Abstract
A striking but poorly understood feature of many diseases is the unique involvement of neural tissue. One example is the CNS-specific disorder DYT1 dystonia, caused by a 3-bp deletion (“ΔE”) in the widely expressed gene TOR1A. Disease mutant knockin mice (Tor1aΔE/ΔE) exhibit disrupted nuclear membranes selectively in neurons, mimicking the tissue specificity of the human disease and providing a model system in which to dissect the mechanisms underlying neural selectivity. Our in vivo studies demonstrate that lamina-associated polypeptide 1 (LAP1) and torsinB function with torsinA to maintain normal nuclear membrane morphology. Moreover, we show that nonneuronal cells express dramatically higher levels of torsinB and that RNAi-mediated depletion of torsinB (but not other torsin family members) causes nuclear membrane abnormalities in Tor1aΔE/ΔE nonneuronal cells. The Tor1aΔE/ΔE neural selective phenotype therefore arises because high levels of torsinB protect nonneuronal cells from the consequences of torsinA dysfunction, demonstrating how tissue specificity may result from differential susceptibility of cell types to insults that disrupt ubiquitous biological pathways.
Keywords: dystonia, DYT1, nuclear envelope, LAP1
In primary dystonia, dysfunctional CNS motor circuits cause abnormal involuntary twisting movements. DYT1 dystonia, a dominantly inherited form of primary dystonia, is caused by a deletion in TOR1A that removes a single glutamic acid (ΔE) from the protein torsinA. TorsinA is a membrane-associated AAA+ protein that resides within the endoplasmic reticulum/nuclear envelope (ER/NE) endomembrane system (1–3). In vitro studies suggest that torsinA interacts with several proteins within the ER/NE space, including lamina-associated polypeptide 1 (LAP1), LULL1 (luminal domain-like LAP-1), nesprins, and printor (4–7). LAP1 and LULL1 are type-2 transmembrane proteins that interact with torsinA via their homologous C-terminal (luminal) regions (5); these proteins appear to participate in the relative distribution of torsinA between the NE and the main ER (7, 8). The N-terminal regions of LAP1 and LULL1 are highly divergent and are responsible for their differential localization within the NE/ER space: LAP1 concentrates in the inner nuclear membrane, whereas LULL1 is predominantly localized to the main ER (5, 9). However there exist no in vivo studies demonstrating a functional relationship between torsinA and these interacting proteins.
Although the biological function of torsinA is not well understood, we reported previously that Tor1aΔE/ΔE and Tor1a−/− mice exhibit morphological abnormalities of neuronal nuclear membranes, whereas all nonneuronal cell types appear normal (10). Notably, this phenotype mimics the tissue specificity of the human disease symptoms and established that neurons have a unique requirement for nuclear membrane-localized torsinA function.
In general, the neural-specific defect characteristic of torsinA mutant mice could occur because torsinA participates in a biological process that is unique to neurons or, alternatively, in a widespread cellular process that is less vulnerable to disruption in nonneuronal cells. A significant impediment to distinguishing between these possibilities has been the lack of in vivo data identifying proteins that function with torsinA. We now show that mice lacking LAP1 develop neuronal nuclear membrane abnormalities that are indistinguishable from those observed in Tor1aΔE/ΔE and Tor1a−/− mice, establishing a functional role for this protein in the torsinA pathway. Loss of LAP1 function also causes similar nuclear membrane abnormalities in nonneuronal cells, indicating that torsinA and LAP1 participate in a widespread cellular process to which neurons are selectively vulnerable (in torsinA mutant mice). Moreover, we demonstrate that dramatically differing levels of torsinB, another torsin family member, are a key factor in the differential susceptibility of neuronal and nonneuronal tissue. Our unique findings identify in vivo proteins that function with torsinA and implicate LAP1 and torsinB function in the pathogenesis of DYT1 dystonia.
Results and Discussion
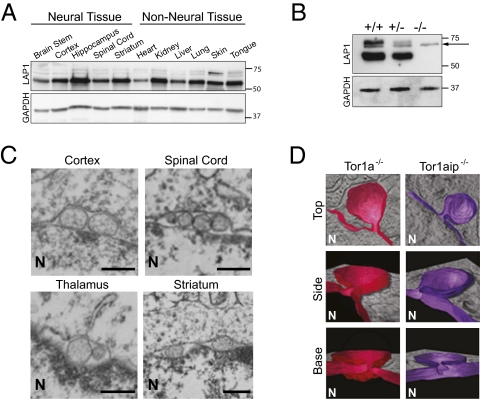
We began our search for torsinA pathway proteins by testing for a functional relationship between torsinA and LAP1. Like torsinA, LAP1 is widely expressed in neuronal and nonneuronal tissue, both during embryogenesis (when torsinA-related nuclear membrane abnormalities begin to form in neurons at E16; Fig. 1A) and in adulthood (5). To examine LAP1 function in vivo, we studied mice in which the Tor1aip1 locus that encodes LAP1 was disrupted with a gene trap cassette (Fig. S1A). Tor1aip1+/− intercrosses generated all expected genotypes with normal Mendelian frequency (300 pups collected at E16–E18: 23.9% WT, 48.5% HET, 27.6% KO). Similar to the Tor1a knockout and ΔE knockin lines (10), Tor1aip1+/− mice were indistinguishable from their wild-type littermate controls into adulthood, whereas Tor1aip1−/− mice exhibit perinatal lethality, typically dying on the last prenatal (E18) or first postnatal day (P0). Western blot analysis on whole brain lysates confirmed the complete loss of the 75-kDa and 60-kDa anti-LAP1 immunoreactive bands (5, 11) (Fig. 1B).
Fig. 1.
LAP1 and torsinA function in a common molecular pathway in neurons. (A) LAP1 is broadly expressed in neural and nonneural murine embryonic tissue. Protein lysates from E16 wild-type tissues probed with anti-LAP1 (Upper) and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Lower) antibodies. (B) LAP1 protein expression is abolished by a gene-trap cassette. Protein lysates from Tor1aip1+/+, Tor1aip1+/−, Tor1aip1−/− E18 mouse brain probed with anti-LAP1 (Upper) and anti-GAPDH (Lower) antibodies. The arrow indicates a nonspecific cross-reactive band. (C) Loss of LAP1 causes nuclear envelope abnormalities in E18 neural tissue. (Scale bar, 0.5 μm.) The nucleus (N) is marked. (D) Loss of torsinA or LAP1 causes the same ultrastructural defect of neuronal nuclear membranes. Reconstituted 3D EM tomographic images (in color) viewed from the top, side, and base of neuronal nuclear membrane abnormality (“bleb”) from Tor1a−/− and Tor1aip1−/− mice. The 3D reconstruction (in color) of the inner nuclear membrane and associated bleb is shown in the context of the EM section from which they reside. The nucleus (N) is marked.
To test whether LAP1 participates in torsinA-related pathways, we examined whether neurons from Tor1aip1−/− mice exhibit ultrastructural changes of nuclear membrane similar to those seen in Tor1a−/− and Tor1aΔE/ΔE animals. Strikingly, the nuclear membranes of Tor1aip1−/− neural tissue or cultured neurons exhibited nuclear membrane abnormalities (Fig. 1C) that appear similar to those of Tor1a mutant mice. Using electron microscopy (EM), membranous vesicle-appearing structures (termed “blebs”) were observed in the perinuclear space of Tor1aip1−/− neurons, and similar to Tor1a mutant mice, these abnormalities were seen in all brain regions examined, including striatum, cortex, thalamus, and spinal cord. No such abnormalities were observed in Tor1aip1+/− mice. The subcellular localization and expression levels of torsinA and all nuclear membrane proteins examined appeared normal in Tor1aip1−/− neurons (Fig. S1 B and C). Thus, nuclear membrane blebs do not appear to result from widespread disruption in the organization of nuclear membrane proteins.
Because different morphological abnormalities of nuclear membrane have been observed in a variety of experimental contexts (12–16) we generated high-resolution, three-dimensional (3D) EM tomographic images to rigorously determine whether the loss of torsinA or LAP1 results in similar ultrastructural defects, which would support a functional relationship between these proteins. Three-dimensional tomographic reconstructions of the nuclear membrane blebs demonstrated that torsinA- and LAP1-related NE blebs are structurally indistinguishable; both are membranous outpouchings derived from the inner nuclear membrane to which they remain attached via a well-defined neck (Fig. 1D). Thus, LAP1 null mice phenocopy the perinatal lethality and neuronal nuclear membrane abnormalities seen in torsinA knockout and disease mutant knockin mice.
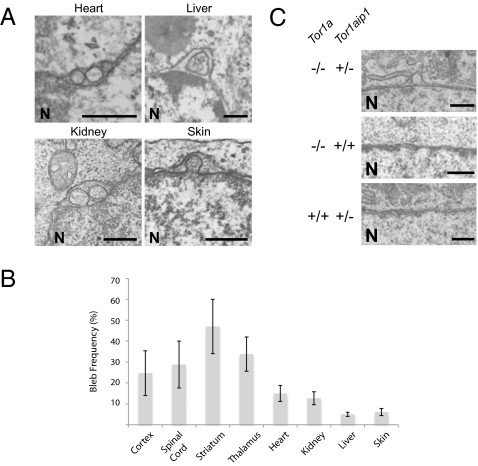
To determine whether neurons exhibit a unique requirement for LAP1 (as they do for torsinA) we next examined the nuclear membrane ultrastructure of nonneural tissues from Tor1aip1−/− embryos. In contrast to Tor1a mutant mice, we observed nuclear membrane blebs in all LAP1 null tissues examined, including liver, striated muscle (cardiac), kidney, and skin (Fig. 2 A and B). To determine whether torsinA functions in this process in nonneuronal cells, we tested for epistasis between Tor1a and Tor1aip1 by intercrossing double heterozygous (Tor1a+/−/Tor1aip1+/−) mice and evaluating the nuclear membrane ultrastructure of mouse embryonic fibroblasts (MEFs) derived from the resultant embryos. Whereas nuclear membrane blebs are never observed in torsinA null or LAP1 heterozygous MEFs on wild-type backgrounds, eliminating torsinA function caused nuclear membrane blebs in LAP1 heterozygous MEFs, supporting a role for torsinA in this pathway in nonneuronal cells (Fig. 2C; blebs are seen in Tor1a−/−/Tor1aip1+/− MEFs, but not in Tor1a−/−/Tor1aip1+/+ or Tor1a+/+/Tor1aip1+/− cells). These findings further support a functional relationship between torsinA and LAP1 and argue against the possibility that these proteins participate in a neural-specific pathway. Rather, these observations indicate that torsinA and LAP1 participate in a widespread biological process to which neurons are more vulnerable.
Fig. 2.
Loss of LAP1 function causes NE blebs in nonneuronal tissues. (A) Absence of LAP1 generates blebs in E18 nonneural tissues. (Scale bar, 0.5 μm.) The nucleus (N) is marked. (B) Summary of bleb frequencies in neural and nonneural tissues in E18 LAP1 null embryos. Error bars = SEM of bleb frequencies from three independent experiments (n = 3) performed on tissues derived from three independent sets of mice. (C) Elimination of torsinA function in Tor1aip1+/− nonneuronal cells (MEFs) leads to bleb formation. (Scale bar, 0.5 μm.) The nucleus (N) is marked.
This cell type-specific susceptibility appears to involve additional proteins that interact with the torsinA–LAP1 pathway. Consistent with this notion are the observations that reducing LAP1 levels causes blebs in torsinA null MEFs (Fig. 2C) and that the frequency of nuclear membrane abnormalities is roughly 6-fold greater in LAP1 than torsinA null neurons (Tor1a−/− 7.1% ± 1.2%; Tor1aip1+/− 46.0% ± 2.0%; n = 3). LULL1 is one such candidate protein. Like LAP1, LULL1 has been implicated in the control of torsinA localization within the NE/ER endomembrane space (7) and, similar to torsinA and LAP1, LULL1 is widely expressed in neural and nonneural tissue (Fig. S2A). Therefore, we tested whether loss of LULL1 function was associated with alterations in nuclear membrane morphology, which would support a functional relationship with torsinA and LULL1. We used four different shRNAs to knock down LULL1 expression in MEFs, each of which markedly reduced the levels of LULL1 protein (Fig. S2B). However, nuclear membrane ultrastructure appeared normal in LULL1-deficient MEFs (Fig. S2C). These experiments do not support a role for LULL1 in the torsinA–LAP1 pathway associated with alterations of nuclear membrane morphology.
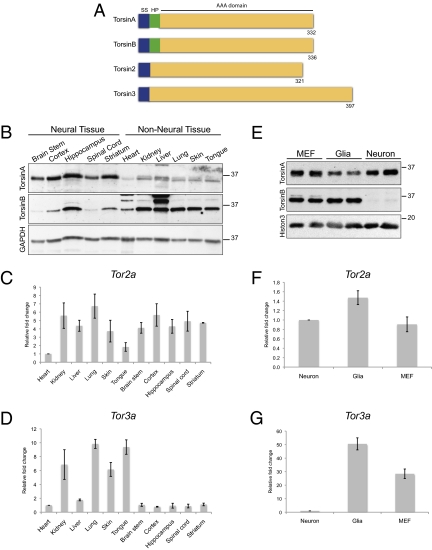
TorsinA is a member of a family of ER luminal AAA+ proteins, making these other family members (torsinB, torsin2, and torsin3; Fig. 3A) attractive candidates for additional proteins that might function with LAP1 and contribute to the neural specificity characteristic of Tor1a mutant mice. Although levels of torsinB protein are lower in neural than nonneural tissue (8), it is unclear whether torsinB levels in neurons or glia—which do not develop NE blebs in Tor1a mutant mice—account for this expression pattern, and no data address whether torsin proteins share redundant functions. To begin exploring these issues, we first measured the neural and nonneural expression levels of all torsin family members at E16, the time when NE blebs begin to be observed (10). TorsinB protein levels were clearly higher in nonneural tissue than in neural tissue, whereas torsinA protein levels were somewhat higher in neural tissue (Fig. 3B). Due to lack of reliable antibodies, we used quantitative RT-PCR for torsin2 and torsin3. Like torsinB, Tor3a mRNA levels were markedly higher in nonneural than in neural tissue (Fig. 3D), whereas Tor2a mRNA levels were similar in both tissues (Fig. 3C). As NE blebs form selectively in neurons in Tor1a mutant mice (10), we assessed relatively pure cultures of neurons, glia, and MEFs to determine whether our findings in whole tissue accurately reflect expression differences between neuronal and nonneuronal cells. For torsinB protein and Tor3a mRNA (Fig. 3 E and G), the expression level differences between neuronal and nonneuronal cells were even more striking than those observed in tissue, whereas Tor2a mRNA levels were similar in the different cell types (Fig. 3F). These findings highlight torsinB and torsin3 as candidate proteins that may act with LAP1 to enable nonneuronal cells to withstand the loss of torsinA function.
Fig. 3.
Torsin family members exhibit distinct expression patterns in neuronal and nonneuronal cells. (A) Cartoon diagram illustrating the torsin protein family. The key features of torsinA, torsinB, torsin2, and torsin3 are shown, including the signal sequence (SS; blue), hydrophobic domain (HP; green), and AAA domain (yellow). The number of amino acids is indicated for each protein. (B) The expression patterns of torsinA and torsinB proteins are opposite in neural and nonneural whole tissue. Levels of torsinB protein are clearly higher in nonneural tissue, whereas levels of torsinA protein are generally higher in neural tissue. Whole tissue protein lysates from E16 wild-type embryos probed with anti-torsinA (Top), anti-torsinB (Middle; 34 kDa), and anti-GAPDH (Bottom) antibodies. The E16 time point was chosen because this is the age when nuclear membrane blebs begin to form. (C) Levels of Tor2a mRNA are similar in neural and nonneural tissue. Quantitative RT-PCR was performed using RNA derived from E16 wild-type embryos. Ct value of Tor2a in each tissue was normalized to the Ct value of GAPDH. Relative fold change was obtained by comparing the normalized Ct value for a designated tissue to the one for heart. Error bars = SEM of fold change from three independent experiments (n = 3) performed on tissue derived from three independent sets of mice. (D) Levels of Tor3a mRNA are markedly higher in nonneural tissue than in neural tissue. Quantitative RT-PCR was performed as in C. (E) Neurons express markedly less torsinB protein than nonneuronal cell types. Whole cell protein lysates from primary cultures of wild-type MEFs, glia, or cortical neurons probed with anti-torisnA (Top), anti-torsinB (Middle), and anti-histone 3 (Bottom, loading control) antibodies. (F) Neurons and nonneuronal cell types express similar levels of Tor2a mRNA. Quantitative RT-PCR was performed using RNA extracted from primary cultures of wild-type MEFs, glia, or cortical neurons. Ct value of Tor2a for each cell type was normalized to the Ct value of β-actin. Relative fold change was obtained by comparing the normalized Ct value for a designated cell type to the one for neurons. Error bars = SEM of fold change from three independent experiments (n = 3) performed on cells derived from three independent sets of cultures. (G) Neurons express markedly less Tor3a mRNA than nonneuronal cell types. Quantitative RT-PCR was performed as in F.
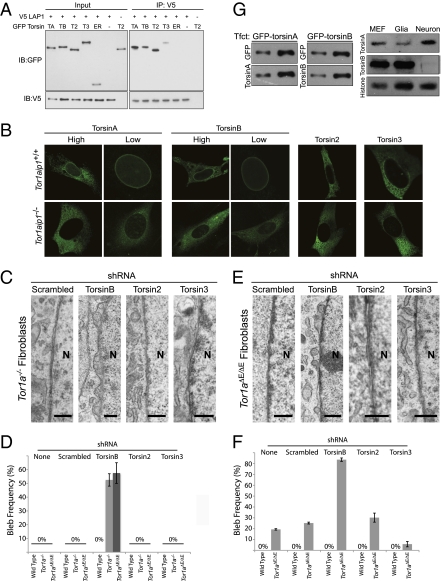
To begin to explore the possibility that other torsin proteins function with LAP1, we first tested whether they interact in coimmunoprecipitation studies, as do torsinA and LAP1 (5). When coexpressed in the mouse CAD cell line, all torsins coimmunoprecipitate with LAP1 (Fig. 4A). Mutating a key residue in the AAA+ domain (E→Q in the Walker B domain) creates a “substrate trap” ATP hydrolysis-deficient form of torsinA that concentrates in the nuclear membrane (2, 17). To test the interaction of LAP1 with torsin family members in a cellular context, we examined subcellular localization of substrate trap torsins in wild-type and Tor1aip1−/− MEFs (Fig. 4B). In wild-type MEFs, EQ forms of torsinA, torsinB, and torsin3 clearly accumulate in the nuclear membrane, whereas EQ-torsin2 is localized to the main endoplasmic reticulum. In Tor1aip1−/− MEF cells, the localization of EQ-torsinA and EQ-torsinB shifts dramatically from the nuclear membrane to the endoplasmic reticulum, demonstrating that LAP1 is necessary for the NE localization of these proteins. In contrast, the subcellular localization of EQ-torsin2 and EQ-torsin3 is similar in wild-type and Tor1aip1−/− MEFs (Fig. 4B). These data are in agreement with LAP1 knockdown studies (8), and are consistent with the possibility that other torsin proteins, particularly torsinB, may enable nonneuronal cells to maintain normal nuclear membrane morphology in the absence of torsinA.
Fig. 4.
TorsinB maintains normal nuclear membrane morphology in torsinA knockout and ΔE-knockin nonneuronal cells. (A) All four torsin family members coimmunoprecipitate (co-IP) with LAP1. CAD cells were cotransfected with V5-LAP1 and each of GFP-tagged torsin family members. LAP1 was immunoprecipitated from whole cell lysates of these transfections with anti-V5 antibody, and the immunoprecipitation (IP) was separated using SDS/PAGE and probed with anti-GFP and anti-V5 antibodies. TA, torsinA; TB, torsinB; T2, torsin2; T3, torsin3; ER, GFP with ER signal sequence and KDEL retention signal. Note that ER-localized GFP (ER) does not co-IP with LAP1, and GFP-torsin does not co-IP in the absence of LAP1 (only torsin2 is shown here). (B) Loss of LAP1 selectively alters the subcellular localization of substrate trap forms of torsinA and torsinB. MEF cells derived from wild-type (Top) or Tor1aip1−/− (Lower) embryos were electroporated with GFP-tagged substrate trapping mutant torsins. Substrate trap forms of torsinA and torsinB exhibit nuclear membrane accumulation that requires LAP1. This is particularly notable for cells expressing low levels (“low”) of torsinA or torsinB. The subcellular localization of substrate trap forms of torsin2 or torsin3 was not altered in Tor1aip1−/− cells. (C) Selective formation of NE blebs following the knockdown of torsinB in Tor1a−/− MEFs. Tor1a−/− and wild-type MEFs were transduced with lentivirus expressing shRNA to knock down torsinB, torsin2, torsin3, or a control shRNA and assessed by EM. Knockdown of torsinB caused NE blebs only in Tor1a−/− cells. Knockdown of torsin2 or torsin3 did not cause NE blebs. The nucleus (N) is marked. (Scale bar, 0.5 μm.) (D) Selective formation of NE blebs following the knockdown of torsinB in Tor1a−/− and Tor1aΔE/ΔE MEFs. Quantification of data from experiments shown in Fig. 4 C and E. Error bars = SEM of bleb frequencies from three independent experiments (n = 3). (E) Selective formation of NE blebs following the knockdown of torsinB in Tor1aΔE/ΔE MEFs. Experiment was performed as in C. The nucleus (N) is marked. (Scale bar, 0.5 μm.) (F) Selective enhancement of NE bleb formation following the knockdown of torsinB in Tor1aΔE/ΔE neurons. Graphical illustration of data described in main text. Error bars = SEM of bleb frequencies from three independent experiments (n = 3). (G) The relative amounts of torsinA and torsinB are strikingly different in neuronal and nonneuronal cell types. Anti-torsinA and anti-torsinB antibodies were normalized using cell lysates transfected with either GFP-torsinA or GFP-torsinB (Left). These normalized conditions were used to assess the relative amounts of torsinA and torsinB in cell lysates derived from primary cultures of wild-type MEFs, glia, or neurons (Right).
To directly address whether other torsin proteins exhibit functional redundancy with torsinA, we used shRNA to acutely down-regulate torsin family members in torsinA null and control MEFs. TorsinA null MEFs do not exhibit NE blebs, so bleb formation following the knockdown of another torsin protein would indicate that the function of this protein maintains normal nuclear membrane morphology in torsinA null nonneuronal cells. Levels of each torsin were markedly and selectively reduced by shRNA-expressing lentivirus compared with control sequences, and the knockdown of these proteins had no effect on the levels of LAP1 (Fig. S3). Knockdown of torsinB led to robust NE bleb formation selectively in torsinA null MEFs (but not wild-type MEFs), causing blebs in ~50% (n = 3) of cells, and these cells frequently exhibited multiple blebs/cell (Fig. 4 C and D). In contrast, the knockdown of torsin2 and torsin3 had no effect on NE morphology in either torsinA null or wild-type MEF cells (Fig. 4 C and D). Paradoxically, overexpression of torsinA or torsinB in Tor1a−/− neurons increased nuclear membrane bleb formation (Fig. S4A; torsinA overexpression increased bleb frequency from 7.15% ± 1.27% to 51.67% ± 10.65%; torsinB overexpression increased bleb frequency from 7.15% ± 1.27% to 16.76% ± 4.25%; n = 3). One possible reason for this finding may be that the nuclear membrane proteome is disrupted in torsinA null neurons, leading torsinA to engage in abnormal protein–protein interactions. To explore this possibility, we overexpressed torsinA in wild-type neurons, which have a normal nuclear membrane organization. Strikingly, this caused NE bleb formation to a similar degree as that seen in torsinA null neurons (Fig. S4B; bleb frequency increased from 0 to 47.33% ± 15.87%; n = 3). These findings suggest that overexpression of nonphysiological levels of torsins disrupt this pathway, precluding rescue-type studies using currently available reagents.
To test the relevance of these results to neurons and to disease mutant ΔE-torsinA, we knocked down torsin family members in Tor1aΔE/ΔE MEFs (which have normal nuclear membranes) and primary cultures of cortical neurons (which show a low frequency of NE blebs). In both cases, a selective role for torsinB was confirmed (Fig. 4 D–F). Knockdown of torsinB (but not torsin2 or torsin3) dramatically increased the frequency of nuclear membrane blebs in Tor1aΔE/ΔE neurons (Fig. 4F; untreated: 19.33% ± 0.67%; scrambled: 25.16% ± 0.87%; torsinB: 83.59% ± 1.59%; torsin2: 30.18% ± 4.18%; torsin3: 5.73% ± 2.46%; n = 3) and caused blebs in ~60% of Tor1aΔE/ΔE MEF cells (Fig. 4 D and E; 57.5% ± 7.5%; n = 3). Knockdown of torsinB (but not of torsin2 or torsin3) also caused NE blebs in Tor1aΔE/ΔE glia (Fig. S4C; 8.15% ± 3.85%; n = 3), supporting a general compensatory mechanism by torsinB in nonneuronal cells. Knockdown of torsinB in Tor1aΔE/+ MEFs, neurons, or glia cells did not alter nuclear membrane morphology, indicating that one wild-type torsinA allele is sufficient to maintain normal nuclear membrane morphology. Quantitative immunoblotting to compare the absolute levels of torsinA and torsinB (Fig. 4G) demonstrated that nonneuronal cells (glia and MEFs) contain strikingly higher levels of torsinB than torsinA, whereas this relationship is reversed in neurons. Taken together, these data argue that torsinA and torsinB share redundant nuclear membrane-localized functions, and that torsinB is responsible for maintaining normal nuclear membrane morphology in the nonneuronal tissues of torsinA null and ΔE-torsinA knockin mice.
We provide unique in vivo genetic evidence demonstrating that LAP1 and torsinA function in a common molecular pathway, one that is also disrupted (in Tor1aΔE/ΔE mice) by a disease mutation underlying human dystonia. Our data also suggest that torsinA and LAP1 act together with torsinB in a biological pathway that is required for normal nuclear membrane morphology in both neuronal and nonneuronal cells, and that the distinct expression patterns of torsinA and torsinB are a key determinant of the neural-specific defect of Tor1a mutant mice. Hanson and colleagues (9) report that the ΔE mutation disrupts the interaction of torsinA with LAP1, consistent with a potential role for LAP1 in the pathogenesis of DYT1 dystonia, and the ΔE mutation also alters the binding of printor (4) and nesprin-3α (6), raising the possibility that these proteins may also participate in the torsinA–LAP1 pathway and contribute to cell-type specific phenotypes caused by torsinA dysfunction. Whereas the role of this pathway at the nuclear membrane is just beginning to be understood, the appearance of torsinA-related nuclear membrane blebs is similar to those that result from null mutations in yeast nucleoporins (18, 19), potentially implicating nuclear pore function in the pathogenesis of DYT1 dystonia. A recent report linking LAP1 function to assembly of the mitotic microtubule spindle, however, underscores the fact that nuclear membrane proteins participate in diverse and sometimes unexpected pathways (20). Finally, linking LAP1 and torsinB function to a pathway disrupted by the DYT1 mutation raises the possibility that polymorphisms in these proteins may affect the penetrance of the DYT1 mutation (which is incomplete) or the severity of symptoms in patients with DYT1 dystonia.
Materials and Methods
Ultrastructural Analysis of Animals and Cells.
Embryos were delivered by caesarian section from timed pregnant female mice housed on a standard light–dark cycle. The day following overnight mating was designated as embryonic day 0 (E0). Tissue was prepared by transcardial perfusion of E18 embryos with 4% paraformaldehyde/3% glutaraldehyde in 10 mM sodium phosphate buffer (pH 7.4), followed by postfixation in the same solution. Cultured monolayer cells were fixed with 2.5% glutaraldehyde in 10 mM sodium phosphate buffer (pH 7.4). All samples were postfixed with 1% osmium tetroxide in 100 mM cacodylate buffer (pH 7.4) on ice for 1 h. Samples were then treated with 0.5% aqueous uranyl acetate, dehydrated in graded alcohol, treated with propylene oxide, and embedded in Embed 812 (Electron Microscopy Sciences). The resin was polymerized in a 60 °C oven for 2–3 days. Sections were cut with a Dupont diamond knife in Reichert-Jung UltraCut E ultramicrotome, collected on copper grids, and doubly stained with saturated aqueous uranyl acetate and lead citrate. Ultrathin sections were viewed using a JEM-1200EX electron microscope manufactured by JEOL. Please see additional details for electron tomography in SI Materials and Methods. All quantitative EM data were obtained from three independent sets of samples (n = 3) and 100 cells were counted in each set by an observer that was blinded to experimental conditions. Only cells for which an entire “ring” of nuclear membrane is visible are counted (i.e., no partial segments of nuclear membrane are analyzed). The entire nucleus is scored as “positive” or “negative” for NE blebs. Positive nuclei are those in which the presence of one or more blebs (clearly arising from the inner nuclear membrane) are observed.
Cell Culture.
CAD cells were maintained in F12/DMEM (Invitrogen) with 8% FBS and penicillin/streptomycin. MEFs were prepared from E13 or E16 embryos and cultured in DMEM media (Invitrogen) containing 15% FBS (HyClone) and 1% penicillin/streptomycin (Invitrogen). Mouse cortical neurons were prepared from cortex of embryos on E16 and maintained in neurobasal media (Invitrogen) with B24, glutamine, and penicillin/streptomycin. Glial cells were prepared from E18 frontal cortex and grown in F12/DMEM media (Invitrogen) with 10% FBS (HyClone) and 1% penicillin/streptomycin (Invitrogen). Please see additional details in SI Materials and Methods.
Transfection and Electroporation.
CAD cells were transfected using Lipofectamine and Plus Reagent (Invitrogen) following manufacturer's instruction. MEF cells were electroporated using Nucleofector (Amaxa). The day before electroporation, MEF cells were split and replated to be 50–60% confluent the next day. On the day of electroporation, cells were trypsinized, centrifuged, and resuspended in 100 μL of complete MEF 1 Nucleofector solution for 3 × 106 cells per reaction. After electroporation with program T-020, cells were immediately transferred to a 6-cm plate containing prewarmed DMEM media with 10% FBS and 1% penicillin/streptomycin. The next day, cells were rinsed to remove dead cells and analyzed after 48 h.
Immunofluorescence.
Cells grown in monolayer on PDL/laminin-coated coverslips (BD Biosciences) were fixed with methanol at −20 °C for 5 min and then rinsed with PBS. Fixed cells were permeabilized and blocked in PBS with 0.25% Triton-X and 10% normalized donkey serum (The Jackson Laboratory) for 1 h at room temperature, followed by overnight incubation with primary antibodies in blocking solution at 4 °C. The next day, coverslips were washed and incubated with secondary antibodies (Molecular Probes, Invitrogen) in blocking solution for 1 h at room temperature. Washed coverslips were mounted on microscope slides (Fisher) with mounting medium containing DAPI (Vector Laboratories) and sealed. Images were obtained using a Leica TCS SP5 system.
Antibodies.
Anti-torsinA, anti-GFP (Ab6556), and anti-NPC (414) antibodies were from Abcam. Anti-LAP1 and anti-LULL1 antibodies have been described (5). Anti-torsinB, anti-LBR, and anti-nesprin 2G antibodies were kind gifts from P. Gonzalez (University of Iowa), H. Herrmann (German Cancer Research Center), and G. Gundersen (Columbia University), respectively. Anti-laminA/C, anti-laminB, and anti-GFP (B-2) antibodies were purchased from Santa Cruz. Anti-V5 antibody was from Invitrogen. Anti-LAP2 and anti-beta actin (AC-74) antibodies were purchased from Sigma. Anti-GAPDH antibody was from Ambion and anti-PDI and anti-histone H3 antibodies were from Cell Signaling. All fluorephore-conjugated secondary antibodies (Alexa Fluor) were purchased from Invitrogen. All HRP-conjugated secondary antibodies were from Pierce.
Quantitative Real-Time PCR.
Total RNA was extracted from tissue and cells using RNeasy kit (Qiagen) following manufacturer's protocol and used for reverse transcription using SuperScript III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen). Quantitative RT-PCR was performed using a Cepheid SmartCycler following the manufacturer's specifications. cDNA was added to a 25-μL volume reaction mix containing OmniMix HS master mix (Cepheid) and SYBR Green I (Invitrogen) together with appropriate primers at 0.2-μM final concentration each. Primer sequences are provided in the SI Materials and Methods. Ct value was read at the peak of second derivative and the relative fold change was calculated using the following equation: R = 2-ΔΔCt, where ΔΔCt = ΔCtsample − ΔCtcalibrator, ΔCt = Cttarget − Ctreference (R, relative fold change; calibrator, the sample that the others are compared with; reference, internal control such as housekeeping genes.)
shRNA Constructs and Lentivirus Use.
shRNA constructs were purchased from Sigma or OpenBiosystems. Further information regarding constructs is provided in the SI Materials and Methods. Lentivirus was generated at the Gene Therapy Center at the University of North Carolina at Chapel Hill. Please see additional details for the lentivirus production and transduction in SI Materials and Methods.
Protein Extraction and Immunoblotting.
Cell pellets or tissue was lysed in 1% Triton-X or 1% SDS lysis buffer with 100 mM Tris-HCL pH 8.0, 10% glycerol, 150 mM NaCl, 2 mM EDTA, protease inhibitor mixture (Sigma), and phenyl methane sulphonyl fluoride (PMSF; Sigma) following standard protocols. Proteins were denatured before SDS/PAGE by boiling with Lammeli's sample buffer (Biorad) containing β-mercaptoethanol for 5 min. After SDS/PAGE, proteins were transferred to nitrocellulose membrane, which was blocked with blocking buffer (5% milk in PBS with 0.2% Tween) for 1 h at room temperature. Blocked membrane was incubated in blocking buffer containing primary antibody overnight at 4 °C. Blot was washed with washing buffer (PBS with 0.2% Tween) and then incubated in blocking buffer with HRP-conjugated secondary antibody for 1 h at room temperature. Washed blot was developed using chemiluminescent substrate for HRP (Pierce) and exposed on films (Kodak X-omat LS films). Quantitative immunoblotting was performed as previously described (5).
Coimmunoprecipitation.
Cells were centrifuged and resuspended in 1 mL of 1% Nonidet P-40 lysis buffer containing 50 mM Tris-HCL pH 7.5, 150 mM NaCl, complete protease inhibitor mixture (Sigma) and PMSF (Sigma). Nonspecific interaction was precleared by incubating the lysate with proteinA sepharose beads (Roche) for 3 h. After preclearing, protein concentration was determined by Braford assay and 500–1,000 μg of proteins was incubated with an appropriate antibody for 1 h, followed by overnight incubation with 50 μL of sepharose proteinA beads. The next day, beads were washed five times, twice with high salt concentration (400 mM NaCl), twice with regular salt concentration (150 mM NaCl), and once with no salt washing buffer. Each wash lasted 20 min. After the final wash, beads were centrifuged and boiled in Lammeli's sample buffer containing β-mercaptoethanol for 5 min at 95 °C to elute bound protein. Beads were centrifuged down and supernatant was used for SDS/PAGE as described above.
Supplementary Material
Acknowledgments
We thank Gil Di Paolo and Lauren Tanabe for their helpful comments regarding the manuscript. We are grateful for the superb technical work performed by Kana Meece, Jamie Twaite, and Hardy Rideout. We also thank Hong Yi and her colleagues at the Emory University Robert P. Apkarian Integrated Electron Microscopy Core for outstanding electron microscopy sample preparation. This work was funded by the Dystonia Medical Research Foundation, the Bachmann Strauss Foundation, the Parkinson's Disease Foundation, and National Institutes of Health R01-HD045708.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0912877107/-/DCSupplemental.
References
- 1.Kustedjo K, Bracey MH, Cravatt BF. TorsinA and its torsion dystonia-associated mutant forms are lumenal glycoproteins that exhibit distinct subcellular localizations. J Biol Chem. 2000;275:27933–27939. doi: 10.1074/jbc.M910025199. [DOI] [PubMed] [Google Scholar]
- 2.Naismith TV, Heuser JE, Breakefield XO, Hanson PI. TorsinA in the nuclear envelope. Proc Natl Acad Sci USA. 2004;101:7612–7617. doi: 10.1073/pnas.0308760101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ozelius LJ, et al. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat Genet. 1997;17:40–48. doi: 10.1038/ng0997-40. [DOI] [PubMed] [Google Scholar]
- 4.Giles LM, Li L, Chin LS. Printor, a novel torsinA-interacting protein implicated in dystonia pathogenesis. J Biol Chem. 2009;284:21765–21775. doi: 10.1074/jbc.M109.004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodchild RE, Dauer WT. The AAA+ protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. J Cell Biol. 2005;168:855–862. doi: 10.1083/jcb.200411026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nery FC, et al. TorsinA binds the KASH domain of nesprins and participates in linkage between nuclear envelope and cytoskeleton. J Cell Sci. 2008;121:3476–3486. doi: 10.1242/jcs.029454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vander Heyden AB, Naismith TV, Snapp EL, Hodzic D, Hanson PI. LULL1 retargets TorsinA to the nuclear envelope revealing an activity that is impaired by the DYT1 dystonia mutation. Mol Biol Cell. 2009;20:2661–2672. doi: 10.1091/mbc.E09-01-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jungwirth M, Dear ML, Brown P, Holbrook K, Goodchild R. Relative tissue expression of homologous torsinB correlates with the neuronal specific importance of DYT1 dystonia-associated torsinA. Hum Mol Genet. 2010;19:888–900. doi: 10.1093/hmg/ddp557. [DOI] [PubMed] [Google Scholar]
- 9.Naismith TV, Dalal S, Hanson PI. Interaction of torsinA with its major binding partners is impaired by the dystonia-associated DeltaGAG deletion. J Biol Chem. 2009;284:27866–27874. doi: 10.1074/jbc.M109.020164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodchild RE, Kim CE, Dauer WT. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron. 2005;48:923–932. doi: 10.1016/j.neuron.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 11.Senior A, Gerace L. Integral membrane proteins specific to the inner nuclear membrane and associated with the nuclear lamina. J Cell Biol. 1988;107:2029–2036. doi: 10.1083/jcb.107.6.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishimura A, Ng JK, Taira M, Young SG, Osada S. Man1, an inner nuclear membrane protein, regulates vascular remodeling by modulating transforming growth factor beta signaling. Development. 2006;133:3919–3928. doi: 10.1242/dev.02538. [DOI] [PubMed] [Google Scholar]
- 13.Kim HY, et al. A novel LMNA gene mutation Leu162Pro and the associated clinical characteristics in a family with autosomal-dominant emery-dreifuss muscular dystrophy. Muscle Nerve. 2008;38:1336–1339. doi: 10.1002/mus.21066. [DOI] [PubMed] [Google Scholar]
- 14.Park YE, et al. Nuclear changes in skeletal muscle extend to satellite cells in autosomal dominant Emery-Dreifuss muscular dystrophy/limb-girdle muscular dystrophy 1B. Neuromuscul Disord. 2009;19:29–36. doi: 10.1016/j.nmd.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Herron AJ, Worman HJ. Pathology and nuclear abnormalities in hearts of transgenic mice expressing M371K lamin A encoded by an LMNA mutation causing Emery-Dreifuss muscular dystrophy. Hum Mol Genet. 2006;15:2479–2489. doi: 10.1093/hmg/ddl170. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, et al. Epidermal expression of the truncated prelamin A causing Hutchinson-Gilford progeria syndrome: Effects on keratinocytes, hair and skin. Hum Mol Genet. 2008;17:2357–2369. doi: 10.1093/hmg/ddn136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodchild RE, Dauer WT. Mislocalization to the nuclear envelope: An effect of the dystonia-causing torsinA mutation. Proc Natl Acad Sci USA. 2004;101:847–852. doi: 10.1073/pnas.0304375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wente SR, Blobel G. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J Cell Biol. 1993;123:275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wente SR, Blobel G. NUP145 encodes a novel yeast glycine-leucine-phenylalanine-glycine (GLFG) nucleoporin required for nuclear envelope structure. J Cell Biol. 1994;125:955–969. doi: 10.1083/jcb.125.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neumann B, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.