI-H446 | 0 | 0 | ++ | 0 |
| Human cervical carcinoma | HeLa | 0 | 0 | 0 | 0 |
| Human breast carcinoma | MCF7 | 0 | 0 | 0 | 0 |
| Hepatocellular carcinoma | HepG2 | + | ++ | +++ | ++ |
Percentage: 0(<15%); +(10–35%); ++(35–60%); +++(60–85%); ++++(>85%).
NSCLC: Non-small-cell lung cancer.
Data from [21].
This specific selectivity makes aptamers ideal candidates to be used as molecular probes in clinical research, especially in cancer studies. In the following part of the review, we will explore the chemical biology approaches for these new aptamers and their applications for cancer diagnosis and therapy in three general areas: cancer cell detection, sorting and enrichment, and targeted drug delivery.
Aptamer-based cancer cell detection
Cancer is a group of diseases that originate from mutations and alterations at the genetic level and, subsequently, at the molecular level. However, in the earliest stages of cancer, the number of tumor cells and, consequently, the number of molecular markers are low, thus limiting detection of the disease. The traditional detection methods, such as computed tomography, MRI and positron emission tomography with radiolabeled 2-fluoro-deoxy-glucose, assess only anatomical changes or nonspecific glucose metabolism [1]. Determination of the molecular characteristics of a cancer, in particular the characteristic proteins associated with a specific cancer, can be of great benefit in early diagnosis [22], particularly in the context of biomarker discovery.
Among the many strategies for aptamer-based cancer detection, this review focuses on two general paradigms that have appeared most frequently in the recent literature: aptamer-based cell detection and aptamer–nanoparticle conjugation for enhanced detection.
■ Aptamer-based cell detection
Aptamers selected by the cell-SELEX method have shown the specificity and high sensitivity required of bioprobes used for the accurate and early diagnosis of tumors. With their high-recognition specificity, aptamers can correctly distinguish different types and even different subtypes, of cancer cells [21]. Moreover, the binding affinity of aptamers to their targets allows the detection of cancer cells at low levels, leading to an early and sensitive diagnosis. In addition, DNA molecules have the advantage of predictable structures and easy site-specific chemical modification. Therefore, aptamers can be conjugated to fluorescent molecules for targeted imaging of the cells, or to nanoparticles for signal enhancement.
In 2008, Medley et al. developed a colorimetric assay for the direct detection of diseased cells using aptamer-conjugated gold nanoparticles [23]. For the initial research design, CCRF-CEM acute leukemia cells were used as the target and Ramos cells, a Burkitt’s lymphoma cell line, were used as control cells. Aptamers specific to CEM cells were conjugated to gold nanoparticles having diameters from 5 to 100 nm. With their specific binding against target cells, aptamer molecules will aggregate on the cell surface membrane. As shown in Figure 2, the association of aptamers with target proteins on the cell surface also brings the aptamer-conjugated gold nanoparticles into close proximity, allowing overlap of their surface plasmon spectra, which in turn leads to a detectable color change in solution. When using the 50-nm gold nanoparticle, this conjugate had the greatest measurable red shift and absorbance past 600 nm on the target cells. This assay has an extremely high sensitivity, with a detection limit of simple absorbance of just 90 cells, while 1000 target cells can lead to a color change that can be observed by the naked eye. This method demonstrates the potential of aptamer-based ana lysis for rapid, simple, direct and, most important, sensitive detection of cancer cells in clinical diagnosis.
Figure 2. Aptamer-conjugated gold nanoparticle-based colorimetric assay.
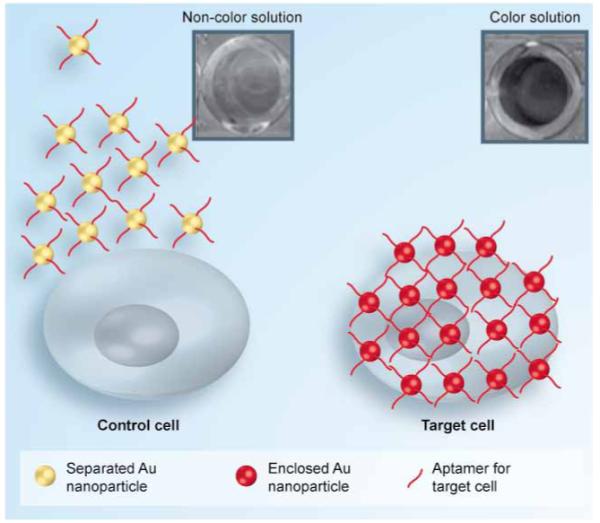
Adapted with permission from [23] © American Chemical Society (2008).
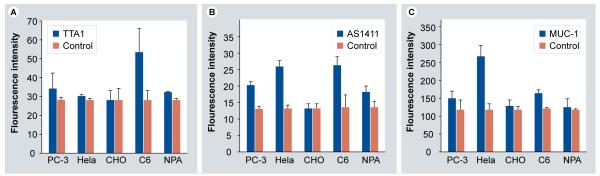
With their ease of chemical manipulation and synthesis, aptamers can be conjugated to fluorophores for the preparation of fluorophore-labeled bioprobes, which have undergone rapid development in recent years [24]. In 2009, Kang et al. demonstrated multiplex detection of cancer cells using quantum-dot (QD)-conjugated aptamers [1]. Three aptamers (TTA1, AS1411 and MUC-1) were conjugated to quantum dots with emission wavelengths of 605, 655 and 705 nm, respectively. For confocal microscopic analysis of multiplex imaging of cancers, healthy and disease cell lines were incubated with each QD-conjugate. QD-TTA1 (Figure 3A) was clearly visualized at 605 nm on C6 cells, but it showed very weak fluorescence signals with PC3, HeLa and NPA cell lines. QDAS1411 (Figure 3B) showed strong fluorescence activity on cellular membranes in HeLa, C6, PC3 and NPA cell lines at 655 nm. QD-MUC-1 (Figure 3B) showed relatively high fluorescence signals on the membrane of HeLa cells at 705 nm. No fluorescence activity was observed from any of the conjugates with the healthy cells, which acted as the negative control.
Figure 3. Fluorescence intensity of (A) QD-TTA1, (B) QD-AS1411 and (C) QD-MUC-1 in the presence of various cancer cells.
Adapted with permission from [1].
As these probes can detect and differentiate different types of cancer cells, as well as produce a visible fluorescence signal in the presence of target cells, they have high potential as a clinical diagnostic tool.
■ Aptamer–nanoparticle conjugation to enhance detection
Cancer cells, especially those in the early stages of disease development, may have a lower density of target on the cell surface than the LOD. To increase the signal and enhance binding affinity, multivalent binding, instead of single-aptamer binding, will greatly help detection. Nanomaterials are particularly advantageous as multivalent ligands owing to their high surface-to-volume ratio.
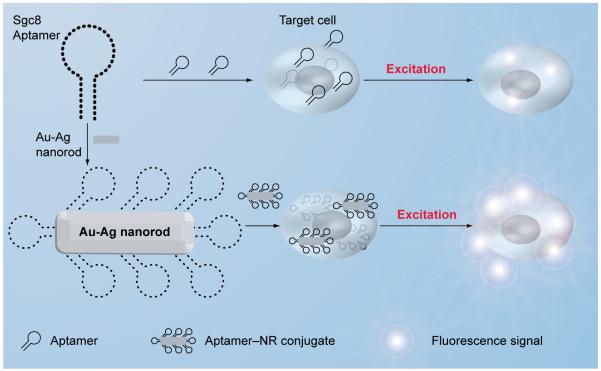
Based on their ease of chemical modification and predictable secondary and tertiary folding structure, aptamers have been conjugated with various nanomaterials to enhance cancer cell detection [22,23,25-28]. Huang et al. have synthesized 12 × 56-nm Au–Ag nanorods (NRs) acting as a nanoplatform for the multivalent binding of aptamers [26]. Up to 80 fluorophore-labeled sgc8 aptamers could be attached on one NR, leading to a 26-fold higher affinity and over 300-fold higher fluorescence signal (Figure 4). The use of nanorods as a nanoplatform for multivalent binding of aptamers increases both the signal and binding strengths of these aptamers in cancer cell recognition. This technique can be used in clinical detection to enhance both binding affinity and signal strength when the concentration of target cells is relatively low.
Figure 4. Aptamer–nanorod signal enhancement.
Aptamer-based cancer cell sorting & enrichment
The ability to diagnose cancer based on the detection of rare cancer cells in blood or other bodily fluids is a significant challenge. Generally, however, in vivo tumors shed only 106 cells per day into the bloodstream [29]. These circulating tumor cells are an extremely rare component within human blood (~10 cells/ml of whole blood), while other more predominant cells occur in high concentration, for example, erythrocytes (~109 cells/ml of whole blood) and leukocytes (~106 cells/ml of whole blood) [30]. To address this challenge, microfluidic cell-affinity chromatography devices can be used to capture cells from any patient sample that contains occult tumor cells.
In 2009, Dharmasiri et al. designed a poly(methyl methacrylate) microchip device for the capture and enumeration of prostate tumor cells [31]. In their study, anti-PSMA aptamers were immobilized onto the surface of a capture bed poised within a PMMA microchip, which was fabricated into a high-throughput micro-sampling unit. The device was able to capture 90% of LNCaP cells (prostate cancer cell line) on the surface of the capture bed, with purity as high as 100%.
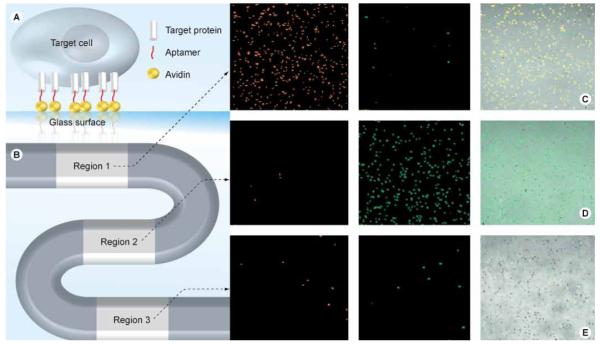
Xu et al. reported an aptamer-based microfluidic system having the unique qualities of simplicity, efficiency and high selectivity for a variety of biological applications [32]. As shown in Figure 5, the S-shaped channel was designed with three cell-capture regions to capture three different types of leukemia cells (CEM, Ramos and Toledo). Aptamers were selected by the cell-SELEX process (Sgc8, TDO5 and Sgd5 for CEM, Ramos and Toledo cells, respectively) and were tandem immobilized on three regions of the device. When the cell mixtures passed through the device, CEM, Ramos and Toledo cells were captured in individual regions with purities of 97, 97 and 88%, respectively. Even without the extra optimization for the cell-capture efficiency, the efficiency was still as high as 83 ± 9% (CEM), 61 ± 14% (Ramos) and 50 ± 10% (Toledo). This enrichment of rare cancer cells makes the device a useful tool for early diagnosis when the concentration of cancer cells in plasma is relatively low.
Figure 5. Microfluidic device and fluorescent images of captured cancer cells.
(A) Microfluidic device, (B) representation of aptamer immobilization, (C–E) enrichment of (C) CEM, (D) Ramos and (E) Toledo cells in their respective regions. Adapted with permission from [32] © American Chemical Society (2009).
After pumping air through the channel, the cells were released from the surface with an efficiency of 45% and subsequent growth of the cells showed that the cells were still viable, indicating that the process does not lead to apoptosis. Therefore, the released cells can still be used for further diagnosis without loss of activity. Furthermore, since there is no physical limit to the number of regions, this device can be developed for the multiplexed detection of more than three types of cancer cells.
The aptamer-based microfludic device can be used for the detection of cancer at an early stage, as well as for multiplexed cancer cell detection. It can discriminate among different cancer cell types with high purity. Simultaneous enrichment of three distinct target cells into independent fractions is also useful for further diagnostic procedures, such as proliferation [33], counting [34] and gene analysis [28]. Finally, since these aptamer-based devices can be stored for long periods and are easily operated, they will be efficient and economical tools for the early diagnosis of cancer.
Aptamer-based targeted drug delivery for cancer therapy
Another challenging problem in cancer therapy is the efficient and selective delivery of pharmaceuticals directly to the tumor cells. Drugs alone do not have sufficient specificity to target cancer cells and, moreover, can seriously damage normal cells. Also, the reticuloendothelial system surveys all entering or circulating antigens and mobilizes an immune response, a sufficient biological defense to antibodies and drugs. In order to address these problems, cytotoxic drugs can be conjugated to a probe molecule having the affinity and selectivity to target only the disease cells. This probe should efficiently recognize abnormal cancer cells, distinguish them from the normal ones and then directly deliver the loaded drug to the tumor cells without causing side effects or harm to the body. This concept was first investigated using antibody–drug conjugates.
In the 1990s, Bristol-Myers Squibb scientists first covalently linked doxorubicin (Dox), an intercalating chemotherapeutic agent that blocks DNA replication, to human mAb BR96 [35]. However, owing to the limited conjugation between the antibody and Dox, as well as the relatively low potency and half-life of the conjugate, the compound failed to show sufficient clinical efficacy [36]. Antibodies also have the disadvantages of immunogenicity [37], poor site specificity for conjugation and inconsistent binding affinities [38].
■ Aptamer-directed conjugation for targeted drug delivery
Fortunately, owing to their specific selectivity, high affinity, easy synthesis and lack of immunogenicity, aptamers have shown great potential for drug delivery in cancer therapy. In recent applications, aptamers have been explored as targeting agents for targeted drug-delivery systems. The specific selectivity of aptamers allows this conjugation to recognize the target cancer cell while reducing damage to normal cells. Furthermore, due to their lack of immunogenicity, aptamers can reduce immune responses from the reticuloendothelial system. Combining the advantages of aptamers with their conjugation potential, these probes hold great promise for chemotherapy, where low toxicity to nontargeted cells is critical.
A novel strategy for targeted drug delivery to cancer cells was reported by Bagalkot et al. via formation of a physical conjugate between Dox and the A10 RNA aptamer [39]. The authors reported that this conjugation was able to target PSMA-expressing prostate cancer LNCaP cells with subsequent intracellular release of doxorubicin at 37°C.
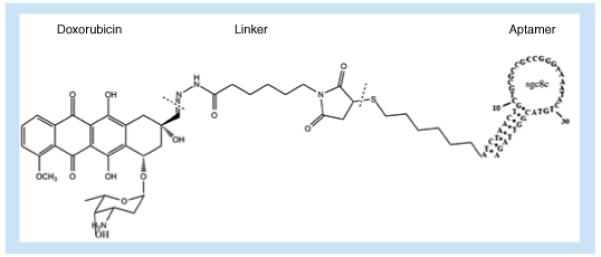
The sgc8–Dox conjugate (Figure 6) was designed by Huang et al. for targeted drug delivery to CCRF-CEM cells (T-cell acute lymphoblastic leukemia, T-cell ALL) [40], combining the advantages of specific recognition of aptamer sgc08 for protein tyrosine kinase (PTK)7 [17,41] and the benefits of doxorubicin in treating acute lymphoblastic and myeloblastic leukemias [42]. Using an indirect fluorescence measurement, the conjugate achieves a dissociation constant (Kd) of Sgc8c–Dox to the CCRF-CEM cells as low as 2.0 ± 0.2 nM. The Sgc8c–Dox conjugates showed a 6.7-fold increase in toxicity to CCRF-CEM cells compared with nontarget NB-4 cells and even higher compared with Ramos cells. This selective killing can reduce the damage to normal cells, thus saving the patient from detrimental side effects. The acid-labile linkage connecting the drug and the aptamer allows the drug to be cleaved in the acidic endosome environment for selective release into the cell.
Figure 6. Conjugation of the drug doxorubicin to aptamer sgc8c for targeted delivery to cancer cells.
■ Aptamer–liposome conjugates for targeted drug delivery
Since most of the existing aptamers cannot directly pass through the cell membrane, finding ways to increase membrane permeation has been an active area of research. In 2009, Cao et al. reported the controlled formulation of aptamer-conjugated, multifunctional liposomes to encapsulate and deliver eisplatin [43]. Because the cell membrane is basically a dynamic lipid bilayer, this liposome nanostructure can efficiently increase cell permeability and enhance drug delivery [44]. The other advantage of liposomes is their ability to increase the plasma residence time of aptamers [45].
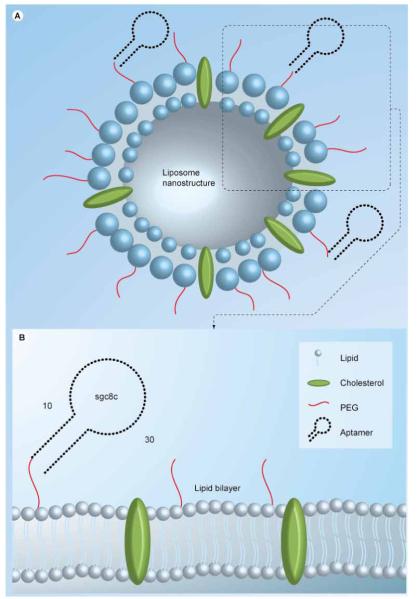
A Sgc8–PEG–liposome nanostructure for drug delivery was reported by Kang et al., as shown in Figure 7 [46]. With approximately 250 aptamers on each liposome, this design leads to multiple aptamer–receptor interactions, provides better binding to the target cells and facilitates translocation through the plasmic membrane. While liposomes are immobilized on the surface membrane of the CEM cells by aptamer recognition, no binding to the non-target NB4 cells is observed.
Figure 7. Multifunctional liposome nanostructure for targeted delivery.
■ Aptamer-micelle conjugates for targeted drug delivery
Based on aptamer-lipid conjugates, the aptamer–micelle nanostructure has been incorporated into recent applications. Micelles are aggregates of surfactant molecules with the hydrophobic parts in the interior and the hydrophilic regions facing outward to the surrounding solvent [47]. In recent applications, micelles constructed of hydrophilic oligonucleotides and a hydrophobic polymer have appeared [48,49] and have shown the ability to carry a variety of cargos to cells, including antisense oligonucleotides and drug molecules [49,50].
Wu et al. reported the development of a self-assembled aptamer–micelle nanostructure [44]. The hydrophilic aptamers were linked to the hydrophobic lipid tail by PEG. In aqueous solutions, this conjugate can self-assemble into a 3D spherical micelle structure. The aptamers formed the hydrophilic surface of the micelle and the lipid tails formed the hydrophobic center. Unlike the bilayer structure of liposomes, micelles only have one layer by the dense packing of the aptamer–PEG–lipid conjugate.
In this aptamer–micelle structure, the aptamer not only acts as the cell-recognition element, but also as the building block for the nano structure. The dense packing of aptamers in such an assembly greatly improves binding affinity. For example, TDO5–micelle has a Kd of 116 nM and a similar size of polymer micelle (60 nm) is estimated to have 1000 copies of units [51]. After constructing the micelle structure, the dissociation constant would be greatly decreased from 88 nM (for free TDO5) to 0.116 nM (for the TDO5–micelle). This multivalent effect from multiple aptamer binding finally leads to an approximate 750-fold increase in binding affinity. After doping TDO5–micelles with a special dye that only fluoresces inside cells (CellTracker™), the process of cell internalization was clearly observed by fluorescent microscopy, indicating good permeability of this structure to the cancer cells (Figure 8). These results indicate that this aptamer–micelle assembly is a promising strategy for clinical applications by increasing effective therapy and reducing the toxicity to normal cells.
Figure 8.
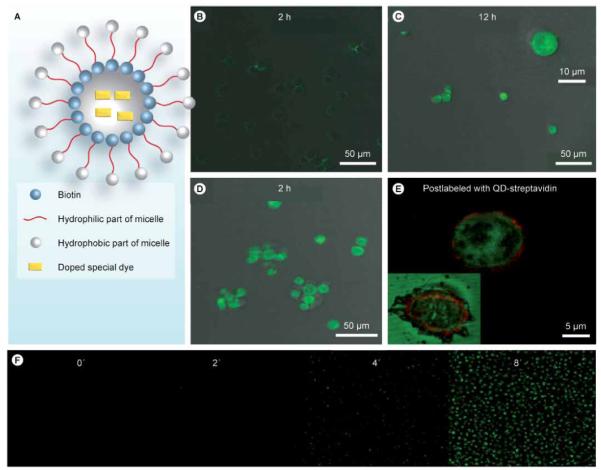
(A) Design scheme of dye-doped micelles. Fluorescent images of Ramos cells for (B) 2 h and (C) 12 h, or incubation with the biotin–TDO5–micelle for (D) 2 h. (E) Enlarged fluorescent image after postlabeling the biotinylated TDO5 aptamer with QD705 streptavidin. The inset is the fluorescent image of the dead cell. (F) Real-time monitoring of doped special dyes released from the core of the micelles and activated by intracellular enzymes.
Adapted with permission from [44] © National Academy of Sciences, USA (2010).
Future perspective
In conclusion, cell-SELEX provides an effective approach for generating a large number of aptamer probes that specifically target a variety of cancer cells whose molecular markers are unknown. The aptamer panels generated from cell-SELEX show the high selectivity required for recognition of different types, or subtypes, of cancer cells. These aptamers can be applied to many areas of cancer cell biology, generating a wide variety of directions for future work, as described in this review. For example, the ease of site-specific chemical modification makes it possible to conjugate aptamers to gold nano-particles or quantum dots for use in colorimetric or fluorescence detection of cancer cells, or to the gold nanorod for the enhancement of the signal. With such high sensitivity, these devices can be applied for the early diagnosis of cancer when the concentration of cells is relatively low. Furthermore, aptamers can be modified for immobilization on a microchip or in a microfluidic device because of their stability and ease of modification under room conditions. Finally, aptamers highly specific to cancer cells can be used as drug-targeting agents. This may result in reducing toxicity while increasing the efficacy of current therapeutic drugs.
Executive summary.
Detection of cancer cells
-
■Colorimetric detection
-
■Fast, simple, sensitive and direct detection for cancer cells.
-
■Direct detection by color change and quantitative detection by absorbance.
-
■Low detection limit: on simple absorbance, 90 cells; 1000 target cells by the naked eye.
-
■
-
■Fluorescence detection
-
■Sensitive and specific detection for different kinds of cancer cells.
-
■Efficiently distinguishes among different types of cancer cells by different wavelengths of fluorescence.
-
■
-
■Aptamer–nanoparticle conjugation to enhance detection
-
■Nanorods act as a nanoplatform for the multivalent binding of aptamers.
-
■80 aptamers attached on one nanorod.
-
■26-fold higher affinity and over 300-fold higher fluorescence signal.
-
■
Sorting & enrichment of cancer cells
-
■Aptamer-based microchip
-
■A small, simple device to capture and enrich rare cancer cells.
-
■Efficiently captures 90% of LNCaP cells.
-
■High purity of 100% gained.
-
■
-
■Aptamer-based microfluidic device
-
■Specific device for the capture, separation, enrichment and release of different kinds of cancer cells.
-
■Selectively separates different kinds of cancer cells in different regions.
-
■Capture efficiencies as high as 83 ± 9% (CEM), 61 ± 14% (Ramos) and 50 ± 10% (Toledo).
-
■Purities as high as 97 (CEM), 97 (Ramos) and 88% (Toledo).
-
■Releases cells with good viability at efficiency of 45%.
-
■
Targeted drug delivery to cancer cells
-
■Aptamer-directed conjugates
-
■Selective drug delivery to cancer cells while reducing side effects to normal cells.
-
■Dissociation constant as low as 2.0 ± 0.2 nM.
-
■6.7-fold increase in toxicity to target cells compared with control cells.
-
■
-
■Aptamer–liposome conjugates
-
■Selective drug delivery with increased membrane permeation.
-
■250 aptamers on each liposome, leading to multiple aptamer–receptor interactions.
-
■
-
■Aptamer–micelle conjugates
-
■Dense packing with 1000 aptamer on each micelle unit.
-
■750-fold increase in binding affinity.
-
■Demonstrated penetration through the cancer cell membrane and drug release inside.
-
■
Continuous development of aptamer selected from cell-SELEX will impact on cancer studies in many ways. In this review, we have demonstrated its great potential in cancer detection, diagnosis and targeted therapy. In theory, all types of cancer cells can be the targets for the cell-SELEX. Even in the same type of cancer cells in different periods, the molecular expressions will be different. In the future, we expect the aptamers selected from cell-SELEX to be further used for the detection of molecular-based changes in tumor progression as well as to distinguish between different subtypes of cancer cells. With the knowledge of molecular profiling, we can not only accurately diagnose the tumor but can also determine the molecular basis of cancer. Selecting aptamers that can distinguish the cancer cells different progressing periods provides the capability to achieve this goal. In summary, the future studies of aptamers will lead to an improved understanding of the biochemistry and molecular basis of cancer and provide a platform for exciting new technologies in detection, diagnosis and treatment of cancer.
Acknowledgements
The authors thank their co-workers, whose work is reported here. The authors also thank Dr Kathryn Williams for helpful discussions and for reading this manuscript.
Key Term
- Fluorophore
Functional group in a molecule that absorbs energy on a specific wavelength and emits energy at a different (but equally specific) wavelength
- Microfluidic system
System that deals with the behavior, precise control and manipulation of fluids that are geometrically constrained to a small, typically sub-millimeter scale
- Reticuloendothelial system
Immune system that consists of the phagocytic cells located in reticular connective tissue; surveys all entering or circulating antigens and mobilizes an immune response
- Doxorubicin
Drug, also known as hydroxydaunorubicin, used in cancer chemotherapy and works by intercalating DNA
- Kd
Dissociation constant. When a complex AxBy breaks down into x A subunits and y B subunits, the dissociation constant is defined as:
- Micelle
Typically a dense aggregate with the hydrophilic ‘head’, which faces the surrounding solvent and hydrophobic single tail crowding in the micelle center
Footnotes
Financial & competing interests disclosure
This work was supported by NIH grants and a Florida State Health Department grant. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
■of interest
■■of considerable interest
- 1.Kang WJ, Chae JR, Cho YL, Lee J, Kim S. Multiplex imaging of single tumor cells using quantum-dot-conjugated aptamers. Small. 2009;22:2519–2522. doi: 10.1002/smll.200900848. [DOI] [PubMed] [Google Scholar]
- 2.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. ■■ One of the first aptamer papers.
- 3.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. ■■ One of the first aptamer papers.
- 4.Osborne SE, Matsumura I, Ellington AD. Aptamers as therapeutic and diagnostic reagents: problems and prospects. Curr. Opin. Chem. Biol. 1997;1:5–9. doi: 10.1016/s1367-5931(97)80102-0. [DOI] [PubMed] [Google Scholar]
- 5.Famulok M, Hartig JS, Mayer G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem. Rev. 2007;107:3715–3743. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- 6.Navani NK, Li Y. Nucleic acid aptamers and enzymes as sensors. Curr. Opin. Chem. Biol. 2006;10:272–281. doi: 10.1016/j.cbpa.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Jiang Y, Zhu C, Ling L, Wan L, Fang X, Bai C. Specific aptamer–protein interaction studied by atomic force microscopy. Anal. Chem. 2003;75:2112–2116. doi: 10.1021/ac026182s. [DOI] [PubMed] [Google Scholar]
- 8.Ireson CR, Kelland LR. Discovery and development of anticancer aptamers. Mol. Cancer Ther. 2006;5:2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- 9.Lupold SE, Hicke BJ, Liny Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002;62:4029–4033. [PubMed] [Google Scholar]
- 10.Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. A tenascin-C aptamer identified by tumor cell SELEX: systematic evolution of ligands by exponential enrichment. Proc. Natl Acad. Sci. USA. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerchia L, Ducongé F, Pestourie C, et al. Neutralizing aptamers from whole-cell SELEX inhibit the RET receptor tyrosine kinase. PLoS. Biol. 2005;3:E123. doi: 10.1371/journal.pbio.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Shamah SM, Healy JM, Cload ST. Complex target SELEX. Acc. Chem. Res. 2008;41:130–138. doi: 10.1021/ar700142z. [DOI] [PubMed] [Google Scholar]
- 13.Blank M, Weinschenk T, Priemer M, Schluesener H. Systematic evolution of a DNA aptamer binding to rat brain tumor microvessels. Selective targeting of endothelial regulatory protein pigpen. J. Biol. Chem. 2001;276:16464–16468. doi: 10.1074/jbc.M100347200. [DOI] [PubMed] [Google Scholar]
- 14.Berezovski MV, Lechmann M, Musheev MU, Mak TW, Krylov SN. Aptamer-facilitated biomarker discovery (AptaBiD) J. Am. Chem. Soc. 2008;130:9137–9143. doi: 10.1021/ja801951p. [DOI] [PubMed] [Google Scholar]
- 15.Guo K, Schafer R, Paul A, Gerber A, Ziemer G, Wendel HP. A new technique for the isolation and surface immobilization of mesenchymal stem cells from whole bone marrow using high-specific DNA aptamers. Stem Cells. 2006;24:2220–2231. doi: 10.1634/stemcells.2006-0015. [DOI] [PubMed] [Google Scholar]
- 16.Raddatz ML, Dolf A, Endl E, Knolle P, Famulok M, Mayer G. Enrichment of cell-targeting and population-specific aptamers by fluorescence-activated cell sorting. Angew. Chem. Int. Ed. 2008;47:5190–5193. doi: 10.1002/anie.200800216. [DOI] [PubMed] [Google Scholar]
- 17.Shangguan D, Li Y, Tang Z, Cao Z, et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl Acad. Sci. USA. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. ■■ Concept and procedure of cell-systematic evolution of ligands by exponential enrichment (SELEX).
- 18.Sefah K, Tang Z, Shangguan D, et al. Molecular recognition of acute myeloid leukemia using aptamers. Leukemia. 2009;23:235–244. doi: 10.1038/leu.2008.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, Medley C, Sefah K, et al. Molecular recognition of small-cell lung cancer cells using aptamers. ChemMedChem. 2008;3:991–1001. doi: 10.1002/cmdc.200800030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang Z, Shangguan D, Wang K, et al. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal. Chem. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Z, Xu L, Shi X, Tan W, Fang X, Shangguan D. Recognition of subtype non-small cell lung cancer by DNA aptamers selected from living cells. Analyst. 2009;134:1808–1814. doi: 10.1039/b904476k. ■ Indicating the specific selectivity of aptamers.
- 22.Smith J, Medley C, Tang Z, Shangguan D, Lofton C, Tan W. Aptamer-conjugated nanoparticles for the collection and detection of multiple cancer cells. Anal. Chem. 2007;79:3075–3082. doi: 10.1021/ac062151b. [DOI] [PubMed] [Google Scholar]
- 23.Medley C, Smith J, Tang Z, Wu Y, Bamrungsap S, Tan W. Gold nanoparticle-based colorimetric assay for the direct detection of cancerous cells. Anal. Chem. 2008;80:1067–1072. doi: 10.1021/ac702037y. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Jia X, Lv X, Deng Y, Xie H. Fluorescent quantum dot-labeled aptamer bioprobes specifically targeting mouse liver cancer cells. Talanta. 2010;81(1–2):505–509. doi: 10.1016/j.talanta.2009.12.031. [DOI] [PubMed] [Google Scholar]
- 25.Chu TC, Shieh F, Lavery LA, et al. Labeling tumor cells with fluorescent nanocrystal–aptamer bioconjugates. Biosens. Bioelectron. 2006;21:1859–1866. doi: 10.1016/j.bios.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 26.Huang Y, Chang H, Tan W. Cancer cell targeting using multiple aptamers conjugated on nanorods. Anal. Chem. 2008;80:567–572. doi: 10.1021/ac702322j. [DOI] [PubMed] [Google Scholar]
- 27.Herr J, Smith J, Medley C, Shangguan D, Tan W. Aptamer-conjugated nanoparticles for selective collection and detection of cancer cells. Anal. Chem. 2006;78:2918–2924. doi: 10.1021/ac052015r. [DOI] [PubMed] [Google Scholar]
- 28.Phillips J, Xu Y, Xia Z, Fan Z, Tan W. Enrichment of cancer cells using aptamers immobilized on a microfluidic channel. Anal. Chem. 2009;81:1033–1039. doi: 10.1021/ac802092j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liotta L, Saidel M, Kleinerman J. The significance of hematogenous tumor cell clumps in the metastatic process. Cancer Res. 1976;36:889–894. [PubMed] [Google Scholar]
- 30.Nagrath S, Sequist L, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dharmasiri U, Balamurugan S, Adams A, Okagbare P, Obubuafo A, Soper S. Highly efficient capture and enumeration of low abundance prostate cancer cells using prostate-specific membrane antigen aptamers immobilized to a polymeric microfluidic device. Electrophoresis. 2009;30:3289–3300. doi: 10.1002/elps.200900141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Y, Philips JA, Yan J, Li Q, Fan ZH, Tan W. Aptamer-based microfluidic device for enrichment, sorting, and detection of multiple cancer cells. Anal. Chem. 2009;81(17):7436–7442. doi: 10.1021/ac9012072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu W, Zhu X, Futai N, Cho BS, Takayama S. Computerized microfluidic cell culture using elastomeric channels and braille displays. Proc. Natl Acad. Sci. USA. 2004;101:15861–15866. doi: 10.1073/pnas.0404353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcus JS, Anderson WF, Quake SR. Microfluidic single-cell mRNA isolation and analysis. Anal. Chem. 2006;78:3084–3089. doi: 10.1021/ac0519460. [DOI] [PubMed] [Google Scholar]
- 35.Trail PA, Willner D, Lasch SJ, et al. Cure of xenografted human carcinomas by BR96–doxorubicin immunoconjugates. Science. 1993;261:212–215. doi: 10.1126/science.8327892. [DOI] [PubMed] [Google Scholar]
- 36.Laurent D, Bernhard S. Antibody–drug conjugates: linking cytotoxic payloads to monoclonal antibodies. Bioconjugate Chem. 2010;21:5–13. doi: 10.1021/bc9002019. [DOI] [PubMed] [Google Scholar]
- 37.Kirpotin DB, Drummond DC, Shao Y, et al. Antibody targeting of long-circulating lipidic nanoparticles does not increase tumor localization but does increase internalization in animal models. Cancer Res. 2006;66:6732. doi: 10.1158/0008-5472.CAN-05-4199. [DOI] [PubMed] [Google Scholar]
- 38.Sapra P, Allen TM. Ligand-targeted liposomal anticancer drugs. Prog. Lipid Res. 2003;42:439. doi: 10.1016/s0163-7827(03)00032-8. [DOI] [PubMed] [Google Scholar]
- 39.Bagalkot V, Zhang L, Levy-Nissenbaum E, et al. Quantum dot–aptamer conjugates for synchronous cancer imaging, therapy, and sensing of drug delivery based on bi-fluorescence resonance energy transfer. Nano Lett. 2007;7:3065. doi: 10.1021/nl071546n. [DOI] [PubMed] [Google Scholar]
- 40.Huang Y, Shangguan D, Liu H, et al. Molecular assembly of an aptamer-drug conjugate for targeted drug delivery to tumor cells. ChemBioChem. 2009;10:862. doi: 10.1002/cbic.200800805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shangguan D, Cao Z, Meng L, et al. Cell-specific aptamer probes for membrane protein elucidation in cancer cells. J. Proteome Res. 2008;7:2133–2139. doi: 10.1021/pr700894d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiss RB, Sarosy G, Clagett-Carr K, Russo M, Leyland-Jones B. Anthracycline analogs: the past, present, and future. Cancer Chemother. Pharmacol. 1986;18:185–197. doi: 10.1007/BF00273384. [DOI] [PubMed] [Google Scholar]
- 43.Cao Z, Tong R, Mishra A, et al. Reversible cell-specific drug delivery with aptamer-functionalized liposomes. Angew. Chem. Int. Ed. 2009;48:6494–6498. doi: 10.1002/anie.200901452. [DOI] [PubMed] [Google Scholar]
- 44.Wu Y, Sefah K, Liu H, Wang R, Tan W. DNA aptamer–micelle as an efficient detection/delivery vehicle toward cancer cells. PNAS. 2010;107(1):5–10. doi: 10.1073/pnas.0909611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willis MC, Collins B, Zhang T, et al. Liposome-anchored vascular endothelial growth factor aptamers. Bioconjugate Chem. 1998;9:573. doi: 10.1021/bc980002x. [DOI] [PubMed] [Google Scholar]
- 46.Kang H, O’Donoghue MB, Liu H, Tan W. A liposome-based nanostructure for aptamer directed delivery. Chem. Commun. 2010;46:249–251. doi: 10.1039/b916911c. [DOI] [PubMed] [Google Scholar]
- 47.Seddon JM, Templer RH. Reversible Cell-Specific Drug Delivery with Aptamer-Functionalized Liposomes (Volume 1) Elsevier Science; Oxford, UK: 1995. [Google Scholar]
- 48.Ding K, Alemdaroglu FE, Borsch M, Berger R, Herrmann A. Engineering the structural properties of DNA block copolymer micelles by molecular recognition. Angew. Chem. Int. Ed. Engl. 2007;46:1172–1175. doi: 10.1002/anie.200603064. [DOI] [PubMed] [Google Scholar]
- 49.Alemdaroglu FE, Alemdaroglu NC, Langguth P, Herrmann A. Cellular uptake of DNA block copolymer micelles with different shapes. Macromol. Rapid Commun. 2008;29:326–329. [Google Scholar]
- 50.Jeong JH, Park TG. Novel polymer–DNA hybrid polymeric micelles composed of hydrophobic poly (d,l-lactic-co-glycolic acid) and hydrophilic oligonucleotides. Bioconjugate Chem. 2001;12:917–923. doi: 10.1021/bc010052t. [DOI] [PubMed] [Google Scholar]
- 51.Chen H, Kim S, Li L, Wang S, Park K, Cheng J. Release of hydrophobic molecules from polymer micelles into cell membranes revealed by Forster resonance energy transfer imaging. Proc. Natl Acad. Sci. USA. 2008;105:6596–6601. doi: 10.1073/pnas.0707046105. [DOI] [PMC free article] [PubMed] [Google Scholar]