Abstract
Although a key factor in Alzheimer’s disease etiology is enrichment of Zn2+ in aggregates, and there are data suggesting that zinc promotes aggregation, how Zn2+-Aβ coordination promotes aggregation is elusive. Here we probe the structures and mechanisms through which Zn2+ can affect amyloidosis. By covalently linking fragments (that have experiment-based coordinates) we observed that, in oligomeric Zn2+-Aβ42, Zn2+ can simultaneously coordinate intra- and intermolecularly, bridging two peptides. Zinc coordination significantly decreases the solvation energy for large Zn2+-Aβ42 oligomers and thus enhances their aggregation tendency. Zn2+ binding does not change the β-sheet association around the C-terminal hydrophobic region; however, it shifts the relative population of the preexisting amyloid polymorphic ensembles. As a result, although a parallel β-sheet arrangement is still preferred, antiparallel and other less structured assemblies are stabilized, also becoming major species. Overall, Zn2+ coordination promotes Aβ42 aggregation leading to less uniform structures. Our replica exchange molecular dynamics simulations further reproduced an experimental observation that the increasing Zn2+ concentration could slow down the aggregation rate, even though the aggregation rates are still much higher than in Zn2+-free solution.
Keywords: conformational selection, energy landscape, metal ions, modeling amyloid assemblies, seed polymorphism
Alzheimer’s disease (AD) is a progressive neurodegenerative disease and the most common cause of dementia in millions of people worldwide (1). This disease accounts for the majority of clinical senile dementia associated with the formation of senile plaques (2, 3). The primary constituent of the plaques is the aggregated Aβ40/Aβ42 peptides in the brain; therefore, factors that influence the aggregation are of high interest.
In vivo studies reported that Zn2+, Cu+2, and Fe+3 are markedly enriched in Aβ plaques (4, 5), suggesting that these ions may act as seeding factors. Oligomerization of the Aβ peptides can be rapidly induced in the presence of Zn2+ ions under physiological conditions (4, 6, 7). Noy et al. (8) have followed Zn2+ ion-enhanced Aβ aggregation. Studies suggested that H6, H13, and H14 at the N-terminal domain of Aβ coordinate with Zn2+ (9–26). Solution NMR of Zn2+-Aβ1-16 showed that Zn2+ is bound to these three histidines and E11 (18). A recent NMR study of Zn2+-Aβ1-28 proposed that Zn2+ binds to H6, E11, H14, and D1 of rat Aβ1-28 and to H6, E11, H13, H14, and D1 of human Aβ1-28 (22). In addition, X-ray absorption spectroscopy revealed that Zn2+ coordinates with four histidines, H13 and H14 of two adjacent monomers (23). Experimental structural data for Aβ40/Aβ42 oligomers complexed with Zn2+ are unavailable.
Although it is believed that reducing zinc-induced Aβ aggregation can decrease toxicity (27), it was also postulated that zinc can lower Aβ toxicity by selectively precipitating aggregation intermediates (28). Key questions on the nature of the interactions of Aβ oligomers with Zn2+, such as (i) what are the dominant structures of Aβ amyloids in the presence of Zn2+, (ii) why Zn2+ promotes Aβ amyloid aggregation, and (iii) what are the biological consequences of the Zn2+-Aβ oligomer interactions, are still unanswered. These questions necessitate exploration of the structural features through which Zn2+ interacts with Aβ40/Aβ42 oligomers.
In this work, we constructed a range of potential oligomers of Aβ42 complexed with Zn2+ (Fig. 1 and Figs. S1–S3) by covalently linking available experiment-based coordinate sets of Aβ fragments of the N and C termini (18, 22, 23, 29) using molecular dynamics (MD) simulations to investigate their stabilities. Using replica exchange molecular dynamics (REMD) simulations, we further explored the relative stabilities and populations of the ordered aggregates as compared to the vast conformational ensembles. We observed that intra- and intermolecular zinc coordination in Aβ oligomers decrease the solvation of the oligomer and therefore promote Zn2+-Aβ aggregation. Amyloids are highly polymorphic, and Zn2+ selects among this polymorphic population monomers and small organizations whose intra- and intermolecular N-terminal conformations are geometrically and chemically favored for coordination. This leads to a population shift redistributing the polymorphic states (30).
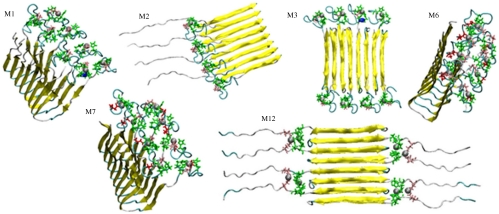
Fig. 1.
Solution NMR-based conformers of the Zn2+ complexed with residues 1–16 (the N-terminal region) fit well with the experiment-based structure for the remainder of the Aβ (residues 17–42), and consequently can be linked to create models of the full-length Aβ42 complexed with Zn2+. Zn2+-coordinated N-terminal coordinates are taken from refs. 18, 22, and 23. Coordinates of residues 17–42 are taken from ref. 29. Models M1–M3 are with the Zirah coordinates (18) linked to the Lührs coordinates (29). In M1 and M2, the Aβ monomers are arranged in parallel; in M1 with eight Zn2+ ions; in M2 with four Zn2+ ions; in M3, the monomers are antiparallel with eight Zn2+ ions. M6 and M7 are obtained by joining the Gaggelli (22) and Lührs coordinates (29) with eight Zn2+ ions, in a parallel organization; M12 is based on the Miniccozzi–Lührs (23, 29) coordinates complexed with eight Zn2+ ions in antiparallel organization. The figure presents the two most highly populated conformers of the Zirah–Lührs model: M1 and M3; the two most highly populated conformers of Gaggeli–Lührs model: M6 and M7; and the highly populated conformer based on Miniccozzi–Lührs model.
Results
Zn2+-Aβ42 Interactions Enhance the Aggregates’ Structural Variability as Compared to Zn2+-Free Aβ42.
The Aβ zinc-binding site may be located in two peptide domains: the disordered N-terminal domain (residues 1–16) and the C-terminal domain (residues 17–42). In the Aβ sequence, His, Glu, and Asp are potential residues to coordinate metal ions. The C-terminal domain contains E22 and D23, whereas seven potential residues (D1, E3, H6, D7, E11, H13, and H14) appear in the N-terminal domain. Experimental data indicate that Zn2+ most likely binds to the N-terminal region in the Aβ peptide, in at least three distinct ways (18, 22, 23).
Based on experimental data and assuming that Zn2+ binds to the N-terminal region also in the full-length Aβ peptide, we considered the U-turn and the two β-sheets forming C-terminal region (17-42/17-40). Two experiment-based amyloid structures are available for the C-terminal region: the Aβ17-42 Lührs model [Protein Data Bank (PDB) ID: 2BEG)] (29); and the solid-state NMR-based model of Aβ9-40 by Tycko and coworkers (31). The latter model is not compatible with the Zn2+-coordinated N-terminal segment (SI Text). However, the Aβ17-42 (29) has an excellent fit with the Zn2+ binding 1–16 region. Thus, we generated octamers of Aβ17-42 using the 2BEG coordinate file (SI Text). We linked the L17 of each Aβ17-42 segment to the K16 of each N-terminal Aβ1-16 segment complexed with Zn2+.
The number of possible combinations for the Zn2+-Aβ complex is large, and the stability and the population distribution of the possible conformers could change depending on concentration, temperature, and pH. Here we selectively constructed 12 polymorphic Zn2+-Aβ models, considering three possible ways of Zn2+-Aβ coordination, based on experimental data and both parallel and antiparallel β-sheet arrangements (Table 1). Among these models, we highlight four that are used in the subsequent REMD simulations to explore the vast variability of the conformational ensemble. Models M1 and M3 were constructed directly from a combination of the N-terminal [Zirah model (18)] and C-terminal (PDB: 2BEG) structures: M1 was arranged in parallel whereas M3 in antiparallel organization (Fig. 1). Similarly, M8 and M10 were constructed from a different Zn2+-Aβ1-16 conformation [Gaggelli model (22)] and C-terminal structure (2BEG), with M8 in parallel and M10 in antiparallel.
Table 1.
Constructed Zn2+-Aβ1-42 oligomers based on Lührs’s model (29), Zirah’s model (18), Gaggelli’s model (22), and Minicozzi’s model (23)
| Model no. | Model* | Residues coordinate to Zn2+ | Conformational organization | Conformational energy†, kcal/mol | Population‡, % | Fig. |
| M1 | Zirah–Lührs | H6, E11, H13, H14 | Parallel | −12065.4 (160.2) | 15 | 1 and S1 |
| M2 | Zirah–Lührs | H6, E11, H13, H14 | Parallel | — | — | 1 and S1 |
| M3 | Zirah–Lührs | H6, E11, H13, H14 | Antiparallel | −11941.6 (177.6) | 13 | 1 and S1 |
| M4 | Zirah–Lührs | H6, E11, H13, H14 | Antiparallel | −11886.4 (170.8) | 11 | S1 |
| M5 | Zirah–Lührs | H6, E11, H13, H14 | Antiparallel | −11517.7 (188.2) | 5 | S1 |
| M6 | Gaggelli–Lührs | D1, H6, E11, H14 | Parallel | −11813.2 (159.3) | 11 | 1 and S2 |
| M7 | Gaggelli–Lührs | D1, H6, E11, H13, H14 | Parallel | −11791.6 (170.9) | 11 | 1 and S2 |
| M8 | Gaggelli–Lührs | D1, H6, E11, H13, H14 | Parallel | −11481.4 (196.5) | 5 | S2 |
| M9 | Gaggelli–Lührs | D1, H6, E11, H13, H14 | Parallel | −11396.3 (183.6) | 5 | S2 |
| M10 | Gaggelli–Lührs | D1, H6, E11, H13, H14 | Antiparallel | −11351.4 (187.0) | 5 | S2 |
| M11 | Gaggelli–Lührs | D1, H6, E11, H13, H14 | Antiparallel | −11551.9 (171.2) | 6 | S2 |
| M12 | Minicozzi–Lührs | E11, H13, H14 | Antiparallel | −11933.1 (200.0) | 13 | 1 and S3 |
*Zirah–Lührs model is constructed from experimental data of Zirah et al. (18) and Lührs et al. (29). The Gaggelli–Lührs model is constructed from experimental data of Gaggelli et al. (22) and Lührs et al. (29). Minicozzi–Lührs model is constructed from experimental data of Minicozzi et al. (23) and Lührs et al. (29).
†Conformational energies were computed using the GBMV calculations (48, 49). Standard deviation values are presented in parenthesis.
‡The percentage of populations or the relative probabilities of the conformational structures were computed using Monte Carlo simulations.
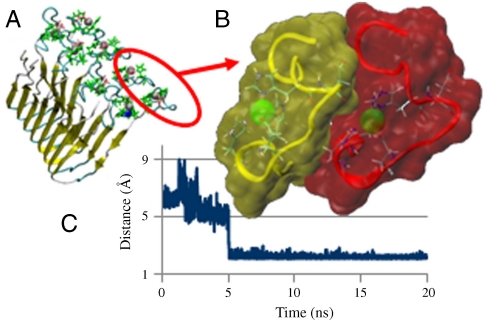
While testing possible arrangements of the Zn2+-Aβ1-16 β-strands for the Zirah (18) and Gaggelli (22) models, we found that for the Zirah model (18) two nearby Zn2+-Aβ1-16 peptides form complementary shapes with tightly packed geometry. The optimal orientation between two nearby Zn2+-Aβ1-16 peptides (Fig. 2A) was obtained using the docking program PatchDock (32) (SI Text). The two highest ranking docking poses have similar values and in both the backbone carbonyl of Asp7 points to Zn2+, with a distance of 3.8 Å (Fig. S4). The Zirah model (18) exhibits a chain rotation that allows the Asp7-COO- to interact with Zn2+ (Fig. 2B). Thus, Zn2+ exhibits intra- and Asp7 intermolecular interactions. Fig. 2C illustrates one of the six averaged distances between the two COO- groups of Asp7 and Zn2+ in model M1 during the simulation. The interactions are formed after 5 ns and stabilize. In the antiparallel M3 model, all six Asp7-COO--Zn2+ distances present strong interactions (Fig. S5).
Fig. 2.
Zn2+ can simultaneously coordinate intra- and intermolecularly, bridging two peptides, consequently promoting aggregation. (A) Model M1, parallel organization, based on the Zirah–Lührs (18, 29) coordinates (Fig. 1). (B) Zn2+ (green balls) can coordinate the N terminals of two neighboring peptides: In the yellow N terminal, the intramolecular coordination of Zn is via three His side chains and one Glu. Zn2+ in the red N terminal can also form intermolecular coordination with Asp7 in the yellow region via either its backbone carbonyl group or side chain. (C) Averaged distance between the two carboxyl groups of Asp7 and Zn2+ during the simulations of M1, indicating that the side chains of Asp7 replace backbone carbonyl group.
Zn2+ Complexed Aβ42 Oligomers Could Exist as Stable Ordered Fibril-Like Structures.
Experimentally, although zinc promotes Aβ peptide aggregation, the resulting aggregates have irregular shapes with no fibril formation (8). The question arises as to whether Zn2+-containing Aβ peptides can form stable fibril-like oligomers. Our simulations indicated that the tested oligomers are structurally stable, suggesting that they could exist under appropriate conditions. For all 12 parallel and antiparallel models, both the N termini and the U-turn rmsds indicate the stability of these regions (SI Text and Figs. S6–S8).
For the complex kinetics of amyloid formation, the 12 constructed conformers are likely to represent only a very small percentage of the ensemble. Nevertheless, the carefully selected models probe the most likely fibril-like organizations. Using all 11 conformers (not including M2) and 11,000 conformations (1,000 for each conformer) generated from the MD simulations, we estimate the overall stability and populations for each conformer based on Monte Carlo simulations with the energy landscape computed with Generalized Born Method with Molecular Volume (GBMV) for all conformers (Table 1).
Among all oligomers studied here, the model M1 is the most stable conformer and is expected to have the highest population. In the case of the N-terminal truncated peptide p3 (Aβ17-42), the in-register parallel conformations are much more highly populated than other arrangements. However, here the other conformers can also have similar probabilities as the most stable M1. For example, the antiparallel organizations of M3 and M12 have fairly large population (13%) compared to M1. The relative stability of M12 is due to the contribution of solvation energy in the oligomeric Zn2+-Aβ complexes. M12 has relatively large solvation energy compared to all constructed models. Two conformers, M6 and M7, also have relatively high populations (11%). Surprisingly, the antiparallel M11 is more stable and has higher population than the parallel models M8 and M9. Comparisons of the energies of these conformers reflect a polymorphic behavior, with Zn2+ coordinating differently in the N-terminal domain. Considering the potential large number of competing conformations, the most stable model M1 could be less preferred due to entropy effects.
Solvation Energy Decreases for Zn2+-Aβ42 Oligomers.
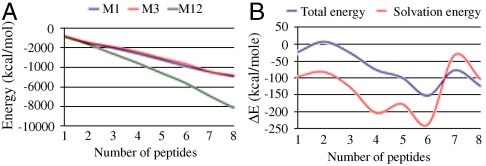
The fact that Zn2+ promotes Aβ peptide aggregation could be the outcome of increased hydrophobicity following Zn2+ coordination to the highly polar N-terminal, which can be expected to reduce the side chains’ solvation. To test the zinc-coordination effects on the solubility of Aβ peptide oligomers, we calculated the solvation energies for various oligomeric sizes. The three most stable models were chosen for comparison: The M1 and M3-Zn2+ coordinated models were used for comparison between parallel and antiparallel oligomers, and because the N termini in M12 are free in solution, the non-Zn2+ coordinated M12 was used as a reference state. Peptides were removed from M1, M3, and M12 one-by-one to form dimer, trimer, tetramer, peptamer, hexamer, and heptamer conformational ensembles for each model. The total energies and the solvation energies of all conformational ensembles were calculated with the GBMV method. At the monomer and dimer levels, M1, M3, and M12 have similar solvation energies (Fig. 3A). However, at the trimer to the octamer levels, the solvation energy of M1 and M3 significantly decreases as compared with the free solvated N termini of M12. The solvation energy gap between M1/M3 and M12 increases with oligomer size, which indicates that larger oligomers of models M1 and M3 are insoluble, and therefore more prone to aggregate than M12.
Fig. 3.
Zinc coordination significantly decreases the solvation energy for large Zn2+-Aβ42 oligomers, enhancing their aggregation tendency. (A) Solvation energy of different number of peptides complexed with Zn2+ for models M1 and M3 and different number of peptides uncomplexed with Zn2+ for model M12. (B) Total energy difference and solvation energy difference between M1 and M3 for various numbers of peptides.
Finally, the relative conformational energies of the monomers and the various oligomeric sizes of models M1 and M3 present a similar trend as their relative solvation energy differences (Fig. 3B). The differences in the solvation energy for the various oligomer sizes could affect the populations of different Zn2+-Aβ complex forms.
Energy Landscape of Aβ42 Aggregation Changes with Zn2+ Concentration.
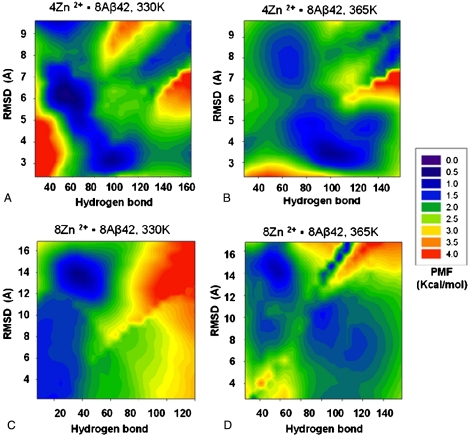
Zn2+ binding to Aβ peptides changes the energy landscape of Aβ peptide aggregation. Our results were based on a limited number of models. To get a more complete picture, we conducted REMD simulations for 8Zn2+-8Aβ42 and 4Zn2+-8Aβ42 systems. We used 64 replicas with temperatures ranging from 330 to 600 K. The starting conformations were generated from four models (M1, M3, M8, and M10). This allows sampling and comparison with random conformations as well as ordered states with different β-strands arrangements, which may be hard to reach otherwise. To have the simulations as close to equilibrium as possible, the first 5-ns simulation in each replica were not used in the analysis. We used two indices to measure conformational similarity: the total number of interpeptide hydrogen bonds and rmsd. In the rmsd calculations, (i) reference conformers are the β-sheet oligomers used in each replica, (ii) because edge strands are very flexible, we used the four core peptides in the octamer, and (iii) the rmsds were calculated using the Cα carbons in the most hydrophobic region (residues 17–36). The population densities for the sampled conformers were computed and the corresponding potential of mean force (PMF) calculated. Fig. 4 compares the energy landscape of Aβ42 peptide aggregation at two Zn2+∶Aβ42 ratios, and at 330 and 365 K. In each plot in Fig. 4, the lower right corner (lower rmsd, higher hydrogen bond counts) represents conformations closer to β-sheet oligomers. A darker color indicates higher populations; red represents higher energy, lower populations.
Fig. 4.
Energy landscape of Aβ aggregation changes with zinc concentration. Energy landscapes near 330 K were obtained from all conformation in all 64 replicas at 330 and 333 K. The PMFs at 365 K are from conformations at 363 and 366 K. (A) The PMFs of 4Zn2+-8Aβ42 near 330 K point to highly ordered β-sheet oligomers separated from the less ordered conformers by a low barrier, indicating that aggregation can readily take place. (B) At the higher temperature (365 K), the energy landscape favors this ordered state with a higher β-sheet population for the 4Zn2+-8Aβ42 system. (C) The 8Zn2+-8Aβ42 system represents the higher zinc concentration with 1∶1 Zn2+∶Aβ42 ratio. At 330 K, the energy landscape indicates that the highly ordered β-sheet oligomers are less populated. (D) At the higher temperature (365 K), the highly ordered β-sheet oligomers are more populated for the 8Zn2+-8Aβ42 system.
The 4Zn2+-8Aβ42 system corresponds to the experimental 1∶2 Zn2+∶Aβ42 ratio (8). The two most populated regions are at around rmsd 6 Å—Hydrogen bonds 60, and 3 Å—100 (Fig. 4A). The first is a “molten globule”-like state with similar shape but fewer hydrogen bonds. The second is closer to the highly ordered β-sheet oligomers. The two regions are connected with a low barrier, suggesting that the amyloid can be readily formed. At the higher temperature (365 K, Fig. 4B), the energy landscape favors higher β-sheet population. However, the overall energy landscape shapes of the 4Zn2+-8Aβ42 system are similar.
The 8Zn2+-8Aβ42 system simulates higher zinc concentration with 1∶1 Zn2+∶Aβ42 ratio. At 330 K (Fig. 4C), the energy landscape undergoes a remarkable change as compared to lower zinc concentration (Fig. 4A). Highly ordered β-sheet oligomers are rarely populated, with most conformations in molten globule states of β-sheet oligomers (low rmsd, but fewer hydrogen bonds). At the higher temperature (365 K, Fig. 4D), the highly ordered β-sheet oligomers are more populated, and the energy landscape is similar to that of lower zinc concentration (4Zn2+-8Aβ42).
These results imply that Aβ aggregation does not increase linearly with zinc concentration. At certain concentration, increasing zinc not only prevents fibril formation, but also decreases aggregation. This scenario is consistent with experimental observation when mixing rapidly Zn and Aβ (8). In the presence of zinc, the Aβ peptide aggregation rates are always higher than in the Zn2+-free solution. However, at 50 μm peptide concentration, the aggregation rate is higher for 1∶2 Zn2+∶Aβ42 than in 2∶1 Zn2+∶Aβ42 ratios (see figure 1 in ref. 8).
Discussion
So far, only few computational studies investigated metal ion-Aβ peptide interaction. Raffa and Rauk (33) simulated a divalent copper interaction with Aβ42 monomer. Jiao and Yang (34) investigated the divalent Cu2+–Aβ10-21 oligomer interaction, proposing that Cu2+ may distort β-strands and thus inhibit amyloid formation. Li et al. (35) studied the zinc-binding effect on the monomeric conformations of Aβ1-28. Experimental structures for Zn2+ ions coordinated to full-length Aβ42 oligomers and simulations of the potential architectures are not yet available. Here we present the conformational stabilities and populations of various Zn2+-Aβ42 oligomers with the goal of understanding the structural basis of zinc-promoted Aβ-aggregation and related energy landscape.
Using all-atom MD simulations in explicit solvent, we investigated polymorphic architectures of Zn2+ ions (enriched in in vivo Aβ plaques) coordinated to full-length Aβ42 oligomers. The 12 studied oligomers differ in organizations, locations, and coordinations of the Zn2+ ions in the peptides. Each model presents a different architecture in which Zn2+ coordinates with the N-terminal domain of Aβ42. This allows us to simulate experiment-based polymorphic organizations of Aβ42. Overall, combined with the putative model-built aggregates, our REMD simulations broaden our understanding of the complex Zn2+ effects on Aβ aggregation.
Several conclusions emerged from this study: First, domain–domain interactions between Asp7 in one peptide and Zn2+ coordinated to a neighboring peptide can play an important role in oligomer stability. Prior to zinc binding, the N-terminal region is likely to be in a disordered state, which is typical in protein polymerization (36). The preorganized, constrained Zn2+-coordinated N-terminal conformation docks into a geometrically matching conformation (30), conceptually similar to the proposed constrained anchoring side chains in protein–protein interaction mechanism (37). This effect is observed for the most stable conformers based on the Zirah–Lührs model and the most stable conformers based on some of the Gaggelli–Lührs models; the intermolecular interaction proposed by Syme and Viles (17) and recently suggested by Minicozzi et al. (23) may have different patterns.
Our simulations also present a stable model (M12) in which Zn2+ coordinates with residues of two different peptides. This study tests this hypothesis and provides architecture in which Zn2+ shares two peptides in Aβ42. Second, dehydration is known to be important for amyloid formation (38), and zinc coordination decreases the solvation energy for tightly packed oligomers. These two findings explain why zinc can promote amyloid formation and its role in Aβ aggregation. The intermolecular interactions of zinc in Aβ oligomers can decrease the lag time of amyloid seed formation. Thermodynamically, zinc coordination decreases both the entropic penalty of the N-terminal region in aggregation and the solvation energy, increasing the precipitation rate of zinc-complexed oligomers. Our study also offers an explanation why zinc ions are able to precipitate Aβ oligomers, but monomeric Aβ is relatively resistant to precipitation (39, 40). The decrease of solvation energy becomes more significant for larger oligomers, and the difference is small between the monomer and dimer (Fig. 3). Our observation from the REMD that highly ordered β-sheet oligomers are preferred at high temperature also supports the role of the hydrophobic effect, which is more pronounced at high temperatures.
Based on experimental data, our simulations show that Zn2+ ions can coordinate at different locations in the N-terminal domain leading to conformational variability with a range of different populations. This extensive polymorphism explains the lower fibril state for zinc-triggered amyloid formation. The energy landscape obtained from the REMD simulation confirmed the entropic effects of competing conformers in the vastly diversified ensemble. Under certain conditions, increasing zinc concentration not only prevents fibril formation, but also decreases aggregation rate. Different zinc concentrations may selectively stabilize different oligomeric forms, and different oligomeric sizes are expected to have different stabilities. This complexity rendered by zinc coordination explains zinc selectivity in precipitating aggregation intermediates. Amyloids are highly polymorphic, and population shifts redistribute polymorphic states (Fig. S9). Zinc coordination and the subsequent change of solvation energy modify the energy landscape of the amyloid β-peptide elongation (41) and the relative stability of the oligomers.
The amyloid landscape presents a broad range of aggregated conformational states. Which species prevail is the outcome of conditions: Different conditions will shift the landscape (30, 42). For an in vivo truncated Aβ variant (Aβ17-4217–42, produced by α-secretase cleavage), we have already shown a range of states, including parallel and antiparallel organizations (43) determined by alternative turn conformations. The observed changing aggregation energy landscape with zinc concentration suggests that toxicity may not linearly correlate with ion concentration. It has been suggested that, in the human brain, zinc concentration is not sufficiently high to promote amyloidosis (8); however, our study implies that trace amounts of free zinc should not be neglected. On a similar note, it was reported that trace amount of Cu2+ can decrease rat memory (44). We further note that toxicity could also relate to the mechanism. Species conformations and solubility could be important for Zn2+-mediated toxicity.
Materials and Methods
Classical MD Simulations.
MD simulations of solvated 8Zn2+-8Aβ42 and 4Zn2+-8Aβ42 oligomers were performed in NPT (N, number of particles; P, pressure; and T, temperature) ensemble at 1 atm and 300 K using the NAMD program (45) and the CHARMM package (46) with the all-atom CHARMM27 force field. The oligomers were explicitly solvated with a TIP3P water box with a minimum distance of 10–15 Å from any edge of the box to any Aβ atom. Long-range electrostatic interactions were calculated using the particle mesh Ewald method with a cutoff of 12.0 Å for all simulations. Counterions (Na+) were added at random locations to neutralize the charge of 8Zn2+-8Aβ42 and 4Zn2+-8Aβ42. The hydrogen atoms were constrained to the equilibrium bond using the SHAKE algorithm (47) (SI Text).
To obtain the relative structural stability and populations of the 8Zn2+-Aβ1-42 oligomers, the Aβ trajectories of the last 5 ns were first extracted from the explicit MD simulation excluding water molecules. The solvation energies of all systems were calculated using the Generalized Born Method with Molecular Volume (GBMV) (48, 49) (SI Text).
Replica Exchange MD Simulations.
REMD simulations were performed using the NAMD program for 8Zn2+-8Aβ42 and 4Zn2+-8Aβ42 systems. Each REMD simulation consists of 64 replicas. The simulation temperature ranges from 330 to 600 K with an exponential distribution. The exchange time among the replicas is 0.25 ps with the averaged acceptance ratio of 18%. The initial conformations for the REMD simulations are generated from the four models based on the “classical” MD simulations (M1, M3, M8, and M10). Thus the 64 replicas are initially evenly distributed with four sets of 16 conformers from the four models. Among the 16 replicas for each model, eight start from conformers near those β-sheet oligomers obtained from 20 ns MD simulations, and eight from random conformers generated from unfolding of the oligomers at 700 K. Each replica runs for 18 ns (total simulation time 1.152 μs), and the last 13 ns of the trajectories were used for analysis. The PMFs were evaluated as PMF = -RT log(pi), where R is the gas constant and pi is the relative populations obtained from the REMD simulations.
Supplementary Material
Acknowledgments.
We thank the Nussinov group members at the Center for Cancer Research Nanobiology Program, National Cancer Institute–Frederick. All simulations had been performed using the high-performance computational facilities of the Biowulf cluster at the National Institutes of Health (NIH). We thank Drs. E. Molteni and F. Stellato for providing the atomic coordinates of the models obtained in their labs. This project has been funded in whole or in part with Federal funds from the National Cancer Institute, NIH, under Contract HHSN261200800001E. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Note Added in Proof.
Since the completion of this work, a review focusing on polymorphism in Aβ organization, describing also polymorphism in metal-binding sites in Aβ, has been published by Miller et al. (50).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0913114107/-/DCSupplemental.
References
- 1.Honig LS, Mayeux R. Natural history of Alzheimer’s disease. Aging. 2001;13:171–182. doi: 10.1007/BF03351476. [DOI] [PubMed] [Google Scholar]
- 2.Masters CL, et al. Amyloid plaque core protein in Alzheimer-disease and Down syndrome. Proc Natl Acad Sci USA . 1985;82(12):4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardy J, Selkoe DJ. Medicine—The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 4.Bush AI, et al. Rapid inductionof Alzheimer A beta amyloid formation by zinc. Science. 1994;265:1464–1467. doi: 10.1126/science.8073293. [DOI] [PubMed] [Google Scholar]
- 5.Stoltenberg M, et al. Amyloid plaques arise from zinc-enriched cortical layers in APP/PS1 transgenic mice and are paradoxically enlarged with dietary zinc deficiency. Neuroscience. 2007;150:357–369. doi: 10.1016/j.neuroscience.2007.09.025. [DOI] [PubMed] [Google Scholar]
- 6.Ali FE, Separovic F, Barrow CJ, Yao SG, Barnham KJ. Copper and zinc mediated oligomerisation of A beta peptides. Int J Pept Res Ther. 2006;12:153–164. [Google Scholar]
- 7.Lim KH, Kim YK, Chang YT. Investigations of the molecular mechanism of metal-induced Abeta (1-40) amyloidogenesis. Biochemistry. 2007;46:13523–13532. doi: 10.1021/bi701112z. [DOI] [PubMed] [Google Scholar]
- 8.Noy D, et al. Zinc-amyloid beta interactions on a millisecond time-scale stabilize non-fibrillar Alzheimer-related species. J Am Chem Soc. 2008;130:1376–1383. doi: 10.1021/ja076282l. [DOI] [PubMed] [Google Scholar]
- 9.Liu ST, Howlett G, Barrow CJ. Histidine-13 is a crucial residue in the zinc ion-induced aggregation of the A beta peptide of Alzheimer’s disease. Biochemistry. 1999;38:9373–9378. doi: 10.1021/bi990205o. [DOI] [PubMed] [Google Scholar]
- 10.Yang DS, McLaurin J, Qin K, Westaway D, Fraser PE. Examining the zinc binding site of the amyloid-beta peptide. Eur J Biochem. 2000;267:6692–6698. doi: 10.1046/j.1432-1327.2000.01767.x. [DOI] [PubMed] [Google Scholar]
- 11.Miura T, Suzuki K, Kohata N, Takeuchi H. Metal binding modes of Alzheimer’s amyloid beta-peptide in insoluble aggregates and soluble complexes. Biochemistry. 2000;39:7024–7031. doi: 10.1021/bi0002479. [DOI] [PubMed] [Google Scholar]
- 12.Curtain CC, et al. Alzheimer’s disease amyloid-beta binds copper and zinc to generate an allosterically ordered membrane-penetrating structure containing superoxide dismutase-like subunits. J Biol Chem. 2001;276:20466–20473. doi: 10.1074/jbc.M100175200. [DOI] [PubMed] [Google Scholar]
- 13.Kozin SA, Zirah S, Rebuffat S, Hoa GH, Debey P. Zinc binding to Alzheimer’s Abeta(1-16) peptide results in stable soluble complex. Biochem Biophys Res Commun. 2001;285:959–964. doi: 10.1006/bbrc.2001.5284. [DOI] [PubMed] [Google Scholar]
- 14.Zirah S, et al. Zinc binding properties of the amyloid fragment A beta(1-16) studied by electrospray-ionization mass spectrometry. Int J Mass Spectrom. 2003;228:999–1016. [Google Scholar]
- 15.Zirah S, et al. Zinc binding agonist effect on the recognition of the beta-amyloid (4-10) epitope by anti-beta-amyloid antibodies. Biochem Biophys Res Commun. 2004;321:324–328. doi: 10.1016/j.bbrc.2004.06.150. [DOI] [PubMed] [Google Scholar]
- 16.Mekmouche Y, et al. Characterization of the Zn-II binding to the peptide amyloid-beta(1-16) linked to Alzheimer’s disease. Chembiochem. 2005;6:1663–1671. doi: 10.1002/cbic.200500057. [DOI] [PubMed] [Google Scholar]
- 17.Syme CD, Viles JH. Solution H-1 NMR investigation of Zn2+ and Cd2+ binding to amyloid-beta peptide (A beta) of Alzheimer’s disease. BBA—Proteins Proteom. 2006;1764:246–256. doi: 10.1016/j.bbapap.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 18.Zirah S, et al. Structural changes of region 1-16 of the Alzheimer disease amyloid beta-peptide upon zinc binding and in vitro aging. J Biol Chem. 2006;281:2151–2161. doi: 10.1074/jbc.M504454200. [DOI] [PubMed] [Google Scholar]
- 19.Stellato F, et al. Metal binding in amyloid beta-peptides shows intra- and inter-peptide coordination modes. Eur Biophys J Biophy. 2006;35:340–351. doi: 10.1007/s00249-005-0041-7. [DOI] [PubMed] [Google Scholar]
- 20.Talmard C, Guilloreau L, Coppel Y, Mazarguil H, Faller P. Amyloid-beta peptide forms monomeric complexes with Cu-II and Zn-II prior to aggregation. Chembiochem. 2007;8:163–165. doi: 10.1002/cbic.200600319. [DOI] [PubMed] [Google Scholar]
- 21.Danielsson J, Pierattelli R, Banci L, Graslund A. High-resolution NMR studies of the zinc-binding site of the Alzheimer’s amyloid beta-peptide. FEBS J. 2007;274:46–59. doi: 10.1111/j.1742-4658.2006.05563.x. [DOI] [PubMed] [Google Scholar]
- 22.Gaggelli E, et al. NMR studies of the Zn2+ interactions with rat and human beta-amyloid (1-28) peptides in water-micelle environment. J Phys Chem B. 2008;112:100–109. doi: 10.1021/jp075168m. [DOI] [PubMed] [Google Scholar]
- 23.Minicozzi V, et al. Identifying the minimal copper- and zinc-binding site sequence in amyloid-beta peptides. J Biol Chem. 2008;283:10784–10792. doi: 10.1074/jbc.M707109200. [DOI] [PubMed] [Google Scholar]
- 24.Faller P, Hureau C. Bioinorganic chemistry of copper and zinc ions coordinated to amyloid-beta peptide. Dalton T. 2009:1080–1094. doi: 10.1039/b813398k. [DOI] [PubMed] [Google Scholar]
- 25.Talmard C, Leuma Yona R, Faller P. Mechanism of zinc(II)-promoted amyloid formation: Zinc(II) binding facilitates the transition from the partially alpha-helical conformer to aggregates of amyloid beta protein(1-28) J Biol Inorg Chem. 2009;14:449–455. doi: 10.1007/s00775-008-0461-9. [DOI] [PubMed] [Google Scholar]
- 26.Dong J, Shokes JE, Scott RA, Lynn DG. Modulating amyloid self-assembly and fibril morphology with Zn(II) J Am Chem Soc. 2006;128:3540–3542. doi: 10.1021/ja055973j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen TT, et al. Effects of cyclen and cyclam on zinc(II)- and copper(II)-induced amyloid beta-peptide aggregation and neurotoxicity. Inorg Chem. 2009;48:5801–5809. doi: 10.1021/ic900025x. [DOI] [PubMed] [Google Scholar]
- 28.Garai K, Sahoo B, Kaushalya SK, Desai R, Maiti S. Zinc lowers amyloid-beta toxicity by selectively precipitating aggregation intermediates. Biochemistry. 2007;46:10655–10663. doi: 10.1021/bi700798b. [DOI] [PubMed] [Google Scholar]
- 29.Luhrs T, et al. 3D structure of Alzheimer’s amyloid-beta(1-42) fibrils. Proc Natl Acad Sci USA. 2005;102:17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S, Ma B, Tsai CJ, Sinha N, Nussinov R. Folding and binding cascades: Dynamic landscapes and population shifts. Protein Sci. 2000;9:10–19. doi: 10.1110/ps.9.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s beta-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raffa DF, Rauk A. Molecular dynamics study of the beta amyloid peptide of Alzheimer’s disease and its divalent copper complexes. J Phys Chem B. 2007;111:3789–3799. doi: 10.1021/jp0689621. [DOI] [PubMed] [Google Scholar]
- 34.Jiao Y, Yang P. Mechanism of copper(II) inhibiting Alzheimer’s amyloid beta-peptide from aggregation: a molecular dynamics investigation. J Phys Chem B. 2007;111:7646–7655. doi: 10.1021/jp0673359. [DOI] [PubMed] [Google Scholar]
- 35.Li W, et al. Effects of zinc binding on the conformational distribution of the amyloid-beta peptide based on molecular dynamics simulations. J Phys Chem B . 2007;111:13814–13821. doi: 10.1021/jp076213t. [DOI] [PubMed] [Google Scholar]
- 36.Uversky VN. Intrinsic disorder in proteins associated with neurodegenerative diseases. Front Biosci. 2009;14:5188–5238. doi: 10.2741/3594. [DOI] [PubMed] [Google Scholar]
- 37.Rajamani D, Thiel S, Vajda S, Camacho CJ. Anchor residues in protein-protein interactions. Proc Natl Acad Sci USA. 2004;101:11287–11292. doi: 10.1073/pnas.0401942101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukherjee S, Chowdhury P, Gai F. Effect of dehydration on the aggregation kinetics of two amyloid peptides. J Phys Chem B . 2009;113:531–535. doi: 10.1021/jp809817s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bush AI, Pettingell WH, Jr, Paradis MD, Tanzi RE. Modulation of A beta adhesiveness and secretase site cleavage by zinc. J Biol Chem. 1994;269:12152–12158. [PubMed] [Google Scholar]
- 40.Garai K, Sengupta P, Sahoo B, Maiti S. Selective destabilization of soluble amyloid beta oligomers by divalent metal ions. Biochem Biophys Res Commun. 2006;345:210–215. doi: 10.1016/j.bbrc.2006.04.056. [DOI] [PubMed] [Google Scholar]
- 41.Massi F, Straub JE. Energy landscape theory for Alzheimer’s amyloid beta-peptide fibril elongation. Proteins. 2001;42:217–229. doi: 10.1002/1097-0134(20010201)42:2<217::aid-prot90>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 42.Tsai CJ, Kumar S, Ma B, Nussinov R. Folding funnels, binding funnels, and protein function. Protein Sci. 1999;8:1181–1190. doi: 10.1110/ps.8.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller Y, Ma B, Nussinov R. Polymorphism of Alzheimer’s Abeta17–42 (p3) oligomers: The importance of the turn location and its conformation. Biophys J. 2009;97:1168–1177. doi: 10.1016/j.bpj.2009.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bush AI, Masters CL, Tanzi RE. Copper, beta-amyloid, and Alzheimer’s disease: Tapping a sensitive connection. Proc Natl Acad Sci USA. 2003;100:11193–11194. doi: 10.1073/pnas.2135061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kale L, et al. NAMD2: Greater scalability for parallel molecular dynamics. J Comput Phys. 1999;151:283–312. [Google Scholar]
- 46.Brooks BR, et al. Charmm—A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 47.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-integration of Cartesian Equations of motion of a system with constraints—Molecular-dynamics of N-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 48.Lee MS, Feig M, Salsbury FR, Brooks CL. New analytic approximation to the standard molecular volume definition and its application to generalized born calculations. J Comput Chem. 2003;24:1348–1356. doi: 10.1002/jcc.10272. [DOI] [PubMed] [Google Scholar]
- 49.Lee MS, Salsbury FR, Brooks CL. Novel generalized Born methods. J Chem Phys. 2002;116:10606–10614. [Google Scholar]
- 50.Miller Y, Ma B, Nussinov R. Polymorphism in Alzheimer Abeta amyloid organization reflects conformational selection in a rugged energy landscape. Chem Rev. 2010 doi: 10.1021/cr900377t. PMID: 20402519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.