Abstract
Differences in brain region size among species are thought to arise late in development via adaptive control over neurogenesis, as cells of previously patterned compartments proliferate, die, and/or differentiate into neurons. Here we investigate comparative brain development in ecologically distinct cichlid fishes from Lake Malawi and demonstrate that brains vary among recently evolved lineages because of early patterning. Divergence among rock-dwellers and sand-dwellers in the relative size of the telencephalon versus the thalamus is correlated with gene expression variation in a regulatory circuit (composed of six3, fezf2, shh, irx1b, and wnt1) known from model organisms to specify anterior-posterior (AP) brain polarity and position the shh-positive signaling boundary zona limitans intrathalamica (ZLI) in the forebrain. To confirm that changes in this coexpression network are sufficient to produce the differences we observe, we manipulated WNT signaling in vivo by treating rock-dwelling cichlid embryos with temporally precise doses of LiCl. Chemically treated rock-dwellers develop gene expression patterns, ZLIs, and forebrains distinct from controls and untreated conspecifics, but strongly resembling those of sand-dwellers. Notably, endemic Malawi rock- and sand-dwelling lineages are alternately fixed for an SNP in irx1b, a mediator of WNT signaling required for proper thalamus and ZLI. Together, these natural experiments in neuroanatomy, development, and genomics suggest that evolutionary changes in AP patterning establish ecologically relevant differences in the elaboration of cichlid forebrain compartments. In general, variation in developmental patterning might lay the foundations on which neurogenesis erects diverse brain architectures.
Keywords: cichlid, development, evolution, forebrain, WNT signaling
Arguably the most-studied vertebrate organ, the brain has played an important role in the evolution of our own species. Modifications of brain structure are responsible for novel behaviors that galvanized evolutionary radiation of the major vertebrate groups (1). Following decades of research in model organisms, we now know a great deal about how the process of development makes a brain (2). We know much less about evolutionary mechanisms of brain diversification.
The brain develops under the iterative influence of antagonistic anterior and posterior signaling molecules, inductive and repressive transcription factors that receive those signals, and lineage restriction boundaries that define compartments (2, 3). Just after gastrulation, the initial anterior-posterior (AP) polarity of the brain is established by a tug-of-war between posteriorizing signals (e.g., wnt1) secreted from the midbrain–hindbrain boundary (MHB) and WNT antagonists (e.g., six3, tlc) expressed from the anterior neural ridge (ANR). The MHB develops to demarcate the hindbrain from the fore- plus midbrain (Fig. S1). With the subsequent formation of the diencephalon–midbrain boundary and the zona limitans intrathalamica (ZLI), the forebrain and midbrain begin to follow separate paths of development.
These initial boundaries and signaling centers (i.e., ANR, MHB, ZLI) continue to direct additional patterning and morphogenesis within the three major brain regions. For example, the forebrain differentiates into rostral (e.g., telencephalon, hypothalamus) and caudal (e.g., thalamus) domains, mediated in part by the ZLI (4). As brain compartments are patterned, proliferating cells within each compartment undergo neurogenesis, maturing and differentiating into functional neurons. Because brain patterning demarcates one region from another, specific compartments may initiate, prolong, and/or terminate neurogenesis independently (1, 5, 6). Given the continuum of patterning and neurogenesis in brain development, vertebrate lineages might evolve brain diversity by (i) varying the strength or timing of signals from the ANR and/or MHB; (ii) shifting the position of early patterning boundaries; (iii) altering the timing, rate, or extent of neurogenesis; or (iv) some combination thereof. Expectations from the field of evolutionary developmental biology suggest a focus on early patterning events, because such differences prefigure the diversity of animal body plans (7), jaws (8, 9), and dentitions (10). In contrast, our understanding of the brain departs from this notion, as an extensive literature highlights the role of neurogenesis in brain diversification. Most prominently, the “late equals large” model explains how the neocortex (i.e., telencephalon) has evolved to dominate the mammalian brain, and how individual lineages (e.g., primates) have further elaborated this region by tipping the balance among neural cell proliferation, differentiation, and apoptosis (5, 6, 11, 12). Addressing the genetic and developmental mechanisms of brain diversification in nature has been difficult, however, because few systems offer the necessary combination of a wide range of brain phenotypes and tractable experimentation with embryos, against a background of genomic and developmental similarity.
Results and Discussion
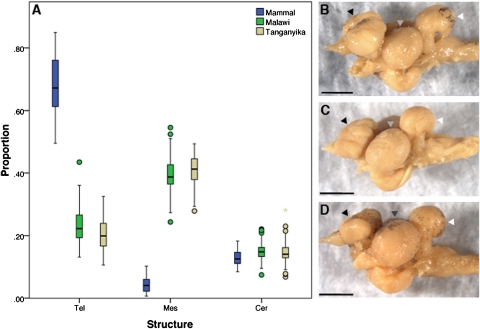
We used cichlid fishes from Lake Malawi to ask when and how brains develop diversity in recently evolved lineages. Cichlid adult brain variation is appreciable (Fig. 1A) and is correlated with ecology and behavior (13–15). For example, algal scrapers exhibit small optic lobes and large telencephala (and olfactory bulbs; Fig. 1B), whereas planktivores have enlarged optic lobes (Fig. 1D). “Sonar” hunters—species that feed by sensing vibrations—have large telencephala and cerebella. This diversity, similar to that observed across seven orders of mammals, including primates, insectivores, marsupials, cetaceans, and bats (16), has evolved rapidly. Hundreds of Lake Malawi cichlid species have diverged from a common ancestor in the last 500,000 years; their genomes are highly similar and retain ancestral polymorphism (17). Malawi cichlid brains are thus as different as those of long-diverged mammals [~150 million years (18)], but their genomes are comparable to those of any two humans.
Fig. 1.
Cichlid brains are diverse. (A) Cerebrotype box plots of mammals (blue), Lake Malawi cichlids (green), and Lake Tanganyika cichlids (tan), grouped by brain proportions of the telencephalon (forebrain), mesencephalon (midbrain), and cerebellum (hindbrain). The heavy line in the middle of the box is the median value, the box itself is the 25–75% interval around the median, the bars are the 10–90% interval, and dots represent data points outside the 95% interval. Mammals have invested heavily in the neocortex (tel); cichlid brains are more proportional in their regional allocations. Despite an order of magnitude difference in divergence time, the range of variation in brain proportions is comparable from mammals to cichlids. Brains of rock-dwelling (mbuna) cichlids. (B) L. fuelleborni (LF, algivore). (C) M. zebra (MZ, generalist). (D) C. afra (CA, planktivore) in lateral view, anterior is to the left. Black arrowheads, telencephala; gray arrowheads, optic tecta; white arrowheads, cerebella. (Scale bar: 2 mm.) Note that the major midbrain structure of fishes (optic tectum) is of uncertain homology to mesencephalic derivatives in mammals.
We focused on species representing the range of ecotypes in Lake Malawi. We studied brain development of three rock-dwelling (mbuna) cichlids—Labeotropheus fuelleborni (LF; obligate algal scraper), Maylandia zebra (MZ; generalist), and Cynotilapia afra (CA; planktivore)—as well as three sand-dwelling nonmbuna—Copadichromis borleyi (CB; planktivore), Mchenga conophorus (MC, generalist), and Aulonocara jacobfreibergi (AJ; sonar hunter). Mbuna and nonmbuna represent distinct evolutionary groups, each containing hundreds of species, with generally contrasting lifestyles, body forms, visual systems, pigment patterns, and trophic adaptations (19–21).
Cichlid Forebrains Differ Early in Development.
By stage 16 (Fig. S1D), Lake Malawi cichlid forebrains have been partitioned into several compartments, visualized in the parasagittal section with the greatest dorsoventral extent (designated “parasagittal section” hereinafter). This is the first stage at which these regions can be reliably measured (Methods and Fig. S2). We quantified the area of forebrain compartments in replicate embryos of the mbuna LF, MZ, and CA and of the nonmbuna CB, MC, and AJ. Embryonic brains show clear divergence between rock-dwelling and sand-dwelling groups (Table 1 and Fig. S3). Rock-dwellers exhibit forebrains with relatively larger telencephala and smaller thalami, and sand-dwellers display the converse pattern. Adult rock-dwelling cichlids have larger telencephala than other habitat specialists on average, perhaps because they spend their lives navigating complex 3D habitats and/or engaging in complex social interactions (13–15). Thalami have been less well studied in fishes, but the vertebrate thalamus is a well-known “relay station” integrating sensory (particularly visual) stimuli (22). Our data demonstrate that differences in cichlid forebrains are apparent early in development, and that these differences might represent a trade-off between rostral and caudal compartments corresponding to adult structures evolved for contrasting ecological demands.
Table 1.
Composition of the cichlid embryonic forebrain at stage 16
| % Tel | % Thal | % Prethal | % Hypothal | |
| MZ (n = 5) | 35.5 ± 0.8 | 18.1 ± 0.7 | 15.3 ± 1.4 | 31.1 ± 1.5 |
| CA (n = 4) | 33.9 ± 2.2 | 19.4 ± 0.4 | 16.2 ± 0.5 | 30.5 ± 2.5 |
| LF (n = 5) | 32.8 ± 0.5 | 20.9 ± 0.5 | 15.3 ± 2.1 | 30.9 ± 2 |
| LFDMSO (n = 3) | 31.7 ± 0.6 | 20.6 ± 0.6 | 16.1 ± 1.2 | 31.6 ± 0.6 |
| LFLiCl (n = 5) | 27.7 ± 1.3 | 24.7 ± 0.9 | 15.7 ± 0.3 | 31.9 ± 0.8 |
| MC (n = 5) | 28.2 ± 0.7 | 24.9 ± 0.9 | 15.6 ± 1.3 | 30.8 ± 1.2 |
| CB (n = 5) | 28 ± 0.8 | 27.7 ± 0.4 | 15.4 ± 1.5 | 28.8 ± 1.3 |
| AJ (n = 4) | 28.4 ± 0.4 | 26.0 ± 0.7 | 15.5 ± 0.8 | 30.1 ± 0.8 |
Mbuna (LF, MZ, and CA) have larger telencephala and smaller thalami than nonmbuna (CB, MC, and AJ). Control (DMSO) and treated (LiCl) LF embryos exhibit similar proportions to mbuna and nonmbuna cichlids, respectively. The area of each compartment is expressed as a percentage (± 1 SD) of the total forebrain area.
Variation in Forebrain Patterning Prefigures Morphological Differences.
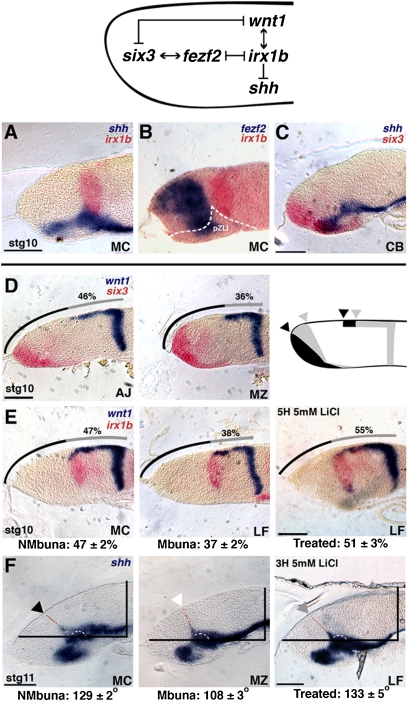
We sought to identify the developmental signals that initiate differences in Malawi cichlid forebrains. Studies in developmental models—zebrafish, frog, and mouse—set the context for our experiments (Fig. 2; network). We focused on a gene circuit known to (i) establish anterior (e.g., telencephalon) versus posterior (e.g., thalamus) fate and (ii) position the signaling boundary ZLI within the forebrain. The transcription factor six3 is a WNT antagonist expressed from the ANR, required for the formation of the telencephalon and ZLI (23). wnt1 is a posteriorizing signal expressed from the MHB; knockout of Wnt1 in mouse results in a smaller thalamus, a posterior shift in the angle of the ZLI, and a larger telencephalon (23). six3 and wnt1 direct the activity of mutually repressive transcription factors fezf2 and irx1, which in turn set the AP position of the shh-positive ZLI (23–25). Knockdown of irx1 in zebrafish produces a posterior expansion of the ZLI at the expense of thalamus, along with a shortening of the wnt1 forebrain domain (25, 26). WNT signaling might induce or be mediated by Irx1 to specify posterior fate (26, 27). We hypothesized that differences between rock-embryonic and sand-embryonic forebrains are produced by temporal and/or spatial shifts in AP forebrain patterning.
Fig. 2.
The forebrain-patterning network differs between rock-dwellers and sand-dwellers. (A) Double in situ hybridization (ISH) of genes shh (blue) and irx1b (red). (B) Double ISH of fezf2 (blue) and irx1b (red); the presumptive ZLI (pZLI) is shown by the dotted white line. (C) Double ISH of genes shh (blue) and six3 (red). A and B are embryos of nonmbuna M. conophorus (MC); C is nonmbuna C. borleyi (CB); A–C are stage 10 embryos. (Scale bar: 100 μm.) (D) From left to right, double ISH of wnt1 (blue) and six3 (red) in A. jacobfreibergi (AJ; nonmbuna), M. zebra (MZ, mbuna), and a schematic summarizing expression differences between nonmbuna and mbuna. Arrowheads mark the relative positions of wnt1 and six3 expression in mbuna (gray) and nonmbuna (black), respectively. (E) From left to right, double ISH of wnt1 (blue) and irx1b (red) in MC (nonmbuna), L. fuelleborni (LF, mbuna), and LF treated for 5 h with 5 mM LiCl. For all panels in D and E, the line above the embryo represents the total length of the dorsal brain anterior to the MHB, and the gray portion of the line represents the rostral extent of wnt1 expression (the wnt1 percentage). The measured wnt1 percentage for each embryo, in each panel, is given. Below row E, from left to right, are the average wnt1 percentages for nonmbuna, mbuna, and LiCl-treated LF, respectively. All embryos in D and E are at stage 10; the scale bars represent 100 μm. (F) ISH for the gene shh demonstrating the angle of the ZLI at stage 11. The ZLI is marked by the black arrowhead in nonmbuna (MC), the white arrowhead in mbuna (MZ), and the gray arrowhead in LiCl-treated LF. The dotted red and white lines show the ZLI angle (Methods). The values below row F show the average ZLI angles for nonmbuna, mbuna, and LiCl-treated LF, respectively. (Scale bar: 100 μm.) All images from all panels are parasagittal sections, with anterior to the left.
Using two-color in situ hybridization (Methods), we observed expected gene expression patterns of six3, wnt1, fezf2, irx1b, and shh in the cichlid embryonic brain. The ZLI begins to form as an initial wedge of shh at stage 10; fezf2 is expressed rostral to the wedge, and irx1b is positioned caudal to the wedge, in the presumptive thalamus (Fig. 2 A–C). The antagonists six3 and wnt1 are initially localized to the ANR and MHB, respectively, at the neurula stage. wnt1 extends rostrodorsally from the MHB as development proceeds, encompassing the presumptive midbrain and the caudal forebrain, where it is coexpressed with irx1b. At stage 10, six3 and wnt1 show contrasting distributions between mbuna and nonmbuna cichlids (Fig. 2D). Mbuna are characterized by a shortened wnt1 rostrodorsal domain and more caudodorsal expression of six3; nonmbuna exhibit the opposite pattern. Despite species-specific brain shapes, the wnt1 rostral domain marks a greater proportion of the dorsal brain anterior to the MHB in stage 10 nonmbuna CB, MC, and AJ (47 ± 2%) than in mbuna LF, MZ, and CA (37 ± 2%; Student's t test, two-tailed; t = 11.78; P < 0.0001; between three and seven individuals of each species, n = 32 embryos) (Fig. 2 D and E).
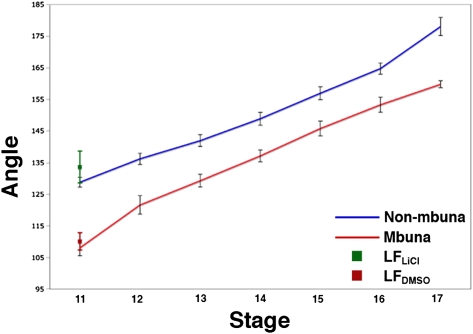
By stage 11, the ZLI is a narrowing finger of shh expression within the diencephalon, forming a characteristically obtuse angle with the alar domain (Fig. 2F). We observed that the ZLI angle is greater in nonmbuna CB, MC, and AJ than in mbuna LF, MZ, and CA (129° ± 2° vs. 108° ± 3, Student's t test, two-tailed; t = 18.24; P < 0.0001; between two and four individuals of each species; n = 20 embryos). The larger ZLI angle in nonmbuna, as measured at stage 11, matches the more rostrodorsal expression of wnt1-irx1b and reduced six3 domain at stage 10. We tracked the angle of the shh-positive ZLI from stage 11 to stage 17 (encompassing 3 days of development and the time point of forebrain compartment measurements; Table 1) in replicate embryos of the three mbuna and nonmbuna species (between two and five embryos per species per stage; n = 120 embryos). In both mbuna and nonmbuna, the angle of the ZLI increases as thalamic and tectal structures grow and proliferate, yet the nonmbuna maintain a greater ZLI angle throughout (Fig. 3 and Fig. S4).
Fig. 3.
The angle of the signaling boundary ZLI impacts brain regionalization during neurogenesis and growth. The graph presents the average ZLI angle (± 1SD; between two and five specimens per species per stage; n = 120) over seven ontogenetic stages for nonmbuna CB, MC, and AJ (blue) and mbuna LF, MZ, and CA (red) (also see Fig. S4). The green and red data points at stage 11 represent LiCl-treated and DMSO control LF embryos, respectively.
Manipulation of WNT Signaling Mimics Natural Variation Among Cichlid Forebrains.
The brain and gene expression phenotypes that we observed to differentiate mbuna cichlids (reduced rostrodorsal extent of wnt1-irx1b, more acute ZLI angle, smaller thalamus, larger telencephalon) from nonmbuna cichlids (greater rostrodorsal extent of wnt1-irx1b, more obtuse ZLI angle, larger thalamus, smaller telencephalon) (Table 1 and Fig. 2) partially phenocopy zebrafish irx1 knockdown versus control embryos (23) and Wnt1 null versus control mice (26). Thus, we manipulated WNT signaling in vivo by treating cichlid embryos with nonlethal doses of the chemical agonist LiCl (Methods). This approach does not allow the genetic specificity of other methods, such as morpholinos, but does provide the temporal precision critical to our experiments. We bathed stage 9 embryos (during which wnt1 expression “moves” rostrodorsally from the MHB; Fig. S1) of the mbuna LF in a 5 mM solution of LiCl or a vehicle control (DMSO), for 3 or 5 h. We then washed the embryos, returned them to fish water, and cultured them until sacrifice (i) at stage 10 to measure the rostrodorsal extent of the wnt1-irx1b expression domain, (ii) at stage 11 to measure the angle of the shh-positive ZLI, or (iii) at stage 16 to measure the relative area of forebrain compartments. We predicted that up-regulation of WNT signaling would transform the treated mbuna brains of LF to resemble those of nonmbuna.
At stage 10, LiCl-treated LF (n = 6; 3 and 5 h treatments combined) exhibited wnt1-irx1b expression domains that covered a greater proportion of the dorsal brain anterior to the MHB (51 ± 3%) compared with DMSO controls (n = 4, 37 ± 1%; Student's t test, two-tailed; t = 9.05; P < 0.0001) (Fig. 2E, Right). At stage 11, LiCl-treated LF (n = 4; 3 and 5 h treatments combined) exhibited greater ZLI angles (133° ± 5°) than did DMSO controls (n = 2; 110° ± 3°; Student's t test, two-tailed; t = 5.96; P = 0.004) (Fig. 2F, Right). Finally, at stage 16, LiCl-treated LF (n = 5; 3 and 5 h treatments combined) exhibited smaller telencephala (27.7 ± 1.3%) and larger thalami (24.7 ± 0.9%) compared with DMSO controls (n = 3; 31.7 ± 0.6% for telencephala and 20.6 ± 0.6% for thalami; Table 1). The areas of prethalami and hypothalami did not differ between treatment and control embryos (Table 1). Our measurements for LF bathed in DMSO (controls) fell within the statistical distribution for untreated mbuna, and the LiCl-treated LF embryos had wnt1-irx1b gene expression patterns, ZLIs, and forebrains that strongly resembled those of nonmbuna (Table 1 and Figs. 2 and 3). These data demonstrate that manipulation of WNT signaling during early embryogenesis is sufficient to produce distinct cichlid forebrains, which nearly exactly mimic the natural developmental differences between rock-dwellers and sand-dwellers.
An SNP in irx1b Is Alternately Fixed Between Rock-Dwellers and Sand-Dwellers.
We wanted to identify genetic differences between mbuna and nonmbuna that might contribute to variation in gene expression and brain phenotypes. We did not use the typical approach to map quantitative trait loci responsible for phenotypic variance (9, 28) because we could not effectively cross chosen rock-dweller with sand-dweller individuals. Therefore, we explored an alternative strategy. Because mbuna and nonmbuna evolutionary groups diverged recently, the genomes of individuals share polymorphism across the lineage boundary (17). Loci that are strongly genetically differentiated between lineages are statistical outliers. For instance, only 1 of 96 independent SNPs is alternately fixed in a large sample of Malawi mbuna versus nonmbuna. The SNP is a replacement change in the 3′ coding sequence of the transcription factor irx1b, fixed between endemic Malawi mbuna (25 species, 140 alleles) and nonmbuna (52 species, 230 alleles) (Fig. S5 and SI Methods). This represents a signature of divergent selection against a background of shared polymorphism (17), and suggests that genetic variation in the irx1b cistron might play a role in the differentiation of Malawi cichlid forebrains. At this point, we are uncertain whether this substitution itself is causative or linked (physically and/or epistatically) to other causal mutations, but the central position of irx1b in the forebrain-patterning network (Fig. 2) implies that mbuna and nonmbuna alleles might interact differently with fezf2, wnt1, and/or shh. These data, coupled with the discovery that two amino acid changes between the human versus chimpanzee gene Foxp2 can drive considerable transcriptional variation in the brain (29), make cichlid irx1b a prime target of future study.
Brain Diversity by Patterning Differences.
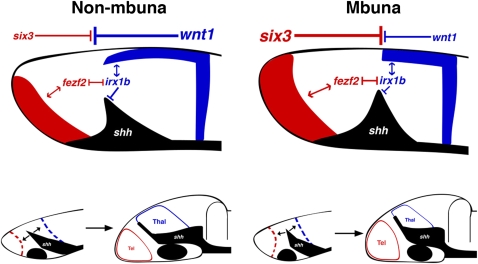
We interpret our natural experiments in comparative neuroanatomy, development, and genomics, coupled with functional information from model organisms, to indicate that evolutionary modifications in a gene circuit composed of six3, fezf2, irx1b, wnt1, and shh establish two distinct modes of AP forebrain patterning in mbuna versus nonmbuna Malawi cichlids (Fig. 4). Nonmbuna embryonic forebrains are dominated by posterior signals (e.g., wnt1) and ultimately elaborate a posterior structure (thalamus), whereas their mbuna counterparts are under greater influence of the ANR (e.g., six3) and elaborate the telencephalon. Anterior and posterior signaling gradients converge on the transcription factors fezf2 and irx1b, which integrate these cues to position the angle of the ZLI. This angle, once set, is important because it (i) apportions cells to the anterior versus the posterior forebrain and (ii) represents a boundary restricting further AP cross-talk between ANR and MHB. Our data highlight early patterning variation that initiates contrasting forebrain bauplans in rock-dwelling versus sand-dwelling Malawi cichlids. Species- and ecotype-specific adult brains (e.g., Fig. 1) are likely the result of this initial difference, modified throughout ontogeny by neurogenesis. Thus, the initial angle of the signaling boundary ZLI might impact subsequent forebrain patterning and neurogenesis in ecologically distinct Malawi cichlids.
Fig. 4.
Brain diversity develops at the boundaries. A model summarizes the evolutionary developmental differences between nonmbuna and mbuna forebrains. At stage 10 (Upper), the allocation of forebrain structures is determined via the competing influence of posteriorizing factors from the MHB (e.g., wnt1, shown in blue) versus WNT antagonists expressed from the ANR (e.g., six3, shown in red). This in turn sets the position and angle of the presumptive ZLI (black). In nonmbuna, posterior factors dominate the forebrain, establishing a greater (i.e., more obtuse) ZLI angle relative to mbuna. This results in the differential allocation of cells to anterior versus posterior forebrain compartments. During subsequent stages 11–16 (Lower), the initial difference in ZLI angle set during early patterning persists, with the consequence of a smaller telencephalon (tel; red) but larger thalamus (thal; blue) in nonmbuna versus mbuna.
We suggest a new interpretation of how organisms evolve brain diversity: Differences in the earliest signaling and patterning centers establish divergent blueprints for elaboration during growth and neurogenesis. Such comparisons are difficult to make in mam-mals, because deep evolutionary distances separate lineages and considerable size variation obscures brain adaptations (11, 16, 30). Our conclusion extends those from studies of cavefishes and birds (31, 32), and suggests that evolution might capture any point along the continuum of patterning and neurogenesis as neural systems diversify.
Methods
Cerebrotype Analysis.
Mammalian brain data (multiple individuals of 75 species spanning seven taxonomic orders) are from the supplementary material of Clark et al. (16). The Malawi data (222 individuals from 113 species) represent a combination of published material (33) and new measurements made on preserved, wild-caught (from 2005) adult specimens for this study. The Tanganyika data (58 individuals, 53 species) are entirely from reference 33. Volumes for the Malawi and Tanganyika measurements were generated using the ellipsoid model, (L × W × H)π/6 (14,33). All cerebrotypes were calculated using the method of Clark et al. (16). Boxplots were generated using SPSS version 16.0.
Embryo Staging.
Embryos were removed from the mouths of brooding females ~24 h after identification and, if required, maintained for further development in culture flasks at 28 °C. Embryos were observed every 8–12 h until they reached the desired stage. Stages were determined for Malawi cichlids by comparing embryonic morphology with zebrafish (34) and tilapia (35) descriptions. Identification of fish brain anatomy followed reference 36.
Embryonic Forebrain Measurements.
We measured the areas of forebrain compartments in mbuna [L. fuelleborni (LF), M. zebra (MZ), and C. afra (CA)] and in nonmbuna [C. borleyi (CB), M. conophorus (MC), and A. jacobfreibergi (AJ)]. We chose these species (LF, algal scraper; MZ, generalist; CA, planktivore; CB, planktivore; MC, generalist; AJ, sonar hunter) to represent the range of the ecological diversity within these evolutionary lineages. Measurements were made at the first developmental stage (stage 16) in which compartments are demarcated by cellular restriction boundaries and/or cellular behavior (Fig. S2). All measurements were made on scaled digital images of parasagittal sections using ImageJ software. In addition, we used anatomical landmarks and gene expression patterns to ensure that measurements were taken from comparable serial sections (e.g., Figs. S2–S4). We defined the total forebrain area as the region anterior to the posterior commissure, including the hypothalamus (36, 37). The pretectum was not included in our measurements, because it could not be consistently visualized in parasagittal section. The prethalamus included the preoptic region, and the hypothalamus included the presumptive posterior tuberculum (36). To eliminate the confounding effect of oblique sections, we typically sectioned 10–20 embryos per species (often from different broods) and selected 4 or 5 for measurement. The area of each compartment was expressed as a percentage of total forebrain area.
In Situ Hybridization.
ISH experiments were based on published protocols (10), with modification for double ISH. Gene sequences were derived from partial genome assemblies of Lake Malawi cichlids (17). Probes were constructed from cDNA sequences identical in the species examined. In general, Lake Malawi cichlids exhibit genetic variation comparable to that observed across laboratory strains of zebrafish (17). Embryos were hybridized with both fluorescein-labeled (Roche) and digoxigenin-labeled RNA probes. The fluorescein was visualized first, by treating the embryos with anti–fluorescein-AP sheep antibody (Roche), followed by FastRed tablets (1 tablet every 2 mL of 0.1M Tris-HCl; Roche). Once the color reaction was complete, the antibody was inactivated with 0.1 M glycine-HCl (Polysciences). Embryos were then fixed briefly in 4% PFA, and the digoxigenin-labeled probes were visualized as described previously (10). All ISH experiments were performed with multiple specimens (multiple individuals fixed at regular intervals, within single broods, then repeated at least twice with alternative broods) to fully characterize expression patterns within and across species. Embryos were embedded in gelatin and chick albumin with 2.5% gluteraldehyde. The gelatin-albumin blocks were postfixed in 4% PFA before sectioning. Thin sections were cut at 15 μm using a Leica Microsystems VT1000 vibratome and imaged using a Leica Microsystems DM2500 compound microscope.
Measuring the Rostrodorsal Extent of wnt1 Expression.
At stage 10, wnt1 is expressed in the MHB, as well as rostrally along the dorsal surface of the cichlid embryonic prosencephalon (midbrain plus forebrain). wnt1 is a posteriorizing signal that functions across the dorsal embryonic brain (3, 23). As such, we wanted to calculate and compare the percentage of the dorsal prosencephalon under the influence of the wnt1 signal in mbuna and nonmbuna. From photographs of embryos in parasagittal sections, we used Image J to measure the length of a curved line from the MHB to the rostral-most tip of the embryo, generally identified by a noticeable ‘lip’ demarcating dorsal from ventral (Fig. 2). This represents the total length of the dorsal prosencephalon. We next measured the rostral extent of wnt1 expression and calculated the percentage of the dorsal prosencephalon covered by wnt1 expression. We performed these measurements in replicate embryos of mbuna versus nonmbuna species, as well as in LiCl treatment versus control LF embryos.
Measuring the Angle of the ZLI.
At stage 11, shh expression in the ZLI forms a characteristic angle with shh expression in the alar domain; this angle persists during subsequent stages. The ZLI angle was measured from stage 11–17 using Image J, from photographs of parasagittal sections. To standardize measurement across developmental time points, two guidelines were added to each image (e.g., Fig. 2, Fig. S4). The first marks the position of the vertical midbrain-hindbrain boundary (MHB) and the second is perpendicular to the first, and typically parallel to shh expression in the alar domain. This second line served as a consistent reference point across all stages to account for any irregularities, in section, of the shh alar domain. The ZLI angle was measured using the second line as the ‘base’ of the angle, and the position of the ZLI as the “arm” of the angle. Multiple (2–5) embryos per species of the mbuna LF, MZ, and CA, as well as nonmbuna CB, MC, and AJ, were measured across the seven developmental stages (n = 120 embryos).
Chemical Treatments.
A 4 M lithium chloride (LiCl) stock solution was made by dissolving 640 mg of high purity LiCl (Alexis Biochemicals) into 15 mL of Dimethyl Sulfoxide (DMSO, MP Biomedicals). The LiCl stock was diluted to a final experimental concentration of 5mM (8.75 μl 4M LiCl in 7 mL of fish water). Embryos were taken from the mbuna, L. fuelleborni (LF), at stage 9, around 36–40 h post fertilization. Approximately 10–12 individuals were placed in separate 5mM LiCl cultures for either 3 or 5 h, at 28 °C. Additionally, 5–7 embryos were placed in 0.125% DMSO (8.75 μl DMSO in 7 mL fish water) for 3 or 5 h, at 28 °C. After treatment, embryos were washed twice with fish water, and placed in fresh fish water, in culture flasks at 28 °C. Embryos were removed from culture and killed (i) at stage 10 to measure the rostro-dorsal extent of the wnt1-irx1b expression domain, (ii) at stage 11 to measure the angle of the shh-positive ZLI, or (iii) at stage 16 to measure the relative area of forebrain compartments. The experiment was repeated twice, with two different LF broods. Treatment and control embryos were postprocessed (ISH, sectioning, measurements) identically to descriptions above for untreated embryos.
Supplementary Material
Acknowledgments
We thank Corinne Houart, Tom Kocher, Karen Carleton, Hans Hofmann, and Craig Albertson for their comments on pre-vious drafts of this manuscript. Our research is supported by awards from the National Science Foundation (IOS 0546423 and IOS 0922964), National Institutes of Health (DE017182), and the Alfred P. Sloan Foundation (BR4499) to J.T.S. J.B.S. is a Georgia Institute of Technology Presidential Fellow. C.A.R. was a Georgia Institute of Technology Institute for Bioengineering and Biosciences Undergraduate Research Scholar.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000395107/-/DCSupplemental.
References
- 1.Striedter GF. Principles of Brain Evolution. Sunderland, MA: Sinauer Associates; 2005. [Google Scholar]
- 2.Kiecker C, Lumsden A. Compartments and their boundaries in vertebrate brain development. Nat Rev Neurosci. 2005;6:553–564. doi: 10.1038/nrn1702. [DOI] [PubMed] [Google Scholar]
- 3.Wilson SW, Houart C. Early steps in the development of the forebrain. Dev Cell. 2004;6:167–181. doi: 10.1016/s1534-5807(04)00027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scholpp S, Wolf O, Brand M, Lumsden A. Hedgehog signaling from the zona limitans intrathalamica orchestrates patterning of the zebrafish diencephalon. Development. 2006;133:855–864. doi: 10.1242/dev.02248. [DOI] [PubMed] [Google Scholar]
- 5.Finlay BL, Darlington RB, Nicastro N. Developmental structure in brain evolution. Behav Brain Sci. 2001;24:263–278. [PubMed] [Google Scholar]
- 6.Takahashi T, Nowakowski RS, Caviness VS., Jr. The leaving or Q fraction of the murine cerebral proliferative epithelium: A general model of neocortical neuronogenesis. J Neurosci. 1996;16:6183–6196. doi: 10.1523/JNEUROSCI.16-19-06183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerhart J, Kirschner M. Cells, Embryos, and Evolution Toward a Cellular and Developmental Understanding of Phenotypic Variation and Evolutionary Adaptability. Malden, MA: Blackwell Science; 1997. [Google Scholar]
- 8.Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin's finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- 9.Albertson RC, Streelman JT, Kocher TD, Yelick PC. Integration and evolution of the cichlid mandible: The molecular basis of alternate feeding strategies. Proc Natl Acad Sci USA. 2005;102:16287–16292. doi: 10.1073/pnas.0506649102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser GJ, Bloomquist RF, Streelman JT. A periodic pattern generator for dental diversity. BMC Biol. 2008 doi: 10.1186/1741-7007-6-32. 10.1186/1741-7007-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finlay BL, Darlington RB. Linked regularities in the development and evolution of mammalian brains. Science. 1995;268:1578–1584. doi: 10.1126/science.7777856. [DOI] [PubMed] [Google Scholar]
- 12.Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- 13.Huber R, van Staaden MJ, Kaufman LS, Liem KF. Microhabitat use, trophic patterns, and the evolution of brain structure in African cichlids. Brain Behav Evol. 1997;50:167–182. doi: 10.1159/000113330. [DOI] [PubMed] [Google Scholar]
- 14.Pollen AA, et al. Environmental complexity and social organization sculpt the brain in Lake Tanganyikan cichlid fish. Brain Behav Evol. 2007;70:21–39. doi: 10.1159/000101067. [DOI] [PubMed] [Google Scholar]
- 15.Shumway CA. Habitat complexity, brain, and behavior. Brain Behav Evol. 2008;72:123–134. doi: 10.1159/000151472. [DOI] [PubMed] [Google Scholar]
- 16.Clark DA, Mitra PP, Wang SSH. Scalable architecture in mammalian brains. Nature. 2001;411:189–193. doi: 10.1038/35075564. [DOI] [PubMed] [Google Scholar]
- 17.Loh YHE, et al. Comparative analysis reveals signatures of differentiation amid genomic polymorphism in Lake Malawi cichlids. Genome Biol. 2008;9:R113. doi: 10.1186/gb-2008-9-7-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 19.Carleton KL, et al. Visual sensitivities tuned by heterochronic shifts in opsin gene expression. BMC Biol. 2008;6:22. doi: 10.1186/1741-7007-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hulsey CD, Mims MC, Streelman JT. Do constructional constraints influence cichlid craniofacial diversification? Proc R Soc B. 2007;274:1867–1875. doi: 10.1098/rspb.2007.0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Streelman JT, Danley PD. The stages of vertebrate evolutionary radiation. Trends Ecol Evol. 2003;18:126–131. [Google Scholar]
- 22.Jones EG. The Thalamus. , 2nd Ed. Cambridge, UK: Cambridge Univ Press; 2007. [Google Scholar]
- 23.Lavado A, Lagutin OV, Oliver G. Six3 inactivation causes progressive caudalization and aberrant patterning of the mammalian diencephalon. Development. 2008;135:441–450. doi: 10.1242/dev.010082. [DOI] [PubMed] [Google Scholar]
- 24.Rodríguez-Seguel E, Alarcón P, Gómez-Skarmeta JL. The Xenopus Irx genes are essential for neural patterning and define the border between prethalamus and thalamus through mutual antagonism with the anterior repressors Fezf and Arx. Dev Biol. 2009;329:258–268. doi: 10.1016/j.ydbio.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 25.Scholpp S, et al. Otx1l, Otx2 and Irx1b establish and position the ZLI in the diencephalon. Development. 2007;134:3167–3176. doi: 10.1242/dev.001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Itoh M, Kudoh T, Dedekian M, Kim CH, Chitnis AB. A role for iro1 and iro7 in the establishment of an anteroposterior compartment of the ectoderm adjacent to the midbrain-hindbrain boundary. Development. 2002;129:2317–2327. doi: 10.1242/dev.129.10.2317. [DOI] [PubMed] [Google Scholar]
- 27.Gómez-Skarmeta JL, de La Calle-Mustienes E, Modolell J. The Wnt-activated Xiro1 gene encodes a repressor that is essential for neural development and down-regulates Bmp4. Development. 2001;128:551–560. doi: 10.1242/dev.128.4.551. [DOI] [PubMed] [Google Scholar]
- 28.Streelman JT, Albertson RC. Evolution of novelty in the cichlid dentition. J Exp Zool B. 2006;306:216–226. doi: 10.1002/jez.b.21101. [DOI] [PubMed] [Google Scholar]
- 29.Konopka G, et al. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462:213–217. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barton RA, Harvey PH. Mosaic evolution of brain structure in mammals. Nature. 2000;405:1055–1058. doi: 10.1038/35016580. [DOI] [PubMed] [Google Scholar]
- 31.Menuet A, Alunni A, Joly JS, Jeffery WR, Rétaux S. Expanded expression of Sonic Hedgehog in Astyanax cavefish: Multiple consequences on forebrain development and evolution. Development. 2007;134:845–855. doi: 10.1242/dev.02780. [DOI] [PubMed] [Google Scholar]
- 32.Striedter GF, Charvet CJ. Developmental origins of species differences in telencephalon and tectum size: Morphometric comparisons between a parakeet (Melopsittacus undulatus) and a quail (Colinus virgianus) J Comp Neurol. 2008;507:1663–1675. doi: 10.1002/cne.21640. [DOI] [PubMed] [Google Scholar]
- 33.van Staaden MJ, Huber R, Kaufman LS, Liem KF. Brain evolution in cichlids of the African great lakes: Brain and body size, general patterns, and evolutionary trends. Zoology. 1994;98:165–178. [Google Scholar]
- 34.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 35.Fujimura K, Okada N. Development of the embryo, larva and early juvenile of Nile tilapia Oreochromis niloticus (Pisces: Cichlidae): Developmental staging system. Dev Growth Differ. 2007;49:301–324. doi: 10.1111/j.1440-169X.2007.00926.x. [DOI] [PubMed] [Google Scholar]
- 36.Wullimann MF, Puelles L. Postembryonic neural proliferation in the zebrafish forebrain and its relationship to prosomeric domains. Anat Embryol (Berl) 1999;199:329–348. doi: 10.1007/s004290050232. [DOI] [PubMed] [Google Scholar]
- 37.Puelles L, Rubenstein JLR. Forebrain gene expression domains and the evolving prosomeric model. Trends Neurosci. 2003;26:469–476. doi: 10.1016/S0166-2236(03)00234-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.