Abstract
Macrophages respond to cytosolic nucleic acids by activating cysteine protease caspase-1 within a complex called the inflammasome. Subsequent cleavage and secretion of proinflammatory cytokines IL-1β and IL-18 are critical for innate immunity. Here, we show that macrophages from mice lacking absent in melanoma 2 (AIM2) cannot sense cytosolic double-stranded DNA and fail to trigger inflammasome assembly. Caspase-1 activation in response to intracellular pathogen Francisella tularensis also required AIM2. Immunofluorescence microscopy of macrophages infected with F. tularensis revealed striking colocalization of bacterial DNA with endogenous AIM2 and inflammasome adaptor ASC. By contrast, type I IFN (IFN-α and -β) secretion in response to F. tularensis did not require AIM2. IFN-I did, however, boost AIM2-dependent caspase-1 activation by increasing AIM2 protein levels. Thus, inflammasome activation was reduced in infected macrophages lacking either the IFN-I receptor or stimulator of interferon genes (STING). Finally, AIM2-deficient mice displayed increased susceptibility to F. tularensis infection compared with wild-type mice. Their increased bacterial burden in vivo confirmed that AIM2 is essential for an effective innate immune response.
Keywords: inflammasome, innate immunity, interferon, apoptosis-associated speck-like protein containing a caspase recruitment domain
The innate immune system reacts to diverse molecules that are collectively termed pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) (1, 2). These molecules include nucleic acids. RNA, for example, is recognized by several toll-like receptors as well as the RNA helicases retinoic acid-inducible gene-I (RIG-I; also called DDX58), melanoma differentiation-associated gene-5 (MDA5; also called IFIH1), and laboratory of genetics and physiology 2 (LGP2) (1). DNA recognition mechanisms have proved more elusive. Toll-like receptor (TLR) 9 is located in phagosomes and recognizes DNA with unmethylated cytosine-phosphate-guanine (CpG) motifs, leading to NF-κB–dependent inflammatory responses (3).The DNA-dependent activator of IFN-regulatory factors (DAI; also known as DLM-1 and ZBP1), the first identified cytosolic DNA sensor, binds cytosolic dsDNA and leads to the production of IFN-I, although the lack of proven relevance in vivo has lead to the hypothesis that redundant cytosolic DNA sensors exist (4, 5). Additionally, the recently identified adapter stimulator of interferon genes (STING) or mediator of interferon regulatory factor 3 (IRF3) activation (MITA; hereafter referred to as STING) mediates IFN-I production in response to DNA transfection as well bacterial and viral infection (6–9).
DNA is also a potent activator of a multiprotein complex known as the inflammasome, which contains a nucleotide-binding oligomerization domain (NOD)-like receptor (NLR), the adapter apoptosis-associated speck-like protein containing a caspase recruitment domain (CARD) (ASC; also known as PYCARD), and the cysteine protease caspase-1 (10). Recent overexpression and knockdown studies in cell lines suggested that DNA engages absent in melanoma 2 (AIM2), an NLR, that then interacts with ASC to promote inflammasome assembly and caspase-1 activation (11, 12). AIM2 is an IFN-I–inducible cytosolic protein containing pyrin (PYD) and hematopoietic interferon-inducible nuclear antigens with 200 amino acid repeats (HIN200) domains (12, 13) (Fig. S1C). The HIN domain facilitates binding of DNA, whereas the pyrin domain allows for the association with ASC and formation of a caspase-1–activating inflammasome, leading to the processing and release of mature IL-1β and IL-18 and host cell death (11, 12, 14). The role of AIM2 in innate immunity is unknown.
The causative agent of tularemia, Francisella tularensis, is a facultative intracellular Gram-negative pathogen that escapes phagosomal degradation in macrophages and replicates in the host cell cytosol. Cytosolic replication is required for bacterial virulence, because F. tularensis mutants that fail to escape the vacuole cannot replicate in macrophages and are avirulent in mice (15–19). Moreover, cytosolic F. tularensis sequentially activates proinflammatory host responses, characterized by the initial production of IFN-I, such as IFN-β, that is required for the subsequent activation of an ASC inflammasome (20, 21). Inflammasome activation is critical to host defense against F. tularensis, because mice lacking inflammasome components are more susceptible to infection (22). The PAMPs produced during F. tularensis infection and the host pattern recognition receptors (PRRs) required for pathogen recognition remain a mystery.
Results
AIM2 Is Essential for Inflammasome Activation in Response to Cytosolic dsDNA.
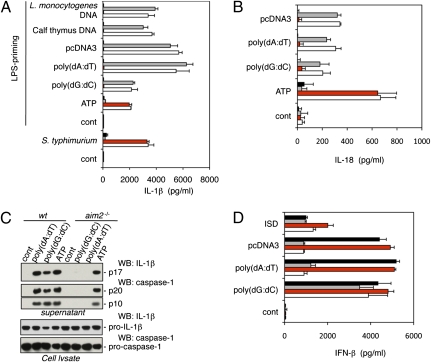
We investigated the role of AIM2 in vivo with gene-targeted aim2−/− mice (Fig. S1 A and B). Western blotting with an antibody raised against amino acids 2–274 of mouse AIM2 confirmed that aim2−/− bone marrow-derived macrophages (BMDMs) lacked AIM2 protein (Fig. S1C). First, we compared dsDNA-induced IL-1β secretion from aim2−/−, asc−/−, nlrp3−/−nlrc4−/−, and wild-type BMDMs. Cells were primed with LPS to induce pro–IL-1β expression and then, were transfected with poly(dA-dT) · poly(dA-dT) [hereafter referred to as poly(dA:dT)], poly(dG-dC) · poly(dG-dC) [hereafter referred to as poly(dG:dC)], pcDNA3 plasmid DNA, calf thymus DNA, or Listeria monocytogenes DNA. All of these dsDNAs induced AIM2- and ASC-dependent IL-1β secretion (Fig. 1A). In contrast, NLR family, pyrin domain containing 3 (NLRP3) (23, 24) (also called Nalp3, cryopyrin, or CIAS1), which engages the caspase-1 adaptor protein ASC in response to a variety of PAMPs and DAMPs, and NLR family, CARD domain containing 4 (NLRC4) (25) (also called Ipaf), which engages ASC in response to Salmonella typhimurium flagellin, were dispensable for dsDNA-induced IL-1β secretion. Loss of AIM2, unlike ASC deficiency, did not cause a general defect in IL-1β secretion, because aim2−/− and wild-type BMDMs secreted equivalent amounts of IL-1β after infection with S. typhimurium or treatment with LPS plus ATP (Fig. 1A). Similar results were obtained with peritoneal macrophages (Fig. S2). AIM2 deficiency also blocked IL-18 secretion in response to dsDNA but not ATP (Fig. 1B). Therefore, AIM2 is essential for IL-18 and IL-1β secretion in response to dsDNA.
Fig. 1.
AIM2 is essential for inflammasome activation in response to cytosolic dsDNA. ■, asc−/−;  , nlrp3−/− nlrc4−/−;
, nlrp3−/− nlrc4−/−;  , aim2−/−; □, wt. (A) IL-1β secretion by LPS-primed BMDMs treated with 5 mM ATP or transfected with 1 μg/mL of the indicated dsDNAs for 16 h. BMDMs were infected with S. typhimurium (multiplicity of infection = 100) without LPS priming. (B) IL-18 secretion by BMDMs treated as in A. LPS priming was used for ATP stimulation only. (C) Upper shows mature IL-1β and cleaved caspase-1 secreted from LPS-primed BMDMs after stimulation with ATP or transfection with dsDNA for 5 h. Lower shows procaspase-1 and pro–IL-1β in the cell lysate. (D) IFN-β secretion by BMDMs treated as in A but without LPS priming. Graphs show the mean ± SD of triplicate wells and are representative of three independent experiments.
, aim2−/−; □, wt. (A) IL-1β secretion by LPS-primed BMDMs treated with 5 mM ATP or transfected with 1 μg/mL of the indicated dsDNAs for 16 h. BMDMs were infected with S. typhimurium (multiplicity of infection = 100) without LPS priming. (B) IL-18 secretion by BMDMs treated as in A. LPS priming was used for ATP stimulation only. (C) Upper shows mature IL-1β and cleaved caspase-1 secreted from LPS-primed BMDMs after stimulation with ATP or transfection with dsDNA for 5 h. Lower shows procaspase-1 and pro–IL-1β in the cell lysate. (D) IFN-β secretion by BMDMs treated as in A but without LPS priming. Graphs show the mean ± SD of triplicate wells and are representative of three independent experiments.
Processing of pro–IL-1β and pro–IL-18 by caspase-1 is necessary for secretion of biologically active IL-1β and IL-18 (26, 27), and therefore, we compared caspase-1 activation in wild-type and aim2−/− BMDMs by Western blotting for the p20 and p10 caspase-1 subunits that are generated by autocatalytic cleavage and released from the cell by a poorly defined mechanism. Consistent with AIM2 promoting caspase-1 activation in response to dsDNA, culture supernatants from LPS-primed wild-type BMDMs contained mature IL-1β plus the caspase-1 p10 and p20 subunits after transfection with poly(dA:dT) or poly(dG:dC), but supernatants from aim2−/− BMDMs did not (Fig. 1C). aim2−/− BMDMs expressed wild-type levels of procaspase-1 and pro–IL-1β, and they released IL-1β and processed caspase-1 normally in response to LPS plus ATP. These data indicate a specific requirement for AIM2 in caspase-1 activation by dsDNA.
Next, we determined if aim2−/− BMDMs produce inflammasome-independent proinflammatory cytokines such as IFN-I and TNF-α in response to dsDNA. Traf family member-associated NF-κB activator (TANK)-binding kinase 1 (TBK1) and the transcription factors IRF3 and IRF7 signal IFN-I synthesis in response to dsDNA (28, 29), but how dsDNA engages this pathway is unclear. Unexpectedly, aim2−/−, asc−/−, and caspase-1−/− BMDMs produced significantly more TNF-α (Fig. S3A) and IFN-β (Fig. 1D and Fig. S3C) than wild-type or nlrp3−/−nlrc4−/− BMDMs after transfection with poly(dA:dT) or pcDNA3. There was little or no difference in IFN-β and TNF-α production, however, when the cells were transfected with poly(dG:dC) or a 45-bp IFN-stimulatory DNA (ISD) (29). In addition, wild-type, aim2−/−, asc−/−, and nlrp3−/−nlrc4−/− BMDMs produced equivalent TNF-α in response to LPS (Fig. S3A). Therefore, AIM2 is dispensable for IFN-β and TNF secretion. Increased IFN-β and TNF-α production by aim2−/− and caspase-1−/− BMDMs in response to poly(dA:dT) or pcDNA3 correlated with enhanced cell survival. Between 35% and 45% of wild-type BMDMs had released lactate dehydrogenase (LDH) at 5 h after transfection with poly(dA:dT) or pcDNA3, whereas most caspase-1−/− and aim2−/− BMDMs remained viable (Fig. S3B). Poly(dG:dC) was less cytotoxic, killing ~15% of wild-type BMDMs. Consistent with AIM2 engaging ASC and caspase-1 in response to dsDNA but not all stimuli, aim2−/− BMDMs were as sensitive as wild-type BMDMs to caspase-1–dependent death after infection with S. typhimurium.
AIM2 Is Required for Inflammasome Activation in Response to F. tularensis.
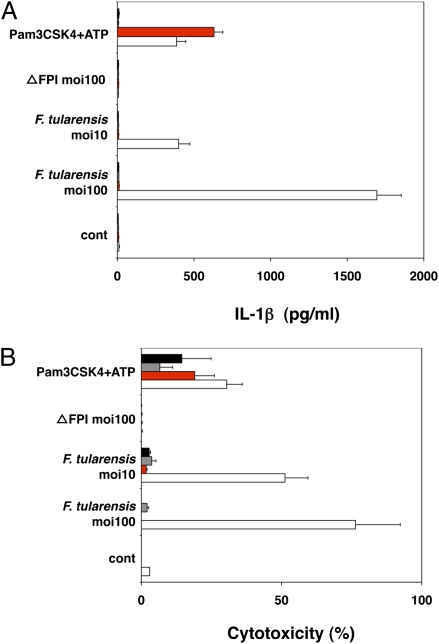
We then explored the contribution of AIM2 to innate immunity to bacterial infection. We focused on inflammasome activation in response to F. tularensis (30, 31), because this intracellular pathogen escapes phagosomal degradation, replicates in the cytosol, and triggers ASC-dependent but NLRP3- and NLRC4-independent caspase-1 activation (22). When wild-type, aim2−/−, asc−/−, and caspase-1−/− BMDMs were infected with F. tularensis subspecies novicida, only wild-type cells secreted IL-1β (Fig. 2A) and died (Fig. 2B), indicating that AIM2, like ASC and caspase-1, is essential for inflammasome activity. We propose that AIM2 recognizes cytosolic F. tularensis, because the avirulent mutant ΔFPI (19), which cannot escape phagocytic vacuoles, failed to stimulate IL-1β secretion (Fig. 2A) or macrophage death (Fig. 2B).
Fig. 2.
AIM2 is required for inflammasome activation in response to F. tularensis. ■, asc−/−;  , casp-1−/−;
, casp-1−/−;  , aim2−/−; □, wt. (A) IL-1β secretion by BMDMs infected with F. tularensis ssp. novidica strain U112 or isogenic mutant ΔFPI for 5 h. Moi, multiplicity of infection. BMDMs treated with 5 mM ATP for 4 h were primed with 500 ng/mL Pam3CSK4 for 16 h. (B) Cytotoxicity as measured by LDH release. Graphs show the mean ± SD of triplicate wells and are representative of three independent experiments.
, aim2−/−; □, wt. (A) IL-1β secretion by BMDMs infected with F. tularensis ssp. novidica strain U112 or isogenic mutant ΔFPI for 5 h. Moi, multiplicity of infection. BMDMs treated with 5 mM ATP for 4 h were primed with 500 ng/mL Pam3CSK4 for 16 h. (B) Cytotoxicity as measured by LDH release. Graphs show the mean ± SD of triplicate wells and are representative of three independent experiments.
AIM2 and ASC Form a Complex with F. tularensis DNA.
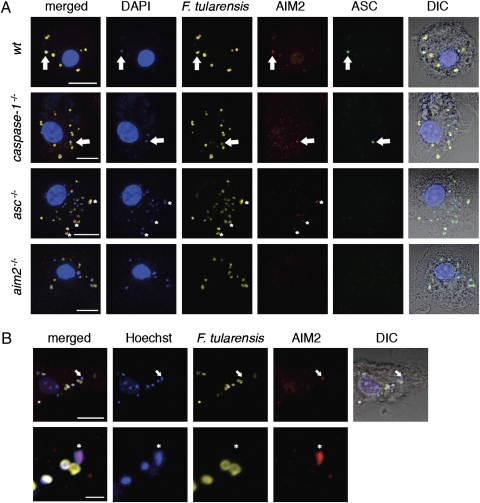
The HIN200 domain of AIM2 recognizes dsDNA, and its pyrin (PYD) domain can engage ASC (11, 14). We visualized inflammasome assembly by immunofluorescence confocal microscopy of BMDMs infected with F. tularensis and stained with antibodies detecting endogenous AIM2, ASC, and F. tularensis. At 5.5 h postinfection, wild-type, asc−/−, and caspase-1−/− BMDMs contained multiple AIM2 specks tightly associated with bright DAPI-staining material, likely reflecting leaked bacterial DNA caused by its proximity to irregular-shaped bacterial remnants (Fig. 3A). Regular-shaped bacteria stained dimly with DAPI. Infections with F. tularensis prelabeled with Hoechst 33342 nucleic acid stain confirmed that AIM2 was recruited to bacterial DNA (Fig. 3B). Merged images revealed that AIM2 overlapped almost completely with the bacterial DNA (Figs. 3 A and B and 4A), indicating that DNA leaked from F. tularensis is likely the PAMP recognized by AIM2 during an infection. Consistent with this notion, IL-1β secretion from wild-type BMDMs primed with Pam3CSK4 and then transfected with F. tularensis extract was abolished when the extract was preincubated with DNase I (Fig. S4A). Purified F. tularensis DNA transfected into Pam3CSK4-primed BMDMs also stimulated AIM2-dependent IL-1β secretion (Fig. S4B).
Fig. 3.
AIM2 and ASC form a complex with F. tularensis DNA. (A) Immunofluorescence microscopy of F. novidica U112-infected BMDMs at 5.5 h postinfection. DIC, differential interference contrast. Arrows indicate colocalization of DNA, degraded bacteria, AIM2, and ASC. Asterisks label diffuse AIM2 accumulation with DNA. Images are representative of at least three independent biological replicates. (Scale bar: 10 μm.) (B) BMDMs were infected with F. novidica U112 prelabeled with Hoechst 33342 nucleic acid stain. Arrows and asterisks indicate colocalization of bacterial DNA and AIM2. (Scale bar: 10 μm for Upper; scale bar: 2 μm for Lower.)
Fig. 4.
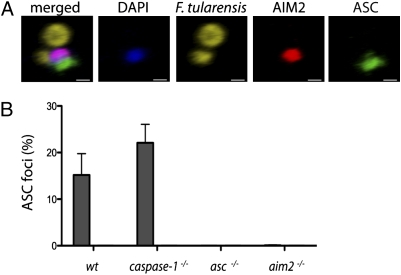
AIM2 is required for the formation of an ASC focus. (A) 3D reconstruction of a confocal image taken of a wild-type BMDM from C. (Scale bar: 0.5 μm.) (B)  , F. tularensis; □, ΔFPI. Graph showing the percentage of infected BMDMs containing an ASC focus from Fig. 3A. Bars represent the mean ± SD of two independent experiments. At least 300 cells of each genotype were examined per infection.
, F. tularensis; □, ΔFPI. Graph showing the percentage of infected BMDMs containing an ASC focus from Fig. 3A. Bars represent the mean ± SD of two independent experiments. At least 300 cells of each genotype were examined per infection.
AIM2 Is Required for the Formation of an ASC Focus.
Despite multiple AIM2 specks forming in an infected cell adjacent to bacterial remnants, ASC was recruited to a single AIM2 speck in ~15–22% of wild-type or caspase-1−/− BMDMs (Figs. 3A and 4B). The vacuole-restricted F. tularensis mutant ΔFPI did not stimulate ASC focus formation, consistent with its inability to stimulate IL-1β secretion (Fig. 2A). Importantly, ASC focus formation required AIM2, because no foci were detected in aim2−/− BMDMs. Others (32) have shown that similar ASC foci are formed in macrophages infected with S. typhimurium on activation of NOD-like receptors. Our data suggest that although AIM2 recognizes cytosolic DNA at multiple sites, only one of these sites will form the platform on which the ASC-containing inflammasome is built.
IFN-I Increases AIM2 Protein Levels and Inflammasome Activity.
Given that AIM2 protein expression is increased in BMDMs treated with IFN-β (Fig. S1C) or infected with F. tularensis (Fig. 5A) and that IFN-I signaling is required for efficient inflammasome signaling in response to F. tularensis (21), we sought to delineate the signaling pathway(s) driving IFN-I and AIM2 synthesis after F. tularensis infection. We investigated the contribution of STING, because it complexes with TBK1 and mediates IFN-I production in response to DNA (9). Unlike wild-type BMDMs that synthesized IFN-β mRNA in response to F. tularensis but not the avirulent mutant ΔFPI, sting−/− BMDMs did not up-regulate IFN-β gene expression after infection (Fig. 5B). sting−/− BMDMs synthesized IFN-β mRNA normally in response to transfected poly I:C, which engages RIG-I and MDA5 (33), excluding a general defect in IFN-β transcription. We speculate that bacterial DNA leaked from lysing F. tularensis leads to STING-dependent IFN-I production, although the sensor that recognizes the DNA remains unknown. Just as AIM2 was not required for IFN-β secretion from BMDMs transfected with dsDNA (Fig. 1D), AIM2 deficiency did not compromise IFN-β secretion from BMDMs infected with F. tularensis (Fig. S5A).
Fig. 5.
IFN-I increases AIM2 protein levels and inflammasome activity.  , ifnar−/−;
, ifnar−/−;  , sting−/−; □, wt. BMDMs were infected with S. typhimurium (moi = 100), F. tularensis ssp. novidica strain U112, or isogenic mutant ΔFPI for 5 h. (A) Western blot of AIM2 protein expression. (B) IFN-β mRNA expression quantified by RT-PCR. As a control, BMDMs were transfected with 1 μg/mL polyI:C. (C) IL-1β secretion into the culture supernatant. (D) Cytotoxicity as measured by LDH release. Where indicated, BMDMs were treated with 1,000 U/mL recombinant IFN-β at 1 h postinfection. Graphs show the mean ± SD of triplicate wells and are representative of three independent experiments.
, sting−/−; □, wt. BMDMs were infected with S. typhimurium (moi = 100), F. tularensis ssp. novidica strain U112, or isogenic mutant ΔFPI for 5 h. (A) Western blot of AIM2 protein expression. (B) IFN-β mRNA expression quantified by RT-PCR. As a control, BMDMs were transfected with 1 μg/mL polyI:C. (C) IL-1β secretion into the culture supernatant. (D) Cytotoxicity as measured by LDH release. Where indicated, BMDMs were treated with 1,000 U/mL recombinant IFN-β at 1 h postinfection. Graphs show the mean ± SD of triplicate wells and are representative of three independent experiments.
The increased AIM2 expression observed in F. tularensis-infected wild-type BMDMs was not observed in sting−/− or IFN-I receptor-deficient ifnar−/− BMDMs (Fig. 5A). In addition, failure to up-regulate AIM2 correlated with abrogated IL-1β secretion (Fig. 5C) and reduced cell death (Fig. 5D). Forced expression of AIM2 in ifnar−/− BMDMs restored IL-1β secretion in response to F. tularensis (Fig. S5B) and exogenous IFN-β restored cell death in infected sting−/− but not ifnar−/− BMDM cultures (Fig. 5D). We conclude that STING-dependent IFN-I production boosts inflammasome activity during F. tularensis infection by increasing AIM2 expression.
AIM2 Is Required for Host Defense Against F. tularensis.
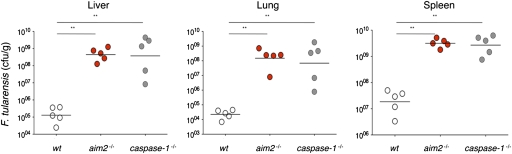
Finally, to extend our findings on the role of AIM2 in cultured macrophages to an in vivo setting, we challenged wild-type, aim2−/−, and caspase-1−/− mice with F. tularensis. caspase-1−/− mice fail to control F. tularensis infections (22), and aim2−/− mice were equally impaired at limiting F. tularensis replication (Fig. 6). Average bacterial loads in liver, lung, and spleen of aim2−/− or caspase-1−/− mice at 36 h postinfection were 120- to 19,000-fold higher than in wild-type mice. These data show that AIM2 is essential for innate immunity to F. tularensis in vivo.
Fig. 6.
AIM2 is required for host defense against F. tularensis. Mice were infected intradermally with 1 × 105 cfu of F. tularensis ssp. novidica strain U112F. Organs were harvested after 36 h and homogenized, and cfu were determined by plating serially diluted tissue extracts on modified Mueller Hinton (MH) agar. Bars indicate the geometric mean cfu per genotype.
Discussion
Collectively, our results provide genetic evidence that AIM2 is an essential DNA sensor of the innate immune system. Furthermore, AIM2 plays a critical role in defense against F. tularensis. We have also visualized an endogenous inflammasome NLR (AIM2) complexed with its ligand (DNA) and the adaptor ASC in the context of an infection. In our model, F. tularensis escapes phagosomal degradation into the macrophage cytosol where some bacteria lyse, which releases DNA into the cytosol. We support this model with visualization of Hoechst prelabeled bacterial DNA observed outside of aberrantly shaped F. tularensis. An unknown sensor(s) recognizes cytosolic bacterial DNA and signals through the adapter STING to produce IFN-I. Autocrine and paracrine signaling through the interferon α/β receptor (IFNAR) leads to an increase in AIM2 protein levels, which accelerates recognition of bacterial DNA by AIM2. We observe colocalization of AIM2 specks with bacterial DNA, one of which acts as a nucleus for formation of an ASC focus. We believe this complex represents a mature inflammasome that leads to secretion of mature IL-1β and host cell death. This model represents coordination of two independent DNA sensing pathways to produce a complete host response to a bacterial infection.
Previous DNA transfection studies have shown inflammasome activation in the absence of IFN-I signaling, suggesting that endogenous levels of AIM2 are sufficient for recognition of transfected DNA (10). Although we observe a dependence on IFN-I signaling for F. tularensis inflammasome activation, we show that we can restore inflammasome activation in the absence of IFN-I signaling through exogenous expression of AIM2, suggesting that AIM2 is sufficient for recognition of F. tularensis. Considering these results, we hypothesize that either the mechanism of DNA delivery or concentration of DNA delivered during F. tularensis infection is insufficient to be recognized by the endogenous levels of AIM2 present in macrophages. The sequential activation of the IFN-I and inflammasome responses also leads us to speculate that the threshold of DNA required to induce IFN-I signaling is less than that required to induce AIM2 inflammasome activation.
Bacterial pathogens can exploit a wide range of niches within a host, yet very few bacteria replicate inside the host cytosol, namely L. monocytogenes, Shigella flexneri, and F. tularensis. The inflammasome has been implicated in innate immunity to all of the aforementioned pathogens, although different NLRs and different bacterial ligands mediate these events. Inflammasome activation by L. monocytogenes has been attributed to production of the pore-forming toxin listeriolysin O (LLO), which is recognized by the NLRP3 inflammasome, as well as bacterial flagellin, which is engaged by the NLRC4 inflammasome (34, 35). Recent studies have elucidated a shared motif between bacterial flagellin and components of the type III secretion system (T3SS) (36); it explains the ability of NLRC4 to recognize S. flexneri, which contains a T3SS and lacks flagellin. It is not clear if the AIM2 inflammasome senses either of these pathogens. However, F. tularensis is nonflagellated, lacks a T3SS, and has not been shown to produce pore-forming toxins. Thus, DNA seems to be the only known inflammasome ligand possessed by F. tularensis.
Cross-talk between the IFN-I, NF-κB, and inflammasome pathways is poorly understood. We observed enhanced TNF-α and IFN-β secretions in asc−/− compared with wild-type BMDMs when transfected with poly(dA:dT) or pcDNA3 but not when transfected with poly(dG:dC) or ISD. One possible explanation for this difference is that there are at least two DNA sensors for IFN (and TNF-α) that recognize different types of DNAs. One such receptor-mediated pathway may receive negative feedback from the inflammasome, whereas the other does not. Additionally, IFN-I positively regulates the AIM2 inflammasome in response to F. tularensis infection. Although a role for the AIM2 inflammasome has not been shown during L. monocytogenes infection, it should be noted that IFN-I signaling also accelerates inflammasome activation in response to this pathogen (37). AIM2 is also likely required for recognition of dsDNA viruses, such as Vaccinia virus (11). Interestingly, Vaccinia actively inhibits the antiviral effects of IFN-I with the viral E3L protein, which may delay inflammasome activation and hence, the innate immune response to the pathogen (38). In addition, AIM2 may contribute to aberrant IL-1β production in response to host DNA, leading to arthritis-like autoimmune disease pathology. The coordination between the IFN-I response and the AIM2 inflammasome in the context of pathogen infection and autoimmunity warrants further investigation and is likely to have broad significance in our understanding of innate immunity.
Materials and Methods
Mice, Bacteria, and Reagents.
asc−/−, caspase-1−/−, ifnar−/−, nlrc4−/−, nlrp3−/−, and sting−/− mice have been described (9, 23, 25, 39). nlrp3−/−nlrc4−/− mice were generated by nlrp3−/− and nlrc4−/− intercrosses. All mice were backcrossed to C57BL/6 for at least 10 generations. Intradermal infections with F. tularensis ssp. novicida strain U112 and isogenic mutant ΔFPI (19) were performed as described (22). The Genentech and Stanford University Animal Care and Use Committees approved all mouse studies. S. typhimurium was from ATCC. Reagents included poly(dA:dT), poly(dG:dC), calf thymus DNA, polyI:C, ATP (Sigma), pcDNA3.1(+) (Invitrogen), IFN-β (R&D Systems), and Pam3CSK4 (Invivogen). Sense (5′TAC AGA TCT ACT AGT GAT CTA TGA CTG ATC TGT ACA TGA TCT ACA) and antisense ISD (5′TGT AGA TCA TGT ACA GAT CAG TCA TAG ATC ACT AGT AGA TCT GTA) (29) were synthesized and annealed at Genentech.
BMDM Cultures.
Bone marrow cells were differentiated in 30% macrophage-colony stimulating factor (M-CSF) conditioned medium for 5–6 days and plated at ~1 × 106 cells/mL for stimulation or infection with S. typhimurium or F. tularensis as described (22). dsDNAs and polyI:C were transfected with Lipofectamine 2000 (Invitrogen). For priming, BMDMs were cultured with 500 ng/mL ultrapure LPS or Pam3CSK4 (Invivogen) for 5 h. Priming was used to induce production of pro–IL-1β in macrophages. Priming with either LPS or Pam3CSK4 induces similar levels of pro–IL-1β; however, this Pam3CSK4 does not induce Trif-dependent IFN-I production as observed with LPS. IL-1β (Meso), IFN-β (Pestka Biomedical Laboratories), and IL-18 (MBL International) were measured by ELISA, TNF-α, and IL-6 by Bioplex-23 cytokine assay (Bio-Rad). Cytotoxicity was measured by LDH release (Promega). Caspase-1 was immunoblotted with 4B4 rat anti-mouse caspase p20 (Genentech) or rabbit anticaspase p10 (sc-514; Santa Cruz) IL-1β with a rabbit polyclonal (GeneTex).
Immunofluorescence Microscopy.
BMDMs (1.25 × 105) were seeded onto glass coverslips in 24-well plates for infection. Where indicated, F. tularensis was incubated for 30 min at 37 °C with 50 μg/mL Hoechst 33342 and then washed seven times before infection. Cells were washed two times with PBS, fixed for 15 min at 37 °C with 4% paraformaldehyde in PBS, washed three times with PBS, and stained with rabbit anti-mouse AIM2 at 1/500 (Genentech), 8E4.1 rat anti-mouse ASC at 1/2,000 (Genentech), and chicken anti-Francisella at 1/2,000 (Monack Laboratory) for 30 min in blocking buffer (3% BSA and 0.1% Saponin in PBS). Cells were washed three times with PBS and incubated for 30 min with Alexa488 anti-rat, Alexa594 anti-rabbit, and Alexa647 anti-chicken antibodies (Invitrogen). Cells washed four times with PBS and stained with DAPI were imaged with a Zeiss LSM700 confocal microscope. DAPI was omitted in samples with Hoechst-stained bacteria.
Quantitative Real-Time RT-PCR.
Primers used for IFN-β mRNA quantification are described (40). Experiments were performed with an iCycler (Bio-Rad) using SYBR green (Applied Biosystems).
Supplementary Material
Acknowledgments
We thank A. Zychlinsky, D. Portnoy, and G. Barber for mice and reagents, J. Liu and S. Mariathansan for discussion, and Q. Phung, D. Arnott, A. Paler Martinez, J. Hongo, and R. Vij for technical assistance. This work was supported by Award 2P01AI063302 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Conflict of interest statement: N.K., K.N., K.O., S.C., J.D., Y.Q., M.R.-G., and V.M.D. are all employees of Genentech, Inc.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003738107/-/DCSupplemental.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Kono H, Rock KL. How dying cells alert the immune system to danger. Nat Rev Immunol. 2008;8:279–289. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hemmi H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 4.Ishii KJ, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 5.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 6.Prantner D, Darville T, Nagarajan UM. Stimulator of IFN gene is critical for induction of IFN-beta during Chlamydia muridarum infection. J Immunol. 2010;184:2551–2560. doi: 10.4049/jimmunol.0903704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muruve DA, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 11.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bürckstümmer T, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 13.Landolfo S, Gariglio M, Gribaudo G, Lembo D. The Ifi 200 genes: An emerging family of IFN-inducible genes. Biochimie. 1998;80:721–728. doi: 10.1016/s0300-9084(99)80025-x. [DOI] [PubMed] [Google Scholar]
- 14.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golovliov I, Baranov V, Krocova Z, Kovarova H, Sjöstedt A. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect Immun. 2003;71:5940–5950. doi: 10.1128/IAI.71.10.5940-5950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nano FE, et al. A Francisella tularensis pathogenicity island required for intramacrophage growth. J Bacteriol. 2004;186:6430–6436. doi: 10.1128/JB.186.19.6430-6436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauriano CM, et al. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc Natl Acad Sci USA. 2004;101:4246–4249. doi: 10.1073/pnas.0307690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brotcke A, Monack DM. Identification of fevR, a novel regulator of virulence gene expression in Francisella novicida. Infect Immun. 2008;76:3473–3480. doi: 10.1128/IAI.00430-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiss DS, et al. In vivo negative selection screen identifies genes required for Francisella virulence. Proc Natl Acad Sci USA. 2007;104:6037–6042. doi: 10.1073/pnas.0609675104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cole LE, et al. Macrophage proinflammatory response to Francisella tularensis live vaccine strain requires coordination of multiple signaling pathways. J Immunol. 2008;180:6885–6891. doi: 10.4049/jimmunol.180.10.6885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J Exp Med. 2007;204:987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mariathasan S, Weiss DS, Dixit VM, Monack DM. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J Exp Med. 2005;202:1043–1049. doi: 10.1084/jem.20050977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mariathasan S, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 24.Martinon F, Pétrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 25.Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 26.Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009;27:519–550. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 27.Kuida K, et al. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science. 1995;267:2000–2003. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 28.Ishii KJ, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 29.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Keim P, Johansson A, Wagner DM. Molecular epidemiology, evolution, and ecology of Francisella. Ann N Y Acad Sci. 2007;1105:30–66. doi: 10.1196/annals.1409.011. [DOI] [PubMed] [Google Scholar]
- 31.Meibom KL, Charbit A. The unraveling panoply of Francisella tularensis virulence attributes. Curr Opin Microbiol. 2010;13:11–17. doi: 10.1016/j.mib.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Fink SL, Bergsbaken T, Cookson BT. Anthrax lethal toxin and Salmonella elicit the common cell death pathway of caspase-1-dependent pyroptosis via distinct mechanisms. Proc Natl Acad Sci USA. 2008;105:4312–4317. doi: 10.1073/pnas.0707370105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sutterwala FS, Ogura Y, Flavell RA. The inflammasome in pathogen recognition and inflammation. J Leukoc Biol. 2007;82:259–264. doi: 10.1189/jlb.1206755. [DOI] [PubMed] [Google Scholar]
- 35.Warren SE, Mao DP, Rodriguez AE, Miao EA, Aderem A. Multiple Nod-like receptors activate caspase 1 during Listeria monocytogenes infection. J Immunol. 2008;180:7558–7564. doi: 10.4049/jimmunol.180.11.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miao EA, et al. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci USA. 2010;107:3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stockinger S, et al. Production of type I IFN sensitizes macrophages to cell death induced by Listeria monocytogenes. J Immunol. 2002;169:6522–6529. doi: 10.4049/jimmunol.169.11.6522. [DOI] [PubMed] [Google Scholar]
- 38.Chang HW, Watson JC, Jacobs BL. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci USA. 1992;89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Müller U, et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 40.Auerbuch V, Brockstedt DG, Meyer-Morse N, O'Riordan M, Portnoy DA. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J Exp Med. 2004;200:527–533. doi: 10.1084/jem.20040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.