Abstract
P63, a p53 family tumor suppressor, is involved in many cellular processes, including growth suppression and differentiation. Thus, p63 activity needs to be tightly controlled. Here, we found that RNPC1, a RNA-binding protein and a target of the p53 family, regulates p63 mRNA stability and consequently p63 activity. Specifically, we showed that overexpression of RNPC1 decreases, whereas knockdown of RNPC1 increases, the half-life of p63 transcript, which leads to altered p63 expression. Consistent with this, we showed that RNPC1 binds the AU-/U-rich elements in p63 3′ UTR in vitro and in vivo and the RRM domain in RNPC1 is required for binding, and regulating the stability of, p63 transcript. Furthermore, we showed that RNPC1 promotes keratinocyte differentiation by repressing p63 expression. Together, we uncovered a previously undetected mechanism by which p63 expression is regulated via mRNA stability and a novel regulatory feedback loop between RNPC1 and p63.
Keywords: the p53 family, RNPC1, RBM38, mRNA stability, p63
P63, a p53 family protein, shares considerable sequence identity with p53, especially in its DNA binding, activation, and tetramerization domains (1–2). Because of the usage of two distinct promoters, p63 is expressed as two major variants, called TAp63 and ΔNp63, both of which have multiple isoforms through alternative splicing at the C-terminus. The TA variant, which is transcribed from the upstream promoter, contains an activation domain (AD) similar to p53 and is able to transactivate p53 target genes. The ΔN variant, which is transcribed from the alternate promoter in intron 3, lacks the N-terminal activation domain and is presumably incompetent for transactivation. Interestingly, p63 contains a second activation domain, which is adjacent to the N-terminal activation domain and present in all of the ΔN isoforms (3). As a result, ΔNp63 is transcriptional active and able to transactivate multiple target genes (4).
Because of its sequence similarity to p53, p63 has many p53-like functions, such as the ability to induce cell cycle arrest, apoptosis, differentiation, and senescence. However, unlike p53, p63 is not a classic tumor suppressor as p63 is rarely mutated in human cancers. In addition, p63 plays a critical role in development. Mice deficient in p63 develop striking epithelial defects, including an almost complete absence of hair, skin, breast, and prostate, and severe limb and craniofacial malformations (5–7). Consistent with this, mutations in the p63 gene are associated with five human syndromes with characteristics of limb malformations, craniofacial clefting, and ectodermal dysplasia (8–11). These differences between p63 and p53 suggest that they are likely to be regulated through distinct mechanisms. Thus, deciphering the mechanism by which p63 expression is regulated is critical for understanding p63 biological function.
Posttranslational modifications are known to regulate p63 expression. For example, p63 protein stability is regulated by several E3 ligases, such as itch, wwp1, and SCFβTrCP1 (12–14). In addition, upon exposure to various stimuli, the level of p63 transcript is regulated by p53 and other transcription factors (15–17). However, whether and how p63 is regulated by other posttranscriptional mechanisms has not been examined. In an effort to examine the role of the p53 family target genes, we found that RNPC1, a RNA-binding protein and a target of the p53 family, negatively regulates p63 mRNA stability and promotes keratinocyte differentiation by repressing p63 expression.
Results
P63 Expression Is Repressed by RNPC1.
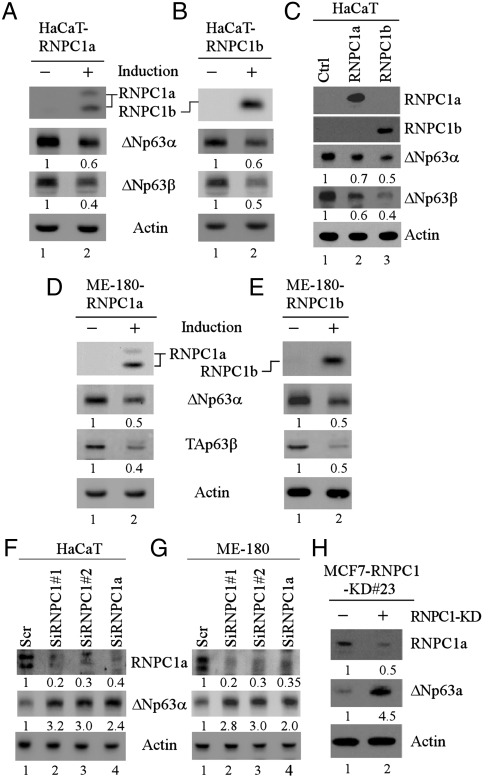
The RNPC1 gene encodes a RNA-binding protein and is expressed as two isoforms, RNPC1a with 239 amino acids and RNPC1b with 121 amino acids (18). Previously, we showed that RNPC1 is a target of the p53 family, including p63, and regulates p21 mRNA stability (18). Since p63 activity needs to be tightly controlled, we investigated the possibility that RNPC1 may regulate p63 expression. Thus, we generated HaCaT cell lines in which HA-tagged RNPC1a or RNPC1b can be inducibly expressed. We showed that upon induction of RNPC1a or RNPC1b, the level of ΔNp63 proteins was markedly reduced under a normal condition (Fig. 1 A and B, ΔNp63α and ΔNp63β panels) as well as DNA-damage-induced conditions (Fig. S1 A and B). Consistent with this, ΔNp63 expression was also inhibited in HaCaT cells upon transient expression of RNPC1a or RNPC1b (Fig. 1C, ΔNp63α and ΔNp63β panels, compare lane 1 with 2 and 3). To rule out potential cell type-specific effects, p63 expression was measured in ME-180 cells and found to be repressed by RNPC1a and RNPC1b under a normal condition (Fig. 1 D and E, ΔNp63α and TAp63β panels) as well as DNA-damage-induced conditions (Fig. S1 C amd D).
Fig. 1.
P63 expression is regulated by RNPC1. (A and B) The level of RNPC1a, RNPC1b, Np63α, ΔNp63β, and actin was measured in HaCaT cells uninduced or induced to express RNPC1a (A) or RNPC1b (B) for 24 h. The relative level of p63 proteins in the absence or presence of RNPC1a or RNPC1b was shown below the lane. (C) The experiment was performed as in (A and B) with HaCaT cells transfected with empty or RNPC1a-/RNPC1b-expressing pcDNA3 for 24 h. (D and E) The experiment was performed as in (A and B) with ME-180 cells uninduced or induced to express HA-tagged RNPC1a (D) or RNPC1b (E) for 24 h. (F and G) The level of RNPC1a, ΔNp63α, and actin was measured in HaCaT (F) and ME-180 (G) cells transfected with scrambled siRNA or siRNAs against total RNPC1 or RNPC1a for 3 d. The relative level of p63 and RNPC1a proteins was shown below the lane. (H) The experiment was performed as in (F and G) with MCF7 cells uninduced or induced to knock down RNPC1 for 3 d.
Next, to determine whether endogenous RNPC1 regulates p63 expression, siRNAs against total RNPC1 (targeting the sequence common to both RNPC1a and RNPC1b) or RNPC1a, or scrambled siRNA were transfected into parental HaCaT and ME-180 cells. qRT-PCR was performed and showed that the level of total RNPC1, RNPC1a, and RNPC1b transcripts was reduced by both siRNAs against total RNPC1 whereas siRNA against RNPC1a specifically decreased the level of RNPC1a but not RNPC1b transcripts (Fig. S1E). Likewise, the level of RNPC1a protein was decreased by siRNAs against total RNPC1 or RNPC1a (Fig. 1 F and G, RNPC1a panel, compare lane 1 with 2–4). Because of the low reactivity of anti-RNPC1, the level of RNPC1b was undetectable. Consistent with observations above, we found that the levels of ΔNp63α were increased upon total RNPC1 or RNPC1a knockdown, but not by scrambled siRNA (Fig. 1 F and G, ΔNp63α panel, compare lane 1 with lanes 2–4). In addition, ΔNp63α was increased by total RNPC1 or RNPC1a knockdown under DNA-damage-induced conditions (Fig. S1F). To further confirm the above findings, we generated a MCF7 cell line in which RNPC1 can be inducibly knocked down. We showed that ΔNp63α expression was markedly increased upon inducible knockdown of total RNPC1 under a normal condition (Fig. 1H, ΔNp63α panel, compare lane 1 with 2) as well as DNA-damage-induced conditions (Fig. S1G). Together, these data suggest that RNPC1 inhibits p63 expression under normal and stress-induced conditions.
P63 mRNA Stability Is Regulated by RNPC1.
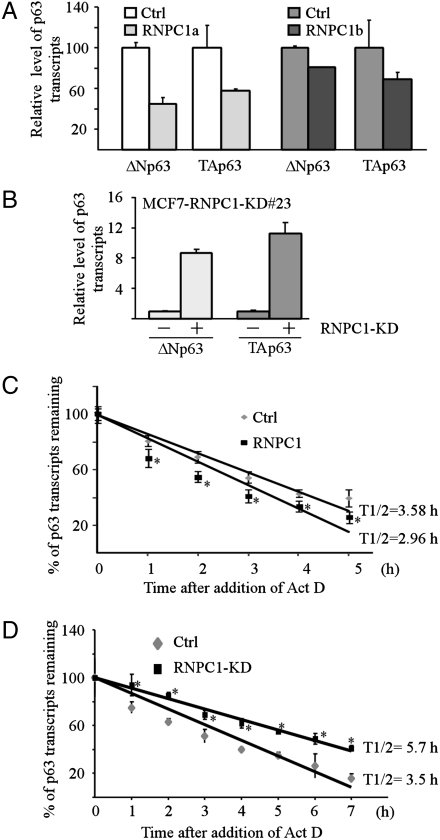
RNA-binding proteins are known to regulate gene expression via posttranscriptional mechanisms, including mRNA stability (19). To explore how RNPC1 inhibits p63 expression, qRT-PCR was performed to measure the level of p63 transcript. We found that overexpression of RNPC1a in MCF7 cells reduced the levels of ΔNp63 and TAp63 transcripts by 55% and 45%, respectively (Fig. 2A, left two columns). In addition, overexpression of RNPC1b had similar effects on p63 expression albeit to a lesser extent (20% reduction for ΔNp63 and 35% reduction for TAp63) (Fig. 2A, right two columns). To rule out potential cell type-specific effects, the levels of p63 transcripts were measured in HaCaT cells and found to be repressed by RNPC1a or RNPC1b under normal and DNA-damage-induced conditions (Fig. S2 A and B). Conversely, we examined whether endogenous RNPC1 is capable of regulating p63 mRNA. We showed that upon knockdown of RNPC1, the level of ΔNp63 and TAp63 transcripts was markedly increased (2.8 ∼ 8 folds for ΔNp63 vs. 4.8 ∼ 12 folds for TAp63) in MCF7 cells under a normal (Fig. 2B) as well as DNA-damage-induced (Fig. S2C) conditions. Next, we determined whether the expression of p63 transcript altered by RNPC1 is due to altered mRNA stability. To test this, HaCaT cells were treated with actinomycin D to inhibit de novo RNA synthesis for various times in the presence or absence of RNPC1a expression, and the level of p63 mRNA measured by qRT-PCR. We showed that the half-life of p63 mRNA was decreased from ∼3.58 h in control cells to ∼2.96 h in RNPC1a-producing cells (Fig. 2C). To further confirm this, the half-life of p63 mRNA was measured in MCF7 cells in which RNPC1 can be inducibly knocked down. We found that the half-life of p63 mRNA was increased from ∼3.5 h in control cells to ∼5.7 h in RNPC1-knockdown cells (Fig. 2D). Together, these data suggest that RNPC1 destabilizes p63 transcript.
Fig. 2.
P63 mRNA stability is regulated by RNPC1. (A) The level of TA and ΔN p63 transcripts was measured by qRT-PCR in HaCaT cells uninduced or induced to express HA-tagged RNPC1a or RNPC1b for 24 h. The level of GAPDH transcript was measured as an internal control. (B) The experiment was performed as in (A) with MCF7 cells uninduced or induced to knock down RNPC1 for 3 d. (C) p63 mRNA half-life is decreased by RNPC1a. The level of p63 transcript was measured by qRT-PCR in HaCaT cells uninduced or induced to express RNPC1a for 24 h, followed by treatment with actinomycin D for various times. Data were presented as Mean ± S.D. (∗P < 0.05; n = 3 per group). (D) The experiment was performed as in (C) with MCF7 cells uninduced or induced to knock down RNPC1 for 3 d. Data were presented as Mean ± S.D. (∗P < 0.05; n = 3 per group).
RNPC1 Binds to AU-/U-rich Elements in p63 3′UTR.
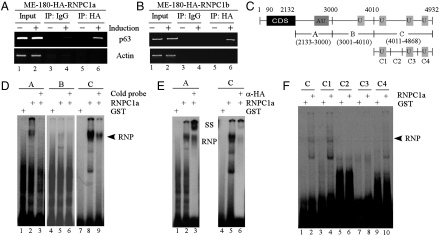
To explore the underlying mechanism by which RNPC1a destabilizes p63 mRNA, we examined whether RNPC1 associates with p63 transcript. Thus, RNA immunoprecipitation assay followed by RT-PCR was performed with extracts from ME-180 cells that can inducibly express HA-tagged RNPC1a or RNPC1b. We found that upon induction of RNPC1a or RNPC1b, p63 mRNA was detected in RNPC1a/RNPC1b, but not control IgG, immunoprecipitates (Fig. 3 A and B, p63 panel, compare lane 4 with 6). As a control, both RNPC1a and RNPC1b were unable to bind actin mRNA (Fig. 3 A and B, actin panel).
Fig. 3.
RNPC1 binds to AU-/U-rich elements in p63 3′UTR. (A and B) RNPC1a (A) and RNPC1b (B) interact with p63 transcript in vivo. ME-180 cells were uninduced or induced to express HA-tagged RNPC1a (A) or RNPC1b (B) for 18 h, followed by immunoprecipitation with anti-HA or mouse IgG as a control. RT-PCR was performed to measure the level of p63 transcript in the control and RNPC1-RNA complexes. (C) Schematic presentation of p63 transcript and the location of probes. AU-/U-rich regions are shown in shaded box. (D) RNPC1a directly binds p63 3′ UTR. 32P-labeled RNA probes were mixed with recombinant GST or HA-RNPC1-GST fusion protein. For competition assay, cold probes A, B, and C were added to the reaction run in lanes 3, 6, and 9, respectively. The arrow indicates RNA-protein complexes. (E) Anti-HA was added in the reaction to “supershift” RNPC1-probe A/C complexes. (F) REMSA assay was performed as in (D) with probes C and C1-4.
To identify RNPC1-binding site(s) in p63 mRNA, RNA electropherotic mobility shift assay (REMSA) was performed using radiolabeled RNA probes (probe A, B, and C), spanning the entire p63 3′ UTR (Fig. 3C). We showed that recombinant GST fusion protein containing HA-tagged RNPC1a, but not GST protein, formed a complex with probes A and C (Fig. 3D, compare lanes 1 and 7 with lanes 2 and 8, respectively). In contrast, RNPC1a did not interact with probe B (Fig. 3D, lanes 5 and 6). The complex formation was inhibited with an excess amount of their respective cold probes (Fig. 3D, lanes 3 and 9). Furthermore, the complex was supershifted with anti-HA, which recognizes HA-tagged RNPC1 (Fig. 3E, lanes 3 and 6). We would like to mention that the formation of RNPC1-probe C complexes was partially inhibited by anti-HA, which is likely due to sterical hindrance of HA antibody or higher affinity of HA-tagged RNPC1 to HA antibody than to probe C (Fig. 3E, compare lanes 5 and 6). Since probe C contains several U-rich elements, four subfragments within probe C (C1, C2, C3, and C4) were made to delineate the region to which RNPC1a binds (Fig. 3C). We found that like full-length probe C, probe C1 showed a strong binding to RNPC1a whereas probe C4 had a weak affinity (Fig. 3F, lanes 2, 4, and 10). However, C2 and C3 probes did not exhibit any binding to RNPC1a (Fig. 3F, lanes 6 and 8). Taken together, these data suggest that RNPC1 can bind the AU-/U-rich elements in p63 3′ UTR.
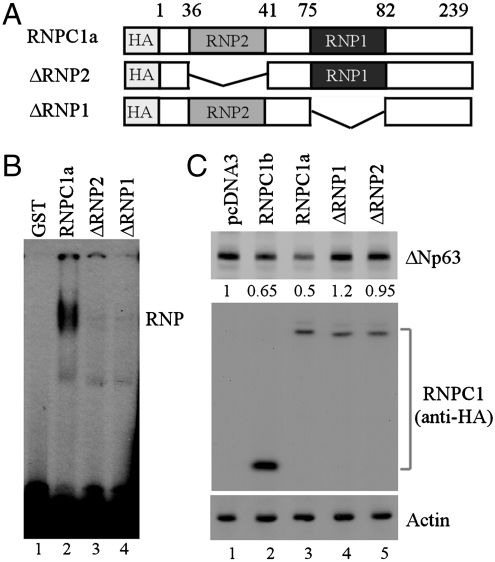
The RNA-binding Domain in RNPC1 Is Required for Binding p63 Transcript and for Inhibiting p63 Expression.
The RNA-binding domain in RNPC1 is composed of two putative RNA-binding submotifs (RNP1 and RNP2) (18). Thus, we examined whether both RNP1 and RNP2 are required for binding p63 transcript using HA-tagged RNPC1 lacking either RNP1 or RNP2 (Fig. 4A). REMSA was performed and showed that neither deletion mutants were capable of binding p63 transcript (Fig. 4B, compare lane 2 with 3 and 4). In line with this, these RNP-deleted mutants were unable to inhibit p63 expression in HaCaT cells compared to RNPC1a and RNPC1b (Fig. 4C, compare lanes 4 and 5 with 2 and 3). Taken together, these data suggest that the RRM domain in RNPC1 is critical for binding p63 transcript and for inhibiting p63 expression.
Fig. 4.
The RNA-binding domain in RNPC1 is required for binding p63 transcript and for inhibiting p63 expression. (A) Schematic illustration of ΔRNP1 and ΔRNP2 mutants. (B) The RNA-binding domain in RNPC1a is required for binding p63 3′UTR. REMSA assay was performed as in Fig. 3A by incubating 32P-labeled probe C with GST, GST-HA-RNPC1, GST-HA-ΔRNP1, or GST-HA-ΔRNP2. (C) RNP1- and RNP2-deletion mutants are unable to inhibit p63 expression. The level of ΔNp63α along with actin was measured in HaCaT cells transfected with an empty vector or a vector expressing HA-tagged RNPC1a, RNPC1b, ΔRNP1, or ΔRNP2. The relative level of p63 proteins was shown below the lane.
RNPC1 Promotes Keratinocyte Differentiation by Repressing p63 Expression.
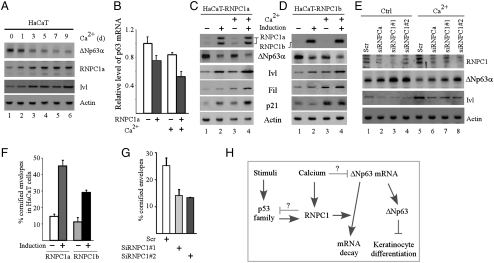
P63 is known to be down-regulated during keratinocyte differentiation and elevated expression of p63 can suppress keratinocyte differentiation (5, 20–22). Thus, to further explore the biological consequence of RNPC1-mediated p63 inhibition, we determined whether RNPC1 plays a role in kerationocyte differentiation. To this end, the level of p63 mRNA and protein was examined in differentiating HaCaT cells over a 9-day period and found to be significantly reduced by calcium, concomitantly with an increased expression of the differentiation marker, involucrin (Fig. 5A and Fig. S3A). This is consistent with previous observations (5, 20–22). Interestingly, we found that the decreased expression of p63 was due to shortened half-life of p63 mRNA (∼3.7 h in control cells vs. ∼2.7 h in calcium-treated cells) (Fig. S3B). In addition, we found that the level of RNPC1 mRNA and protein was increased by calcium in HaCaT cells (Fig. 5A and S3A). Thus, we examined whether RNPC1 regulates p63 expression during keratinocyte differentiation and showed that the level of p63 transcript was reduced by RNPC1a, ranging from 25% in the absence of calcium to 40% in the presence of calcium (Fig. 5B). Similarly, the level of p63 protein was reduced, whereas the level of involucrin and filaggrin was increased, by RNPC1a and RNPC1b in the presence and absence of calcium (Fig. 5 C and D, compare lanes 1 and 3 with 2 and 4, respectively). It should be noted that even though involucrin and filaggrin expression was increased by calcium, their expression was further increased by overexpression of RNPC1a and RNPC1b (Fig. 5 C and D, compare lanes 1 and 2 with 3 and 4, respectively). Additionally, the level of p21 protein, which is required for the cell cycle arrest during differentiation (23), was measured and found to be increased by RNPC1a but not RNPC1b (Fig. 5 C and D, p21 panel, compare lanes 1 and 3 with 2 and 4, respectively), consistent with a previous report (18). To further verify this, HaCaT cells were transiently transfected with scrambled siRNA, siRNAs against total RNPC1, or RNPC1a, followed with or without calcium treatment for 3 d. We showed that the level of RNPC1a was diminished by siRNA against total RNPC1 or RNPC1a (Fig. 5E, RNPC1 panel, compare lanes 1 and 5 with lanes 2–4 and 6–8, respectively). We also showed that upon knockdown of RNPC1, the level of p63 protein was increased in the presence and absence of calcium treatment (Fig. 5E, ΔNp63α panel, compare lanes 1 and 5 with lanes 2–4 and 6–8, respectively). Considering that p63 expression was markedly decreased by calcium (Fig. 5E, compare lane 1 with 5; also see Fig. 5A and Fig. S3 A and B), these results suggest that RNPC1 mediates calcium-induced suppression of p63 expression in HaCaT cells. Furthermore, elevated ΔNp63 induced by knockdown of RNPC1 resulted in decreased expression of involucrin (Fig. 5E, Ivl panel, compare lanes 1 and 5 with lanes 2–4 and 6–8, respectively), consistent with the above observation. Finally, to further demonstrate the role of RNPC1 in terminal keratinocyte differentiation, the formation of cornified envelopes, which are made up of proteins covalently linked together by transglutaminases and function as skin barriers (24), was quantitated. We showed that both RNPC1a and RNPC1b enhanced the formation of cornified envelopes (ranging from 2 to 3 folds) in HaCaT cells cultured for 9 d (Fig. 5F), concomitantly with an increased expression of involucrin and filaggrin (Fig. S3C). Conversely, knockdown of RNPC1 led to about 50% decrease in the number of cornified envelopes in HaCaT cells cultured for 11 d (Fig. 5G), along with an decreased expression of involucrin and filaggrin (Fig. S3D). Taken together, these data suggest that RNPC1 represses p63 expression, leading to enhanced keratinocyte differentiation.
Fig. 5.
RNPC1 promotes keratinocyte differentiation by repressing p63 expression. (A) HaCaT cells grown at confluence were treated with or without 1.5 mM calcium for 0–9 d, and the level of ΔNp63α, RNPC1a, Ivl, and actin was measured by Western blot analysis. (B) The level of p63 transcript was measured in HaCaT cells uninduced or induced to express RNPC1a for 24 h along with or without treatment of 1.5 mM CaCl2 for 3 d. (C and D) The level of Involucrin and filaggrin along with ΔNp63, p21, and actin was measured in HaCaT cells treated as in (B). (E) The experiment was performed as in (C and D) with HaCaT cells transfected with scrambled siRNA or siRNAs against total RNPC1 or RNPC1a for 3 d, followed with or without treatment of CaCl2 for 3 d. (F) Confluent HaCaT cells were uninduced or induced to express RNPC1a or RNPC1b for 24 h, followed by treatment of 1.5 mM calcium for 9 d. Cornified cell envelopes were counted and expressed as percentage of total cells (mean ± S.D.; n = 3). (G) HaCaT cells were transfected with scramble siRNA or siRNA against RNPC1 for 3 d, followed by treatment of 1.5 mM calcium for 11 d. Cornified cell envelopes were counted and expressed as percentage of total cells (mean ± S.D.; n = 3). (H) A model of the RNPC1-p63 feedback loop and the role of RNPC1 in keratinocyte differentiation.
Discussion
Here we showed that overexpression of RNPC1a or RNPC1b inhibits, whereas knockdown of RNPC1 increases, the level of p63 transcript and protein. We also showed that knockdown of RNPC1 leads to prolonged half-life of p63 transcript. Furthermore, we showed that RNPC1 can bind the AU-/U-rich elements in p63 3′ UTR and the RRM domain in RNPC1 is required for binding p63 transcript and for repressing p63 expression. Thus, we uncovered a previously undetected mechanism by which p63 is regulated via mRNA stability. Since RNPC1 is a target of the p53 family, including p63, and can be induced by DNA damage (18), we uncover a feedback regulatory loop between RNPC1 and p63 (Fig. 5H).
Our data showed that RNPC1 binds to multiple regions in p63 3′ UTR (Fig. 3). Upon close examination, we found that RNPC1 prefers to bind the AU-/U-rich elements in p63 3′ UTR (Fig. 3C). However, several questions still remain. For example, a precise RNPC1-binding site in p63 3′UTR needs to be mapped. In addition, the underlying mechanism by which RNPC1 destabilizes p63 transcript is not elucidated and it will be interesting to investigate how RNPC1 cooperates with exosome complexes to facilitate p63 mRNA degradation. Furthermore, considering that one unstable transcript is often regulated by multiple RNA-binding proteins and AU-rich elements are recognized by the embryonic lethal abnormal visual (ELAV) family of RNA-binding proteins, such as HuR (19), it is likely that p63 mRNA stability is cooperatively regulated by RNPC1 along with other RNA-binding proteins. Therefore, it will be interesting to identify other RNA-binding proteins, which interact with RNPC1 and/or directly bind p63 transcript. Finally, since RNPC1 is a target of the p53 family and all p53 family proteins, including p53, p63, and p73, contain AU-rich element in their 3′UTR, further studies are needed to address whether RNPC1 can regulate other p53 family members.
We showed that both RNPC1a and RNPC1b decrease p63 expression, followed by enhanced keratinocyte differentiation in HaCaT cells as evidenced by increased expression of keratinocyte differentiation markers, involucrin and fillagrin, as well as increased formation of cornified envelopes. Importantly, we showed that during the calcium-induced keratinocyte differentiation, RNPC1 is induced, which is required for the repression of p63 expression and consequently, the induction of keratinocyte differentiation (Fig. 5A–G and Fig. S3). Thus, we hypothesize that RNPC1 mediates calcium-induced keratinocyte differentiation at least in part via suppression of ΔNp63 expression (Fig. 5H). We also showed that RNPC1a, but not RNPC1b, can increase p21 expression regardless of calcium treatment, consistent with the previous report that RNPC1a but not RNPC1b can stabilize p21 transcript and induce cell cycle arrest (18). Therefore, our data provided further evidence that the increased keratinocyte differentiation by RNPC1 is due to repression of ΔNp63 expression rather than an indirect consequence of growth arrest via p21. Nevertheless, it is possible that RNPC1 may regulate other factors involved in keratinocyte differentiation since RNA-binding proteins can regulate multiple targets. Furthermore, it is also possible that in nondifferentiating keratinocytes, such as HaCaT cells, ΔNp63 is highly expressed, which may act as a repressor, instead of an activator, of RNPC1 transcription. Thus, upon treatment with calcium or other agents to induce keratinocyte differentiation, RNPC1 is induced, which then represses ΔNp63 expression.
In summary, we uncover a mechanism by which p63 expression is regulated via mRNA turnover. We also uncover a feedback loop between p63 and RNPC1. Thus, our results provide an insight into how to further address p63 function in tumor suppression (25) and maintenance of female germ cell stability (26).
Materials and Methods
Reagents.
Antibodies against p63, Involucrin, Filaggrin, and GAPDH were purchased from Santa Cruz Biotechnology. Anti-HA was purchased from Covance (San Diego, CA). Anti-actin, proteinase inhibitor cocktail, and RNase A were purchased from Sigma. Scrambled siRNA (GGC CGA UUG UCA AAU AAU U), siRNA against RNPC1a (UCC CCU CCT TGU TCC CUG CGG UCT), siRNA#1 against RNPC1 (GTU CTT CGU GGG CTT CGG C), and siRNA#2 against RNPC1 (GCT GUG UGG GCT TGC UUU GUC) were purchased from Dharmacon (Chicago, IL). The iScript cDNA synthesis kit was purchased from Bio-Rad.
Plasmids.
HA-tagged RNPC1a and RNPC1b in pCDNA3 and shRNA against total RNPC1 in pBabe-H1 were generated as described previously (18). To generate constructs expressing HA-tagged ΔRNP1 or ΔRNP2, two-step PCR reactions were performed. The first step was performed to separately amplify cDNA fragments. Fragment #1 was amplified with forward primer, 5′-GAA GCT T GC CGC CAT GGA GTA CCC ATA CGA CGT ACC AGA TTA CGC TAT GCT GCT GCA GCC CGC GCC G-3′, and reverse primer, 5′-GGC GTC GGT AGT GTG GTA CTT GGT GAA CGT GGT GTC C -3′ for RNPC1a(ΔRNP1), or 5′-AGC TGC CGC CCG GTC GGC GGA CTT GCC CGT CTG GCG GT-3′ for RNPC1a(ΔRNP2). Fragment #2 was amplified with forward primer, 5′-GGA CAC CAC GTT CAC CAA GTA CCA CAC TAC CGA CGC C-3′ for RNPC1a(ΔRNP1), or 5′-ACC GCC AGA CGG GCA AGT CCG CCG ACC GGG CGG CAG CT-3′ for RNPC1a(ΔRNP2), and reverse primer, 5′-GGA ATT CTC ACT GCA TCC TGT CAG GCT GC-3′. The second-step PCR reaction was performed using a mixture of fragments #1 and #2 as a template with the forward primer for fragment #1 and the reverse primer for fragment #2, and resulting fragments were separately cloned and confirmed by sequencing. A HindIII–EcoRI fragment containing the coding region for ΔRNP1 and ΔRNP2 was cloned into pcDNA3.
To generate REMSA probes, various regions in p63 3′ UTR were amplified by PCR and cloned into pGEM vectors. The primers for probe A were 5′-GGA TCC TAA TAC GAC TCA CTA TAG GGA GGC CTC ACC ATG TGA GCT CTT CC-3′ and 5′-TTT AAG GGG GTT ACT GAT AT-3′. The primers for probe B were 5′-GGA TCC TAA TAC GAC TCA CTA TAG GGA GTT TAA TAC CAG ATA CCT TAT-3′ and 5′-ACT AAA TGG TAT TTT CAT GA-3′. The primers for probe C were 5′-GGA TCC TAA TAC GAC TCA CTA TAG GGA GAA GAA TAC CAC ATC AAA TAA-3′ and 5′-GCA TGT CCT GGC AAA CAA AA-3′. The primers for probe C1 were 5′-GGA TCC TAA TAC GAC TCA CTA TAG GGA GTG TTC CTT GGT CCT AGT AAG-3′ and 5′-GCT TTC ATT CTT CCC CTT AA-3′. The primers for probe C2 were 5′-GGA TCC TAA TAC GAC TCA CTA TAG GGA GTG AGT AGC CAG GGT AAG GGG-3′ and 5′-TAC ACT CAA GGA GAG TAG GC-3′. The primers for probe C3 were 5′-GGA TCC TAA TAC GAC TCA CTA TAG GGA GTA TGT GGG ATA TTG AAT GTT-3′ and 5′-TAC ACT CAA GGA GAG TAG GC-3′. The primers for probe C4 were 5′-GGA TCC TAA TAC GAC TCA CTA TAG GGA GTA TGT GGG ATA TTG AAT GTT-3′ and 5′-CTG TTA TTT TAG GGG ATT AC-3′.
Cell Culture.
HaCaT, ME-180, and MCF7 cells were grown in DMEM plus 10% FBS. HaCaT and ME-180 cell lines, which can inducibly express HA-tagged RNPC1a or RNPC1b, were generated as previously described (18). To generate inducible RNPC1-KD cell line, pBabe-H1-siRNPC1 was transfected into MCF7 cells in which a tetracycline repressor is expressed by pcDNA6 (27). RNPC1-KD cell lines were selected with puromycin and confirmed by Western blot analysis. When assayed for differentiation, confluent HaCaT cells were uninduced or induced to express protein of interest for 24 h and then switched to DMEM containing 0.1% FBS plus 1.5 mM CaCl2 at indicated time.
RNA Isolation, RT-PCR, and Quantitative PCR (qPCR).
Total RNA was isolated with Trizol reagent as described (3). cDNA was synthesized with iScript kit and used for RT-PCR. qPCR was performed in 20-μl reactions using 2X QPCR SYBR Green Mix (ABgene, Epsom, UK) with 5 μM primers. Reactions were run on a realplex (Eppendorf, Germany) using a two-step cycling program: 95 °C for 15 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s, and 68 °C for 30 s. A melt curve (57–95 °C) was generated at the end of each run to verify the specificity. The primers used to amplify actin were 5'-CTG AAG TAC CCC ATC GAG CAC GGC A-3' and 5'-GGA TAG CAC AGC CTG GAT AGC AAC G-3'. The primers for total p63 were 5′-TCC TGG TCC ACC AGT CC-3′ and 5′-GCA ATT TGG CAG TAG AGT TT-3′. The primers for TAp63 were 5′-AGC CCA TTG ACT TGA ACT T-3′ and 5′-GGA CTG GTG GAC GAG GA-3′. The primers for RNPC1a were 5′-CAA CGT GAA CCT GGC ATA TC-3′and 5′- TAA GTC CGC TGG ATC AAG GT -3′. The primers for GAPDH were 5′- CCC AGC CTC AAG ATC ATC AGC AAT G -3′ and 5′- ATG GAC TGT GGT CAT GAG TCC TT -3′.
Western Blot Analysis.
The assay was performed as previously described (3).
RNA Immunoprecipitation Followed by RT-PCR (RNA-CHIP).
RNA-CHIP was performed as described (28). Briefly, 2 × 107 cells were uninduced or induced to express RNPC1a or RNPC1b. Cell extracts were prepared with immunoprecipitation buffer (100 mM KCl, 5 mM MgCl2, 10 mM Hepes, 1 mM DTT, and 0.5% NP-40 ) and then incubated with 2 μg of anti-HA or mouse IgG at 4 °C overnight. The RNA-protein immunocomplexes were precipitated by protein A/G beads and subjected to RT-PCR.
Recombinant Protein Purification, Probe Labeling, and RNA Electrophoretic Mobility Shift Assay (REMSA).
Recombinant HA-tagged RNPC1-GST and GST proteins were expressed in bacteria BL21 and purified by glutathione sepharose beads. RNA probes were generated and 32P-labeled by in vitro transcription using linearized pGEM vectors containing various regions from p63 3′ UTR as a template. For REMSA, 32P-labeled probes were incubated with GST-tagged RNPC1a in a binding buffer [10 mM HEPES-KOH (pH 7.5), 90 mM potassium acetate, 1.5 mM magnesium acetate, 2.5 mM DTT, and 40U RNase inhibitor (Ambion)] at 30 °C for 30 min. To supershift RNA-protein complexes, 1 μg of anti-HA was added to the reaction mixture and incubated for an additional 30 min. RNA-protein complexes were resolved on a 5% acrylamide gel and radioactive signals were detected by autoradiography.
Cornified Cell Envelope Assay.
Cornified cell envelopes were counted in HaCaT cells that cornified spontaneously during in vitro differentiation as previously described (29). Briefly, cells were trypsinized and resuspended in 1 mL of PBS plus 2 mM EDTA. An aliquot (10 μL) was removed to count total cells. The remainder of the cells were centrifuged, resuspended in 1 mL of cell envelope dissociation buffer [2% SDS, 20 mM DTT, 5 mM EDTA, 0.1 M Tris-HCl (pH 8.5)], and boiled for 5 min. Detergent-insoluble cell envelopes were cooled, centrifuged, and resuspended in 50 μL PBS. Cell envelopes were counted in a hemacytometer via phase-contrast microscopy and the data were expressed as total cornified envelopes/total cells × 100.
Supplementary Material
Acknowledgments.
This work is supported in part by a National Institutes of Health grant (CA102188).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0912594107/-/DCSupplemental.
References
- 1.Yang A, McKeon F. P63 and P73: P53 mimics, menaces and more. Nat Rev Mol Cell Biol. 2000;1:199–207. doi: 10.1038/35043127. [DOI] [PubMed] [Google Scholar]
- 2.Yang A, Kaghad M, Caput D, McKeon F. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet. 2002;18:90–95. doi: 10.1016/s0168-9525(02)02595-7. [DOI] [PubMed] [Google Scholar]
- 3.Dohn M, Zhang S, Chen X. p63alpha and DeltaNp63alpha can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene. 2001;20:3193–3205. doi: 10.1038/sj.onc.1204427. [DOI] [PubMed] [Google Scholar]
- 4.Harms K, Nozell S, Chen X. The common and distinct target genes of the p53 family transcription factors. Cell Mol Life Sci. 2004;61:822–842. doi: 10.1007/s00018-003-3304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang A, et al. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 6.Mills AA, et al. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 7.Yang A, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 8.Celli J, et al. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell. 1999;99:143–153. doi: 10.1016/s0092-8674(00)81646-3. [DOI] [PubMed] [Google Scholar]
- 9.Amiel J, et al. TP63 gene mutation in ADULT syndrome. Eur J Hum Genet. 2001;9:642–645. doi: 10.1038/sj.ejhg.5200676. [DOI] [PubMed] [Google Scholar]
- 10.McGrath JA, et al. Hay-Wells syndrome is caused by heterozygous missense mutations in the SAM domain of p63. Hum Mol Genet. 2001;10:221–229. doi: 10.1093/hmg/10.3.221. [DOI] [PubMed] [Google Scholar]
- 11.Ianakiev P, et al. Split-hand/split-foot malformation is caused by mutations in the p63 gene on 3q27. Am J Hum Genet. 2000;67:59–66. doi: 10.1086/302972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossi M, et al. The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc Natl Acad Sci USA. 2006;103:12753–12758. doi: 10.1073/pnas.0603449103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Zhou Z, Chen C. WW domain-containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin-mediated proteasomal degradation and regulates apoptosis. Cell Death Differ. 2008;15:1941–1951. doi: 10.1038/cdd.2008.134. [DOI] [PubMed] [Google Scholar]
- 14.Gallegos JR, et al. SCF TrCP1 activates and ubiquitylates TAp63gamma. J Biol Chem. 2007;283:66–75. doi: 10.1074/jbc.M704686200. [DOI] [PubMed] [Google Scholar]
- 15.Bakkers J, Hild M, Kramer C, Furutani-Seiki M, Hammerschmidt M. Zebrafish DeltaNp63 is a direct target of Bmp signaling and encodes a transcriptional repressor blocking neural specification in the ventral ectoderm. Dev Cell. 2002;2:617–627. doi: 10.1016/s1534-5807(02)00163-6. [DOI] [PubMed] [Google Scholar]
- 16.Bamberger C, Pollet D, Schmale H. Retinoic acid inhibits downregulation of DeltaNp63alpha expression during terminal differentiation of human primary keratinocytes. J Invest Dermatol. 2002;118:133–138. doi: 10.1046/j.0022-202x.2001.01649.x. [DOI] [PubMed] [Google Scholar]
- 17.Liefer KM, et al. Down-regulation of p63 is required for epidermal UV-B-induced apoptosis. Cancer Res. 2000;60:4016–4020. [PubMed] [Google Scholar]
- 18.Shu L, Yan W, Chen X. RNPC1, an RNA-binding protein and a target of the p53 family, is required for maintaining the stability of the basal and stress-induced p21 transcript. Genes Dev. 2006;20:2961–2972. doi: 10.1101/gad.1463306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Chen X. Posttranscriptional regulation of p53 and its targets by RNA-binding proteins. Curr Mol Med. 2008;8:845–849. doi: 10.2174/156652408786733748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King KE, et al. deltaNp63alpha functions as both a positive and a negative transcriptional regulator and blocks in vitro differentiation of murine keratinocytes. Oncogene. 2003;22:3635–3644. doi: 10.1038/sj.onc.1206536. [DOI] [PubMed] [Google Scholar]
- 21.Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004;18:126–131. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nylander K, et al. Differential expression of p63 isoforms in normal tissues and neoplastic cells. J Pathol. 2002;198:417–427. doi: 10.1002/path.1231. [DOI] [PubMed] [Google Scholar]
- 23.Dotto GP. Signal transduction pathways controlling the switch between keratinocyte growth and differentiation. Crit Rev Oral Biol Med. 1999;10:442–457. doi: 10.1177/10454411990100040201. [DOI] [PubMed] [Google Scholar]
- 24.Candi E, Schmidt R, Melino G. The cornified envelope: A model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6:328–340. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Prives C. Are interactions with p63 and p73 involved in mutant p53 gain of oncogenic function? Oncogene. 2007;26:2220–2225. doi: 10.1038/sj.onc.1210311. [DOI] [PubMed] [Google Scholar]
- 26.Suh EK, et al. p63 protects the female germ line during meiotic arrest. Nature. 2006;444:624–628. doi: 10.1038/nature05337. [DOI] [PubMed] [Google Scholar]
- 27.Yan W, Chen X. GPX2, a direct target of p63, inhibits oxidative stress-induced apoptosis in a p53-dependent manner. J Biol Chem. 2006;281:7856–7862. doi: 10.1074/jbc.M512655200. [DOI] [PubMed] [Google Scholar]
- 28.Peritz T, et al. Immunoprecipitation of mRNA-protein complexes. Nat Protoc. 2006;1:577–580. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
- 29.Ladd PA, Du L, Capdevila JH, Mernaugh R, Keeney DS. Epoxyeicosatrienoic acids activate transglutaminases in situ and induce cornification of epidermal keratinocytes. J Biol Chem. 2003;278:35184–35192. doi: 10.1074/jbc.M301666200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.