Abstract
The 8q24 gene desert contains risk loci for multiple epithelial cancers, including colon, breast, and prostate. Recent evidence suggests these risk loci contain enhancers. In this study, data are presented showing that each risk locus bears epigenetic marks consistent with enhancer elements and forms a long-range chromatin loop with the MYC proto-oncogene located several hundred kilobases telomeric and that these interactions are tissue-specific. We therefore propose that the 8q24 risk loci operate through a common mechanism—as tissue-specific enhancers of MYC.
Keywords: chromosome conformation capture, epigenetic, genetics
Genome-wide association studies (GWAS) have been instrumental in discovering regions of the genome that confer risk to disease. In contrast to highly penetrant disorders, most of the alleles associated with common diseases are located outside of known protein-coding regions (1). Thus, finding the actual targets of these risk regions and determining how they drive the development of complex traits presents a major challenge.
Several independent GWAS have implicated chromosome 8q24 as a hotspot for multiple epithelial cancers (2–7). 8q24 contains at least three independent risk regions for prostate cancer (region 2: 128.14–128.28, region 3: 128.47–128.54, and region 1: 128.54–128.62); one region for colon cancer, which is the same as prostate cancer region 3; and a separate region for breast cancer (128.35–128.51) (4). Each of these loci, however, is in a gene-poor region of chromosome 8q24. The closest annotated protein-coding genes flanking the 8q24 risk loci are FAM84B (centromeric) and MYC (telomeric) (Fig. 1A).
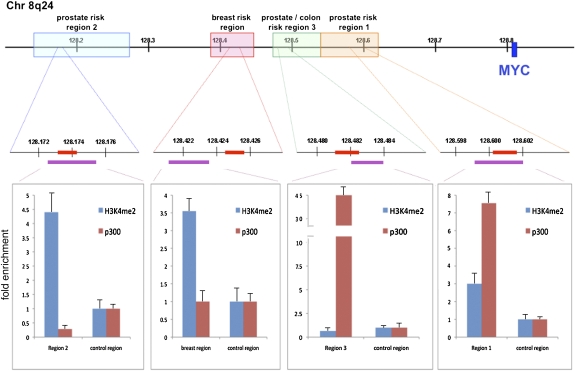
Fig. 1.
Epigenetic annotation of cancer risk loci at chromosome 8q24 is consistent with enhancer activity. (Top) The 800-kb region of 8q24 containing prostate, breast, and colon cancer risk loci and the MYC proto-oncogene. (Middle) Approximately 6-kb segments within the risk loci, which bear the epigenetic marks of histone acetylation (8). The red bars represent the restriction fragments analyzed by 3C for their interactions with MYC. The purple bars encompass the segment interrogated with tiled primers for ChIP-qPCR. (Bottom) Fold enrichment by ChIP-qPCR for histone 3 lysine 4 dimethylation (H3K4me2) and for p300, ±1 SD.
Multiple lines of evidence support the notion that these regions contain regulatory elements, specifically enhancers. The prostate and colorectal regions bear the epigenetic marks of enhancers (8, 9). High-throughput sequencing indicates that there are no microRNAs expressed in normal prostate tissue within the 8q24 prostate risk regions (10). Similarly, high density expression tiling arrays demonstrate that the breast cancer risk region in an MCF-7 breast cancer cell line and the prostate cancer region 1 in LNCaP prostate cancer cell line are devoid of transcriptional activity (8). Prostate cancer region 2 contains a transcript that appears to be expressed in prostate tissue (8); however, no association between the 8q24 prostate cancer risk SNPs and gene expression of the transcript at position chr8:128,168,145–128,168,232 was found when tested in 102 normal prostate tissue by our lab (Fig. S1). Region 3 is devoid of transcription in normal colon cancer tissue (9) and displays minimal transcriptional activity in prostate tissue (8). Therefore, it follows that genetic variation at 8q24 likely confers risk of disease by influencing a distant target gene.
The strongest candidate gene in the region is the MYC proto-oncogene. MYC steady-state RNA and protein expression levels, however, are not associated with risk allele status (6, 9, 10). Despite the negative expression data, we recently demonstrated that the colon cancer risk variant is an enhancer and forms a long-range chromatin loop with MYC more than 300 kb away (9). In the present work, we extend these observations by showing that the colon cancer, breast cancer, and each of the three prostate cancer risk loci at 8q24 bear epigenetic marks of transcriptional regulators most akin to enhancers, and physically interact with MYC in a tissue-specific manner.
Results
Recent studies demonstrate that certain 8q24 risk polymorphisms reside within gene regulatory elements, exerting their effects by influencing distal genes (9, 11–13). Elements such as enhancers can be characterized by their chromatin marks (14–16). Previous work showed that the prostate and colon risk regions disproportionately display histone 3 lysine 4 monomethylation (H3K4me1) compared with histone 3 lysine 4 trimethylation (H3K4me3), a signature shown to be consistent with enhancers (8, 9, 14). To complement and extend these findings, we used two other marks that demonstrate enrichment at enhancers, H3K4me2 and p300 (14, 17, 18). Areas nearby 8q24 risk loci with known histone acetylation marks were evaluated in LNCaP and MCF-7 cells for H3K4me2 and p300 antibodies by ChIP-qPCR (8). Enrichment of either H3K4me2 or p300 was observed at all three prostate cancer risk loci in the LNCaP cell line and at the breast cancer risk locus in the MCF-7 cell line (Fig. 1).
Because these elements displayed the epigenetic marks of enhancers, we tested whether they physically interacted with MYC using the chromosome conformation capture (3C) assay. 3C is a powerful method that allows for the determination of chromatin interactions (19, 20). Through fixation of chromatin, digestion of genomic DNA with a restriction enzyme, and ligation of fragment ends that are in close proximity with each other, a library is created where segments of the genome that are in close proximity in 3D space become ligated to each other. These ligation fragments can then be quantified. Specifically, 3C assesses whether a region of interest, such as an enhancer (herein referred to as the constant fragment) interacts with a series of prespecified target genomic regions (herein referred to as the target fragments). We investigated whether the prostate, breast, and colon cancer risk loci regulatory regions at 8q24 interacted with MYC and whether they did so in a tissue-specific manner.
3C libraries of native chromatin interactions were prepared in prostate (LNCaP), breast (MCF-7), and colon (LS174T) cancer cell lines, as well as in a nontumorigenic breast epithelial line (MCF10A) and a control lung fibroblast cell line (LL24). In each cancer cell line, the interaction between the constant fragment anchored at each cell-specific risk locus was tested for putative DNA interactions with a series of 36 target fragments distributed both centromeric and telomeric to the risk loci. These targets covered a 1.3-Mb region including the two closest flanking genes—MYC and FAM84B (Fig. 1 and 2A). Additionally, in each cancer cell line, a nonrisk locus for that particular cell type also served as a constant fragment and was tested against the same series of 36 target fragments. Thus, tissue specificity between risk loci and MYC interactions could be evaluated. The ligation products of each of these putative constant fragment-target fragment ligation products were quantified using a highly quantitative competitive PCR strategy (Methods) (9).
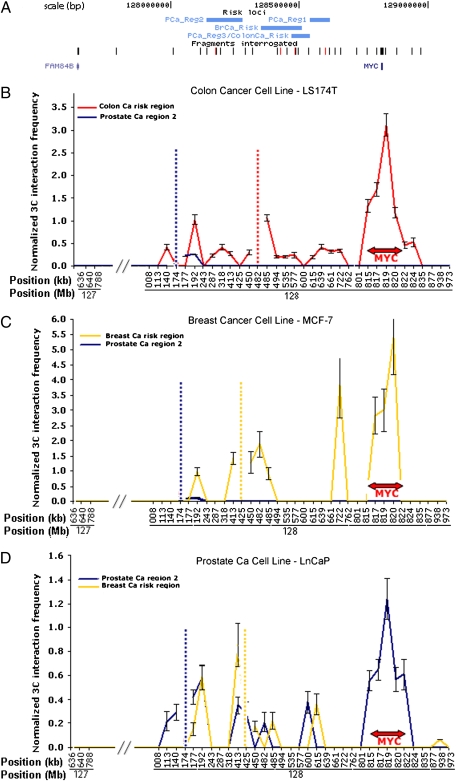
Fig. 2.
8q24 region and 3C interaction frequency of risk loci with MYC: x axis: genomic position (not drawn to scale); y axis: 3C interaction frequency ±1 SEM of the constant fragment with each of the target fragments including MYC, normalized to a 3C interaction within a housekeeping gene, FAM32A. (A) Schematic depicting the 8q24 risk loci in relation to the closest genes, as well as the locations of the constant fragments (red ticks) and target fragments (black ticks) interrogated; (B) normalized 3C interaction frequency of colon cancer risk locus (red line plot) and prostate cancer region 2 (blue line plot) in a colon cancer cell line—LS174T; (C) normalized 3C interaction frequency of breast cancer risk locus (yellow line plot) and prostate cancer region 2 (blue line plot) in a breast cancer cell line—MCF-7; (D) normalized 3C interaction frequency of prostate cancer region 2 (blue line plot) and breast cancer risk region (yellow line plot) in a prostate cancer cell line—LNCaP. Vertical hatched lines denote genomic positions of respective constant fragments (color-coded).
The colon, breast, and each of the three prostate cancer risk regions demonstrated robust interaction with MYC in their respective cancer cell lines (Fig. 2 B–D and Fig. S2). Although the colon cancer risk locus interacted with MYC in a colon cancer cell line and the breast cancer risk locus interacted with MYC in a breast cancer cell line, the region 2 prostate cancer risk locus did not interact with MYC in either the colon or breast cancer cell lines (Fig. 2 B and. C). Similarly, the breast cancer risk locus did not interact with MYC in the LNCaP prostate cancer cell line (Fig. 2D). Thus, the colon, prostate, and breast cancer risk loci form tissue-specific long-range chromatin loops with the MYC proto-oncogene.
Other 3C interactions were also observed in the cancer cell lines. Each of the three prostate cancer risk loci interacted with each other in the LNCaP cancer cell line (Fig. 2D and Fig. S2). Additionally, every risk locus that interacted with MYC also interacted with a fragment located at 128.192 Mb, in each cell line. In contrast to the interactions between respective cancer risk loci and MYC, the interaction between the cancer risk loci and the fragment at 128.192 Mb was not tissue specific.
None of the risk loci interacted with MYC in a lung fibroblast cell line, a nonepithelial cell line (Fig. S3). To evaluate the interaction in the context of an immortalized, nontumorigenic cell line, we tested MCF10A. In this cell line, the breast cancer risk region demonstrated minimal interaction with MYC (Fig. S4).
Discussion
In one of the largest 3C studies to date, we evaluated over 300 potential 3C interactions of multiple risk loci at 8q24 and determined that each of the 8q24 prostate, breast, and colon cancer risk loci interacts with MYC in a tissue-specific manner. Epigenetic annotation of the prostate and breast cancer risk loci, and previously of the colon cancer risk locus, suggests that these risk loci are regulatory elements. The 8q24 risk loci are therefore likely operating (at least in part) through a common mechanism—as tissue specific regulators of MYC.
Others have shown regulatory elements such as enhancers to have cell-type specific effects. Specifically, enhancers are marked with highly cell-type-specific histone modification patterns, strongly correlate to cell-type-specific gene expression on a global scale and are functionally active in a cell-type-specific manner (15, 18). Additionally, the association between genetic variation at enhancers and gene expression was recently found to be largely cell-specific (21).
In prostate cancer, the three regions that independently confer an elevated risk of disease not only interacted with MYC but also with each other. This observation suggests that the prostate cancer 8q24 loci may be part of a gene-regulatory unit, such as an active chromatin hub, where regions of DNA that are involved in regulation of MYC physically interact with each other in 3D nuclear space with looping out of inactive regions (22).
We also observed that a fragment at position 128.192kb interacts with each risk locus in each respective cell line. Based on the consistent presence of interaction with this region and based on known CTCF binding sites annotated in CD4 cells, the fragment at 128.192kb may be an important MYC regulatory element (16). To fully address this hypothesis, CTCF ChIP could be performed in the cell types examined in this study. Further work, however, will be necessary to understand the nature and significance of this locus and any relationship to looping interactions with MYC (23, 24).
Although we have demonstrated looping interaction of these risk regulatory elements with MYC, extensive study in normal and tumor human prostate and colon tissue have failed to show an association between risk allele status and MYC expression levels (9–11). Similarly, there is no clear correlation between 3C interaction frequency with MYC and MYC expression levels in the cell lines that we examined (Fig. S5). For example, the breast cancer risk locus demonstrated only very slight interaction with MYC in a normal breast epithelial line, although showing robust interaction with MYC in a breast cancer line. Despite these differences in interaction frequency, both of these cell lines had equivalent expressions of MYC. Given the complexity of MYC transcription, this discordance is not entirely surprising. It is possible that MYC is differentially expressed by allelic status, but the expression differences are subtle. In support of this point, a recent publication demonstrated that the rs6983267 risk allele expressed approximately twofold more MYC than the nonrisk allele (12). Other possibilities include that the differential expression is occurring in a subset of cells (e.g., stem cells, epithelial, or stromal), or that steady state measures of MYC do not reflect the difference (i.e., the difference would only be observed under nonsteady state conditions).
MYC regulation is extremely complex; even after decades of study, the physiologic transcriptional control of this gene is not fully understood, perhaps because of the lack of full annotation of regulatory elements across different tissue types (25). We have identified four regulatory regions that form looping interaction with MYC in a tissue-specific manner, providing a focus of study for MYC regulation. A more complete picture of the underlying molecular mechanisms of long-range enhancer-target interactions is just beginning to emerge. Study of model loci such as β-globin (26, 27) and T-helper 2 (28) have demonstrated that a variety of proteins are able to form, maintain, and regulate loop formation. Among these are transcription factors, insulators, chromatin remodeling proteins, and nuclear architecture proteins (29). Identifying the key proteins regulating loop formation at the 8q24 enhancer-MYC locus should help further our understanding of MYC transcriptional regulation.
Going from risk allele to function remains a challenge, particularly when the identified risk loci lie in nonprotein coding regions of the genome. Here, we have demonstrated a common mechanism through which four independent risk regions for three epithelial cancers on 8q24 likely contribute to risk—through their tissue-specific long-range chromatin interactions with MYC.
Methods
3C. 3C Library Preparation.
3C library was prepared in batches of 10 million cells, through formaldehyde fixation, digestion with Csp6I, and ligation, as previously described (9). Cell lines were obtained from ATCC and grown according to their recommended culture protocols.
Cells were fixed with 1% formaldehyde on a rocking platform for 10 min, and quenched with a final concentration of 0.125 mM glycine on ice for 5 min. Cells were counted and placed into aliquots 10 × 106 cells, snap frozen, and then stored in −80 °C in storage media (1× PBS with 0.125 mM glycine). Cells were lysed with lysis buffer (500 μL of 10 mM TrisHCl pH 8, 10 mM NaCl, 0.2% Nonidet P-40) including 4× protease inhibitor, and incubated for 15 min on ice while disrupting the pellet by pipetting up and down. Cell nuclei were pelleted and washed with 1× Csp6I restriction enzyme (RE) buffer B freshly made. Cell nuclei were pelleted and resuspended in 200 μL of 1× RE buffer B and distributed into four Eppendorf tubes. 337 μL of 1× RE buffer B were added to each tube. SDS was added to each aliquot, which were vortexed and incubated for 10 min at 65 °C with a final concentration of 0.1%. After incubation, Triton X-100 was added to each aliquot to a final concentration of 1.8% followed by 400 units of Csp6I restriction enzyme (Fermentas) and incubated for 24 hr at 37 °C. Each aliquot received 86% of 10%SDS and was incubated for 30 min at 65 °C.
Ligation mixes were prepared in 15-mL tubes (745 μL 10× T4 ligase Buffer, 10% Triton X 100, 80 μL 10 mg/mL BSA, 6 mL water, 575 μL of cell lysate, 4,000 units of T4 ligase) and incubated for 96 hr in a 16 °C water bath. This was followed by Proteinase K digestion at 65 °C, ethanol precipitation, and cleanup of 3C library.
3C fragment quantification.
A competitive PCR approach was used to quantify the products formed as a result of our constant fragment (or risk locus) of interest ligating to a series of predetermined target fragments of interest, including MYC. Because Csp6I cuts at predicted sites, we are able to construct a host of possible ligation products within this region of interest. We construct competitor oligonucleotide sequences that are identical to the predicted ligation product being measured, except for a single base pair change, which allows us to differentiate between 3C library versus competitor oligonucleotide. PCR with different concentrations of this competitor oligonucleotide allows us to construct a standard curve and determine the concentration at which competitor oligonucleotide equals that of native 3C library. This is the EC50. We normalize the quantity of our ligation products through measurement of the abundance of a ligation product within housekeeping gene that is in high abundance and diploid by cytogenetic analysis across all cell types—FAM32A. Further details are described in Pomerantz et al. (9).
RT-qPCR.
Normalized MYC expression levels were ascertained in LL24, LS174T, LnCap, MCF-7, and MCF10A cell lines. RNA was extracted from pelleted cells (Norgen) and reverse transcribed (Invitrogen; SuperScript III) per manufacturer's protocol. qPCR with SYBR green was performed in a 25-μL reaction on ABI 7300 cyclers. MYC expression was normalized to that of two housekeeping genes: β-actin and FAM32A, and the geometric mean of these taken as an average relative MYC expression. Fold change relative to LL24 was calculated, following the delta-delta Ct method (30).
ChIP qPCR.
ChIP was performed on MCF7 and LNCaP cells using one 15-cm plate at 80% confluence per ChIP. Cells were crosslinked with 1% formalin for 10 min and subsequently incubated with 2 M glycine for 5 min, on ice, to quench crosslinking. Cells, kept on ice, were lysed with 350 μL of lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl pH 8.1, 1× protease inhibitor) and sonicated four times for 15 seconds at 12% amplitude (Fisher Scientific; Sonic Dimembranator, Model 500), allowing suspension to cool on ice for 20 seconds between pulses. Sonication efficiency was evaluated by running 1% of sheared material on an agarose gel. Chromatin was fragmented in a range between 200 bp and 1.5 kb. After centrifugation, 5% of the supernatant was used as input, and the reminder was diluted 5-fold in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl pH 8.1) and immunoprecipitated over night at 4 °C on a rotating platform with antibody-coupled protein A Dynal beads (Invitrogen). Forty microliters of beads per sample was prepared by washing three times in 5 mg/mL BSA/PBS solution, incubated with specific antibody [Anti-dimethyl Histone (Lys-4); Milli Pore; catalog 07–030 and p300; Santa Cruz; catalog number sc-585], for 4 hr at 4 °C on rotating platform. Antibody-coupled beads were subsequently washed three times with 5 mg/mL BSA/PBS solution, and added to the diluted chromatin. Precipitates were washed with RIPA buffer (50 mM Hepes pH 7.6, 1 mM EDTA, 0.7% Na Deoxycholate, 1% Nonidet P-40, 0.5 LiCl) at 4 °C six times, and once with TE buffer (pH 7.6). Immunoprecipitated chromatin was removed from beads by incubating 30 min at 65 °C with 100 μL elution buffer (1% SDS, 0.1 M NaHCO3). Input and immunoprecipitated chromatin were decrosslinked in elution buffer for 5 hr at 65 °C. Samples were subsequently purified and eluted in 50 μL of RNase-DNase free H2O with Qiagen QIAquick PCR purification kit (Qiagen), per manufacturer's protocol. Enrichment was determined by qPCR. Specifically, the primer set used for MCF7 cell line was located in chr8:128,423,748–128,424,352. Primer sets for LNCaP cell line were designed over the following chromosomal locations: chr8:128,173,203–128,174,158, chr8:128,182,013–128,182,949, chr8:128,482,486–128,483,650, chr8:128,574,860–128,575,974, chr8:128,589,642–128,590,670. Quantitative PCR was performed in triplicate using SybrGreen (Applied Biosystems) on a ABI Prism 7300 and 7500 apparatus (Applied Biosystems). At least two biologic replicates were performed for each cell line, antibody, and primer set.
Supplementary Material
Acknowledgments
We thank Keluo Yao and Sarah Kehoe from Center for Cancer Genome Discovery, Dana-Farber Cancer Center, for running Sequenom Chips. M.L.F. is a Howard Hughes Medical Institute Physician-Scientist Early Career Awardee and is a recipient of a 2006 Doris Duke Clinical Scientist Development Award. This work was supported by grants from the National Institutes of Health (R01 CA129435 to M.L.F.), the Mayer Foundation (to M.L.F.), the H.L. Snyder Medical Foundation (to M.L.F.), the Dana-Farber/Harvard Cancer Center Prostate Cancer SPORE (National Cancer Institute Grant 5P50CA90381), the Emerald Foundation, Inc. (to M.L.F.), National Cancer Institute R01 CA 109147 and National Cancer Institute R01 CA 136924 (to G.A.C.), and by the Dana-Farber/Harvard SPORE in breast cancer from the National Cancer Institute (CA089393). M.P. is the recipient of the William L. Edwards Prostate Cancer Foundation Young Investigator Award.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0910668107/-/DCSupplemental.
References
- 1.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 2.Ghoussaini M, et al. UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology. UK ProtecT Study Collaborators Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kiemeney LA, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet. 2008;40:1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haiman CA, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Easton DF, et al. SEARCH collaborators. kConFab. AOCS Management Group Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zanke BW, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 7.Tomlinson I, et al. CORGI Consortium A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 8.Jia L, et al. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet. 2009;5:e1000597. doi: 10.1371/journal.pgen.1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pomerantz MM, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pomerantz MM, et al. Evaluation of the 8q24 prostate cancer risk locus and MYC expression. Cancer Res. 2009;69:5568–5574. doi: 10.1158/0008-5472.CAN-09-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuupanen S, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 12.Wright JB, Brown SJ, Cole MD. Upregulation of c-MYC in Cis through a large chromatin loop linked to a cancer risk-associated SNP in colorectal cancer cells. Mol Cell Biol. 2010;30:1411–1420. doi: 10.1128/MCB.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sotelo J, et al. Long-range enhancers on 8q24 regulate c-Myc. Proc Natl Acad Sci USA. 107:3001–3005. doi: 10.1073/pnas.0906067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 15.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Hon G, Wang W, Ren B. Discovery and annotation of functional chromatin signatures in the human genome. PLOS Comput Biol. 2009;5:e1000566. doi: 10.1371/journal.pcbi.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miele A, Gheldof N, Tabuchi TM, Dostie J, Dekker J. Curr Protoc Mol Biol. 2006. Mapping chromatin interactions by chromosome conformation capture. Chapter 21:Unit 21.11. [DOI] [PubMed] [Google Scholar]
- 20.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 21.Dimas AS, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Laat W, Grosveld F. Spatial organization of gene expression: The active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- 23.Gaszner M, Felsenfeld G. Insulators: Exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 24.Phillips JE, Corces VG. CTCF: Master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levens D. How the c-myc promoter works and why it sometimes does not. J Natl Cancer Inst Monogr. 2008;39:41–43. doi: 10.1093/jncimonographs/lgn004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vakoc CR, et al. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell . 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 27.Song SH, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol Cell. 2007;28:810–822. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 29.Sexton T, Bantignies F, Cavalli G. Genomic interactions: Chromatin loops and gene meeting points in transcriptional regulation. Semin Cell Dev Biol. 2009;20:849–855. doi: 10.1016/j.semcdb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Bookout AL, Cummins CL, Mangelsdorf DJ, Pesola JM, Kramer MF. High-throughput real-time quantitative reverse transcription PCR. Current Protocols in Molecular Biology. 2006 doi: 10.1002/0471142727.mb1508s73. Chap 15, unit 15.8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.