Abstract
Here, we report the development of target-specific binding proteins based on the kringle domain (KD) (∼80 residues), a ubiquitous modular structural unit occurring across eukaryotic species. By exploiting the highly conserved backbone folding by core residues, but using extensive sequence variations in the seven loop regions of naturally occurring human KDs, we generated a synthetic KD library on the yeast cell surface by randomizing 45 residues in the loops of a human KD template. We isolated KD variants that specifically bind to anticancer target proteins, such as human death receptor 4 (DR4) and/or DR5, and that function as agonists to induce apoptotic cell death in several cancer cell lines in vitro and inhibit tumor progression in mouse models. Combined treatments with KD variants possessing different recognition sites on the same target protein exerted synergisitic tumoricidal activities, compared to treatment with individual variants. In addition to the agonists, we isolated an antagonistic KD variant that binds human tumor necrosis factor-α (TNFα) and efficiently neutralizes TNFα-induced cytotoxicity in vitro and in vivo. The KD scaffold with seven flexible loops protruding from the central core was strongly sequence-tolerant to mutations in the loop regions, offering a potential advantage of distinct binding sites for target recognition on the single domain. Our results suggest that the KD scaffold can be used to develop target-specific binding proteins that function as agonists or antagonists toward given target molecules, indicative of their potential use as biotherapeutics.
Keywords: nonantibody scaffold, protein engineering, alternative protein scaffold
Development of proteins that interact specifically with a given target molecule with high affinity is of interest for many pharmaceutical and industrial applications. Antibodies (Abs) have been the primary choice for target-specific binding proteins, which can be isolated by immunization or from synthetic Ab libraries in vitro (1). In addition to Abs, non-Ab proteins of diverse sizes and structural features have recently been exploited as so-called alternative protein scaffolds (reviewed in refs. 2 and 3). Target-specific binding proteins with high affinity can be isolated from a synthetic library of alternative protein scaffolds, in which particular regions on a rigid scaffold core have been randomized (4–10). These scaffolds often have favorable characteristics, such as small size, high stability, ease of modification, and/or cost-efficient production, compared to Abs (2, 3).
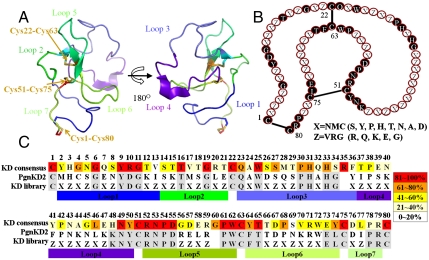
Here, we describe an alternative protein scaffold based on the kringle domain (KD), a ubiquitous domain occurring across eukaryotic species (11). In humans, 39 KDs composed of 78–80 amino acids are present as modular structural units of one to tens of copies in 31 functionally distinct proteins, most of which are present in blood plasma, such as coagulation factors, proteases, apolipoprotein, and growth factors (11–13). Although the exact physiological function of KDs is not yet clear, they are believed to play a role as a binding mediator for various molecules, such as proteins, lipids, and small molecules (12, 13). Single or multiple KDs of human plasminogen (Pgn), which possesses five KDs (KD1–KD5), have inhibitory effects on endothelial cell proliferation and angiogenesis via interactions with various molecules (12–14). KDs share a rigid core structure composed of two short antiparallel β-sheets and three disulfide bonds connected in a characteristic 1–6 (Cys1-Cys80), 2–4 (Cys22-Cys63), and 3–5 (Cys51-Cys75) pattern, and these residues are highly conserved (Fig. 1 and Fig. S1) (12, 13, 15, 16). The core folding of KD constrained by the disulfide bonds presents a structure with seven surface-exposed loops (15, 16), and the residues in these regions can diverge substantially between individual KDs (Fig. 1C and Fig. S1A). Despite low sequence homology in the loops, the overall structures of the KDs are closely superimposed (Fig. S1B). This result suggests that KDs may have evolved functional variability in binding specificities by diversifying the residues in the flexible loop structures, while maintaining the residues in the core that are essential for KD folding. This hypothesis attracted us to engineer target-specific binding proteins by generating a synthetic KD library with mutations in the seven flexible loop structures.
Fig. 1.
Structural characteristics of KD and library generation scheme based on the KD fold. (A) Three-dimensional α-carbon trace of PgnKD2 (PDB entry code 1I5K) (15). The seven loops and three disulfide bonds are indicated by different colors and arrows, respectively. The images were generated using the PyMol software (DeLano Scientific LLC). (B) Schematic of KD architecture with the three disulfide linkages of KD and the positions of conserved and mutated residues in the library. The mutated residues were encoded by one of two degenerate codons, X = NMC and Z = VRG, as described in the text. (C) Amino acid sequence alignment of human KD consensus, PgnKD2, and KD library. The KD consensus sequence and its amino acid frequency (%) at each position (indicated by five grades of color shading) were derived from sequence alignment of 39 human KDs. Amino acid positions at which the original amino acids of the PgnKD2 are conserved in the KD library are shown in gray. The residues were numbered according to the standard KD numbering convention (12, 16).
Earlier engineering studies of non-Ab protein scaffold have primarily isolated only target-specific binding proteins (4–8) or antagonists (9, 10). Here, in addition to an antagonist against human TNFα functioning in vitro and in vivo, we have isolated KD variants that behave as agonists. These KD variants specifically bind to the human TNF-related apoptosis inducing ligand (TRAIL) receptors, death receptor 4 (DR4) and DR5, and exhibit antitumor activity for several tumor-cell types in vitro and in vivo. Our results demonstrate that a non-Ab protein scafffold, KD, can be engineered to generate agonists or antagonists to modulate the biological activity of target molecules.
Results
Design and Construction of a Synthetic KD library.
For the backbone of the KD scaffold library, we used KD2 from human Pgn (PgnKD2), which consists of 78 amino acids, missing the 35th and 60th residues in the conventional KD numbering scheme (Fig. 1C) (13, 15, 16). By primary structural analyses of 39 human KDs, a yeast surface-displayed KD library was generated from the PgnKD2 template by synthetic shuffling using 10 partially overlapping oligonucleotides. The oligonucleotides introduce either of two degenerate codons, NMC (N = A/T/G/C, M = A/C) or VRG (V = A/C/G, R = A/G), at the 45 targeted residues in the surface-exposed loops, while preserving the highly conserved residues for the core folding (Fig. 1C). These two codons were employed to preferentially introduce hydrophilic amino acids at the targeted residues, mimicking residues in naturally occurring human KDs (Fig. S1A), but excluding stop and Cys codons. The KD library (diversity of ∼2 × 106) with expected mutations at the targeted regions was expressed well on the yeast cell surface, but sparsely sampled the theoretical sequence space (> 1036).
Isolation of Target-Specific KD Variants.
For target proteins to evaluate the synthetic KD library, we chose TRAIL receptors, DR4 and DR5, in order to isolate anticancer agonists that induce cell death in tumor cells, and TNFα, in order to isolate antagonists that neutralize TNFα-mediated cytotoxicity. DR4 and DR5 are attractive targets for the development of anticancer therapeutics since the receptor-mediated signaling can induce tumor-selective cell death through the extrinsic apoptotic pathway with little cytotoxicity to normal cells (17–19). TNFα, which is associated with autoimmune inflammatory diseases, such as rheumatoid arthritis and psoriasis, has been extensively targeted by mAbs and Fc-fused receptors (20, 21). Screening with two rounds of magnetic-activated cell sorting followed by three rounds of FACS, using biotinylated-target proteins against the yeast surface-displayed KD library, isolated 10, 10, and 4 unique KD variants against DR4, DR5, and TNFα, respectively (Fig. S2).
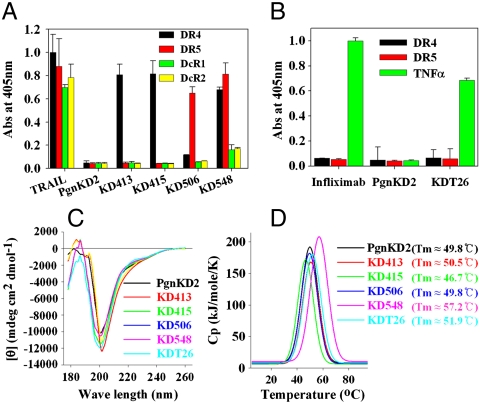
All of the isolated variants were expressed as soluble proteins in Pichia (15) and purified in monomeric form without intermolecular disulfide bonds (Fig. S3), giving purification yields of ∼0.4–8 mg per one-liter flask culture (Table S1). Disulfide bond mapping of representative KD variants by mass spectrometry demonstrated that the KD-specific disulfide bond pattern was retained (Fig. S4). Most of the isolated KD variants showed concentration-dependent specific binding to their cognate target proteins with dissociation constants (KD) ranging from 10-6 to 10-8 M (Fig. 2 A and B and Table S1). Some variants showed cross-reactivity with proteins homologous to the target protein. In particular, KD548 isolated against DR5 showed preferential binding to DR5, but cross-reacted significantly with the homologous DR4 and weakly with the death decoy receptor (DcR) 1 and DcR2 (Fig. 2A), similar to TRAIL (18). The control PgnKD2 did not bind to the target proteins.
Fig. 2.
Biochemical characterization of the selected KD variants. (A, B) Analyses of the binding specificity of the KD variants (1 μM) isolated against DR4 (KD413 and KD415) and DR5 (KD506 and KD548) (A), or TNFα (KDT26) (B), which were determined by ELISA. PgnKD2 (1 μM), TRAIL (0.1 μM), and the anti-TNFα mAb infliximab (0.1 μM) were used as controls. Binding specificities for the other KD variants are shown in Table S1. (C) Far-UV CD spectra of PgnKD2 and the KD variants (100 μg/mL in PBS, pH 7.4) to monitor the secondary structure are shown. (D) DSC thermograms of PgnKD2 and the selected KD variants. The change in heat capacity at constant pressure (ΔCp) was plotted against the temperature. The temperature of maximum heat capacity (Tm) for each sample was indicated in the panel.
Biological Efficacy of the Isolated KD Variants.
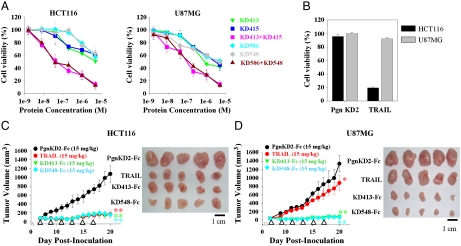
The isolated KD variants were primarily screened for biological activity. Of the 20 KD variants isolated against DR4 and DR5, only 4 variants, DR4-specific KD413 and KD415, DR5-specific KD506, and TRAIL-like KD548, efficiently induced dose-dependent cell death of multiple human cancer cell lines, including TRAIL-sensitive HCT116 (colorectal carcinoma), H460 (nonsmall cell lung carcinoma), Jurkat (acute T cell leukemia), and HL60 (acute promyelocytic leukemia) cells (22) (Fig. 3A and Fig. S5). More strikingly, these 4 KD variants induced cell death at significant levels in U87MG (glioma) and slightly in HepG2 (hepatoma) cells (22), which are resistant to TRAIL treatment (Fig. 3B and Fig. S5E). The cell death-inducing activity of the 4 KD variants differed depending on the cell type, displaying EC50s (effective concentration for 50% cell death after 60 h) ranging from ∼11μM to ∼90 nM (Table S1), without any strict correlation between the binding affinity and the cell death-inducing activity. Jurkat and HL60 cells treated with either KD413 or KD548 were significantly labeled as early apoptotic cells demonstrated by Annexin-V-FITC positive staining (Fig. S5F), similar to TRAIL treatments, suggesting that the KD variants most likely induced apoptotic cell death of the tumor cells (17). Soluble competitor experiments and specific binding activity analyses of the 4 KD variants for cell surface-expressed DR4 and/or DR5 demonstrated that the KD variants killed the tumor cells by specifically binding to the target protein on the cell surface (Fig. S6) (22). Despite binding to their respective target proteins, other KD variants failed to show antitumor activity (Table S1), suggesting that the specific binding site is important for biological activity.
Fig. 3.
The KD variants selected against DR4 and DR5 exhibited antitumor activities in vitro and in vivo. (A) In vitro cell killing activities of KD variant alone (KD413, KD415, KD506, and KD548) and combinations of KD variants (KD413 + KD415, KD506 + KD548) for HCT116 (Left) and U87MG (Right) cells. Cells were incubated with the indicated concentrations of the variants (2 nM–5 μM) for 60 h before cell viability assay. The protein concentration in the combined treatment is the total concentration of KD variants mixed in a 1∶1 molar ratio. (B) The cell death-inducing activities of PgnKD2 (5 μM, 60 h) and TRAIL (30 nM, 60 h) are shown. The tumoricidal activities of the KD variants, PgnKD2, and TRAIL for H460, HepG2, Jurkat, and HL60 cells are shown in Fig. S5. Error bars, ± SD. (C, D) Tumor growth inhibition by intravenous injection of KD413-Fc, KD548-Fc, PgnKD2-Fc, or TRAIL into preestablished HCT116 (C) or U87MG tumors (D) in athymic nude mice. Tumor growth, as measured by the tumor volume during the treatments (Left), and representative tumor sizes isolated at the end of treatment (after 20 days of tumor inoculation) (Right) are shown for comparison. After preestablished HCT116 and U87MG tumors with a volume of ∼50–70 mm3 (seven-day growth), mice (n = 7 per group) were intravenously injected every 2 days (total = 6 doses (white arrowheads)) with each KD-Fc variant (15 mg/kg/injection), TRAIL (15 mg/kg/injection), or PgnKD2-Fc (15 mg/kg/injection). Error bars, ± SD. Significance was determined using the Student’s t test vs. the control PgnKD2-Fc-treated group (∗ P ≈ 0.02, ∗∗ P < 0.000003).
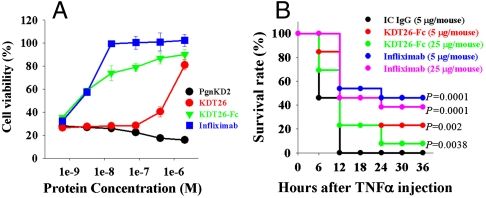
Of the 4 TNFα binding KD variants, biological activity assays using a human TNFα-responsive WEHI 164 cell bioassay (20, 23) revealed that only KDT26 neutralized TNFα-mediated cytotoxicity in a concentration-dependent manner, showing IC50 (50% inhibitory concentration) of ∼0.8 μM (Fig. 4A). The lower efficacy of KDT26 relative to infliximab (Remicade®) (IC50 ∼ 2 nM) may be due to the lower affinity for TNFα [KD ≈ 29 nM for KDT26 vs. KD ≈ 1.9 nM for infliximab (20)]. To increase the affinity by avidity effect, KDT26 was converted into a bivalent molecule by C-terminal fusion to the crystallizable fragment (Fc) of human IgG1. The Fc-fused KDT26 (KDT26-Fc), purified from Pichia in correctly assembled form, retained binding specificity with ∼10-fold increased affinity (KD ≈ 3.8 nM), relative to KDT26 (Table S1). The KDT26-Fc (IC50 ∼ 5 nM) was ∼160-fold more potent than KDT26 (Fig. 4A), suggesting that the bivalent molecule is much more efficient in neutralizing TNFα-mediated cytotoxicity (21).
Fig. 4.
The anti-TNFα KDT26 exhibited neutralizing activity against TNFα-mediated cytotoxicity in vitro and in vivo. (A) Inhibition of TNFα-induced cell death by KDT26 or KDT26-Fc was assessed in mouse WEHI 164 cells, which were cotreated with human TNFα (1 ng/mL) and the indicated concentrations of the KD variants for 20 h. PgnKD2 and infliximab were included as controls. Error bars, ± SD. (B) Protection from TNFα-induced lethality in GalN-sensitized mice (n = 13 per group) by intraperitoneally administered KDT26-Fc (5 or 25 μg/mouse), as compared to IC IgG1 (5 μg/mouse) or infliximab (5 or 25 μg/mouse). Survival was examined every 6 h. A log-rank (Mantel-Cox) test was used for statistical analysis of the survival rates of IC IgG1-treated versus KDT26-Fc/infliximab-treated groups. The P values are indicated at the right.
Biochemical Characterization of the Selected KD Variants.
For the five KD variants (KD413, KD415, KD506, KD548, and KDT26) exhibiting specific biological activity, we performed further biochemical characterization. Investigation of the secondary structures of the selected KD variants by far-UV CD spectroscopy demonstrated that all of the variants exhibited spectra very similar to that of the parent PgnKD2, a negative maximal peak between 202–205 nm with a weak negative shoulder around 220 nm (Fig. 2C), which is typical for the dominant irregular coil (> 50%) and minor β-strand structures of KDs (15, 16). Differential scanning calorimetric (DSC) analyses demonstrated that the selected KD variants possessed comparable thermal stability (Tm ≈ 47–57 °C) to that of PgnKD2 (Tm ≈ 50 °C) (Fig. 2D). When the KD variants were incubated at 37 and 50 °C for up to 14 days, they maintained their specific binding activity for their respective target proteins, retaining ≥84% of the initial binding activity (Fig. S7). These data suggest that the variants are quite stable to thermal stress. Taken together, these results demonstrate that the extensive mutations incorporated into the loop structures of the KD variants did not significantly affect the unique secondary structure or the stability of the native core conformation.
Which region of the target molecule is recognized by the binding protein is a critical factor influencing the biological efficacy, as seen with Ab epitopes (1). Binding region analysis by competition ELISA revealed that anti-DR4 KD413 and KD415, and anti-DR5 KD506 did not compete with TRAIL for the binding to their target proteins, whereas TRAIL-like KD548 partially did (Fig. S8). Further, KD413 and KD415, as well as KD506 and KD548, did not compete with each other for DR4 and DR5 binding, respectively, suggesting that each KD variant recognizes different sites on the target protein (Fig. S8). Combined treatment with KD variants (KD413 + KD415 or KD506 + KD548, mixed at 1∶1 molar ratio) exploiting the polyclonal reactivity on the same target protein exerted synergistic tumoricidal activities. The combined treatments enhanced cell death-inducing activity for HCT116, H460, and U87MG cells by ∼10- to 100-fold, compared with either KD variant alone (Fig. 3A and Fig. S5). This result suggests that concerted stimulation of DR4 and/or DR5 at different regions by the agonists is more effective in transducing cell death signaling through them.
KD structural analysis suggests that the seven loops of the KD scaffold might be grouped into two, dubbed loop cluster region 1 (LCR1) composed of loops 1, 3, and 4 and LCR2 of loops 2, 5, 6, and 7, which protrude from the central core in opposite directions against each other (Fig. 1A and Fig. S8G). Mutants, in which the residues in the LCR1 or LCR2 of KD413 and KD548 were replaced by the corresponding wild-type residues of PgnKD2, efficiently bound to the respective target protein (Fig. S8H), but showed significantly reduced cell death-inducing activity for HCT116 cells compared with the parent variants (Fig. S8K). KD413 and KD548, but not their mutants with substituted LCR1 or LCR2, simultaneously bound to two molecules of the target protein (Fig. S8 I and J). These results indicate that simultaneous binding of the KD variants using the two separate binding sites of LCR1 and LCR2 to two molecules of DR4 and/or DR5 results in the receptor clustering to activate them (18), explaining the efficient agonistic activity of the KD variants even in the monomeric form.
In Vivo Efficacy of the KD Variants.
To increase the size and avidity-mediated affinity, KD413 and KD548 were formatted into the homodimeric Fc-fused form by C-terminal fusion to Fc of human IgG1. Both KD413-Fc and KD548-Fc, expressed in correctly assembled form (Fig. S3 D and H) and maintained target-binding specificity (Table S1), exhibited ∼10-fold higher affinity with the respective target protein and ∼8-fold greater cytotoxicity to both TRAIL-sensitive HCT116 and TRAIL-resistant U87MG cells, compared with the monomeric counterparts (Table S1). To determine whether the Fc-fused KD variants against DR4 and/or DR5 can function in vivo, we assessed the antitumor efficacy by intravenous injections of either KD413-Fc or KD548-Fc into athymic nude mice bearing preestablished solid tumors of HCT116 or U87MG xenografts. The two Fc-fused KD variants almost completely inhibited tumor growth of both tumors, resulting in ∼7-fold or greater reductions in tumor volume by the end of treatment, compared with tumors treated with the control PgnKD2-Fc (Fig. 3 C and D). Similar to a previous result (17), TRAIL intravenously administered significantly retarded tumor propagation in HCT116 xenografts (Fig. 3C). However, TRAIL only slightly inhibited the growth of U87MG tumors (Fig. 3D), consistent with the in vitro response of the cells to TRAIL treatment (Fig. 3A). The ability of the KD variants to inhibit tumor growth of even the TRAIL-resistant U87MG cells may be attributed to their distinct binding sites on the target proteins from those of TRAIL (Fig. S8) (18, 22). Collectively, the above results demonstrate that the agonistic KD variants are potent inhibitors of tumor-cell growth in vivo.
To address the TNFα-neutralizing activity in vivo, KDT26-Fc was injected intraperitoneally into C57BL/6 mice, which were injected 1 h later with a lethal dose of TNFα and D-galactosamine (GalN) (24). KDT26-Fc partially, but significantly, protected the mice from TNFα-induced mortality, as infliximab did, compared to mice treated with the isotype control (IC) IgG1 (Fig. 4B). This result suggests that the neutralizing effect of KDT26-Fc is specific in vivo and is pharmacodynamically effective.
Discussion
Here, we demonstrated that the KD scaffold can generate biologically functional agonists to DR4 and DR5, as well as an antagonist against TNFα, from a synthetic KD library designed by sequence analyses of naturally occurring human KDs. The observed 10-6–10-8 M affinities of the isolated KD variants are comparable to those of target-specific binder from monovalent Ab libraries (1, 25) and other alternative protein scaffold libraries (5–8). Although incorporated by many mutations (up to 45 residues out of 78 residues), the selected KD variants have good thermal stability and retained the typical secondary structure of the parent KD, suggesting that the stability and conformation of the KD scaffold with cystine-knot structural motif is strongly sequence-tolerant for mutations in the loop regions. However, KDs are distinct from other small proteins with three-disulfide bridge structures, such as knottin (∼30 residues) (26) and avimer (∼35 residues) (9), in terms of size as well as the three-disulfide linkage pattern (1–6, 2–4, and 3–5 for KDs vs. 1–4, 2–5, and 3–6 for the latter two proteins).
The reported alternative protein scaffolds have from 5 to 25 residues, which can be varied (2–9), but the KD scaffold has ∼45 residues in the seven loop regions that can be randomized for target recognition, comparable to the six complementarity determining regions (45–82 residues) of Abs (1, 27). Possibly owing to the large sequence space of the KD scaffold, target-specific KD variants against the three target proteins and, at the same time, polyclonal KD variants recognizing different regions on a single molecule were isolated. The KD variants with DR4 and/or DR5 binding sites distinct from those of TRAIL directly induced cell death in a variety of tumor cells, including TRAIL-resistant U87MG glioma cells, and exerted synergistic tumoricidal activity when used in combination. The agonistic activity of some KD variants against DR4 and DR5 even in the monomeric form might be attributed to the simultaneous binding ability to two target molecules using the two LCRs on each opposite side of the structure (Fig. 1A and Fig. S8). Thus, KD scaffold with two potential binding surfaces for target recognition can be developed into bivalent or bispecific, but single domain variants, such as agonists inducing receptor clustering, providing a distinct advantage over the reported non-Ab scaffolds with a single binding surface (2–9). Further, the small and modular structural features of the KD scaffold might offer favorable properties of non-Ab domain scaffolds (2, 3), such as cheap production in a microbial system and easy modifications for fusion with effector molecules (e.g., Fc fusion) and multivalent or multispecific constructs with enhanced functionality by tandem linkages of the same or different KD variants using flexible linkers.
The selected KD variants function in vivo as agonists toward DR4 and/or DR5, or an antagonist against TNFα, like Abs toward the corresponding targets that are in clinical trials (18, 21), suggesting that KD variants have the potential to be developed as biotherapeutics. Because of the extensive mutations in KD variants, however, potential immunogenicity should be addressed during clinical trials. Finally, considering that naturally occurring KDs bind many different molecules, including metabolites, membranes, and carbohydrates (12, 13) that are rarely recognized by Abs, our results may provide an avenue to develop sequence variants of the KD scaffold as target-specific binding proteins for many types of target molecules.
Methods
Details of materials and all methods are described in SI Text.
Synthetic KD Library Construction and Screening.
The gene encoding human PgnKD2 from -2 to 84 residues (12, 15) was subcloned in-frame into the yeast surface display plasmid, pCTCON (20, 25). The KD gene library was created by two-step synthetic shuffling to reconstitute full-size products using 10 partially overlapping oligonucleotides, as described in Fig. 1 and SI Text. The procedures for library transformation and library screening against biotinylated-target proteins were conducted using previously described protocols (25) (also see SI Text for details).
In Vitro and in Vivo Antitumor Assays.
All human cell lines were purchased from the American Type Culture Collect (ATCC) and maintained as described previously (22). Cell viability assays and calculation of the EC50s were conducted as described previously (22). All animal experiments were approved by the internal review committee of the Ajou University Medical School and performed according to the guidelines established by the Institutional Animal Care and Use Committee. For the analysis of tumoricidal activity in vivo, female BALB/c athymic nude mice (Orientbio) were inoculated subcutaneously with 5 × 106 cells of either the HCT116 or U87MG cells in the right thigh (19). When the mean tumor size reached ∼50–70 mm3 (after seven days’ growth), mice were randomized into groups (n = 7 per group) and administered via the tail vein with PgnKD2-Fc, KD413-Fc, KD548-Fc, or TRAIL in a dose/weight-matched fashion every two days (for a total of six treatments).
In Vitro and in Vivo TNFα Neutralization Assay.
In vitro and in vivo neutralizing activities of PgnKD2, the KD variants, KDT26-Fc, and/or infliximab were measured against human TNFα-induced cytotoxicity using mouse WEHI 164 cells (ATCC) and TNFα-induced lethality in C57BL/6 mice (Orientbio), as described previously (20, 23, 24).
Supplementary Material
Acknowledgments.
This work was supported by grants from the basic research (KRF-2007-313-D00248) and the Converging Research Center Program (2009-0093653) from the National Research Foundation of the Ministry of Education, Science and Technology, the “GRRC” Project of Gyeonggi Provincial Government, and the KRIBB Research Initiative Program, Republic of Korea.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001541107/-/DCSupplemental.
References
- 1.Lerner RA. Manufacturing immunity to disease in a test tube: The magic bullet realized. Angew Chem Int Ed Engl. 2006;45:8106–8125. doi: 10.1002/anie.200603381. [DOI] [PubMed] [Google Scholar]
- 2.Skerra A. Alternative non-antibody scaffolds for molecular recognition. Curr Opin Biotechnol. 2007;18:295–304. doi: 10.1016/j.copbio.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Binz HK, Amstutz P, Pluckthun A. Engineering novel binding proteins from nonimmunoglobulin domains. Nat Biotechnol. 2005;23:1257–1268. doi: 10.1038/nbt1127. [DOI] [PubMed] [Google Scholar]
- 4.Binz HK, et al. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat Biotechnol. 2004;22:575–582. doi: 10.1038/nbt962. [DOI] [PubMed] [Google Scholar]
- 5.Schneider S, et al. Mutagenesis and selection of PDZ domains that bind new protein targets. Nat Biotechnol. 1999;17:170–175. doi: 10.1038/6172. [DOI] [PubMed] [Google Scholar]
- 6.Malabarba MG, et al. A repertoire library that allows the selection of synthetic SH2s with altered binding specificities. Oncogene. 2001;20:5186–5194. doi: 10.1038/sj.onc.1204654. [DOI] [PubMed] [Google Scholar]
- 7.Nord K, et al. Binding proteins selected from combinatorial libraries of an alpha-helical bacterial receptor domain. Nat Biotechnol. 1997;15:772–777. doi: 10.1038/nbt0897-772. [DOI] [PubMed] [Google Scholar]
- 8.Beste G, Schmidt FS, Stibora T, Skerra A. Small antibody-like proteins with prescribed ligand specificities derived from the lipocalin fold. Proc Natl Acad Sci USA. 1999;96:1898–1903. doi: 10.1073/pnas.96.5.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silverman J, et al. Multivalent avimer proteins evolved by exon shuffling of a family of human receptor domains. Nat Biotechnol. 2005;23:1556–1561. doi: 10.1038/nbt1166. [DOI] [PubMed] [Google Scholar]
- 10.Schonfeld D, et al. An engineered lipocalin specific for CTLA-4 reveals a combining site with structural and conformational features similar to antibodies. Proc Natl Acad Sci USA. 2009;106:8198–8203. doi: 10.1073/pnas.0813399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Letunic I, et al. SMART 5: Domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao Y, Cao R, Veitonmaki N. Kringle structures and antiangiogenesis. Curr Med Chem Anticancer Agents. 2002;2:667–681. doi: 10.2174/1568011023353705. [DOI] [PubMed] [Google Scholar]
- 13.Castellino FJ, Beals JM. The genetic relationships between the kringle domains of human plasminogen, prothrombin, tissue plasminogen activator, urokinase, and coagulation factor XII. J Mol Evol. 1987;26:358–369. doi: 10.1007/BF02101155. [DOI] [PubMed] [Google Scholar]
- 14.O’Reilly MS, et al. Endostatin: An endogenous inhibitor of angiogenesis and tumor growth. Cell. 1997;88:277–285. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 15.Rios-Steiner JL, Schenone M, Mochalkin I, Tulinsky A, Castellino FJ. Structure and binding determinants of the recombinant kringle-2 domain of human plasminogen to an internal peptide from a group A Streptococcal surface protein. J Mol Biol. 2001;308:705–719. doi: 10.1006/jmbi.2001.4646. [DOI] [PubMed] [Google Scholar]
- 16.Marti DN, Schaller J, Llinas M. Solution structure and dynamics of the plasminogen kringle 2-AMCHA complex: 3(1)-helix in homologous domains. Biochemistry. 1999;38:15741–15755. doi: 10.1021/bi9917378. [DOI] [PubMed] [Google Scholar]
- 17.Ashkenazi A, et al. Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest. 1999;104:155–162. doi: 10.1172/JCI6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashkenazi A, Herbst RS. To kill a tumor cell: The potential of proapoptotic receptor agonists. J Clin Invest. 2008;118:1979–1990. doi: 10.1172/JCI34359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sung ES, et al. A novel agonistic antibody to human death receptor 4 induces apoptotic cell death in various tumor cells without cytotoxicity in hepatocytes. Mol Cancer Ther. 2009;8:2276–2285. doi: 10.1158/1535-7163.MCT-09-0235. [DOI] [PubMed] [Google Scholar]
- 20.Kim MS, et al. Comparative analyses of complex formation and binding sites between human tumor necrosis factor-alpha and its three antagonists elucidate their different neutralizing mechanisms. J Mol Biol. 2007;374:1374–1388. doi: 10.1016/j.jmb.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 21.Tracey D, Klareskog L, Sasso EH, Salfeld JG, Tak PP. Tumor necrosis factor antagonist mechanisms of action: A comprehensive review. Pharmacol Ther. 2008;117:244–279. doi: 10.1016/j.pharmthera.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Park KJ, et al. A human scFv antibody against TRAIL receptor 2 induces autophagic cell death in both TRAIL-sensitive and TRAIL-resistant cancer cells. Cancer Res. 2007;67:7327–7334. doi: 10.1158/0008-5472.CAN-06-4766. [DOI] [PubMed] [Google Scholar]
- 23.Song MY, et al. Characterization of a novel anti-human TNF-alpha murine monoclonal antibody with high binding affinity and neutralizing activity. Exp Mol Med. 2008;40:35–42. doi: 10.3858/emm.2008.40.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angermuller S, Kunstle G, Tiegs G. Pre-apoptotic alterations in hepatocytes of TNFalpha-treated galactosamine-sensitized mice. J Histochem Cytochem. 1998;46:1175–1183. doi: 10.1177/002215549804601009. [DOI] [PubMed] [Google Scholar]
- 25.Lee HW, et al. Construction and characterization of a pseudo-immune human antibody library using yeast surface display. Biochem Biophys Res Commun. 2006;346:896–903. doi: 10.1016/j.bbrc.2006.05.202. [DOI] [PubMed] [Google Scholar]
- 26.Kolmar H. Alternative binding proteins: Biological activity and therapeutic potential of cystine-knot miniproteins. Febs J. 2008;275:2684–2690. doi: 10.1111/j.1742-4658.2008.06440.x. [DOI] [PubMed] [Google Scholar]
- 27.Collis AV, Brouwer AP, Martin AC. Analysis of the antigen combining site: Correlations between length and sequence composition of the hypervariable loops and the nature of the antigen. J Mol Biol. 2003;325:337–354. doi: 10.1016/s0022-2836(02)01222-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.