Abstract
In humans, NH3 transport across cell membranes is facilitated by the Rh (rhesus) family of proteins. Human Rh C glycoprotein (RhCG) forms a trimeric complex that plays an essential role in ammonia excretion and renal pH regulation. The X-ray crystallographic structure of human RhCG, determined at 2.1 Å resolution, reveals the mechanism of ammonia transport. Each monomer contains 12 transmembrane helices, one more than in the bacterial homologs. Reconstituted into proteoliposomes, RhCG conducts NH3 to raise internal pH. Models of the erythrocyte Rh complex based on our RhCG structure suggest that the erythrocytic Rh complex is composed of stochastically assembled heterotrimers of RhAG, RhD, and RhCE.
Keywords: ammonia channel, comparative modeling, membrane protein, rhesus factor, X-ray structure
The five human Rh (Rhesus) family proteins fall into two functionally distinct groups: the ammonia transporting Rh glycoproteins (RhAG, RhBG, and RhCG) and the nontransporting (nonglycosylated) Rh proteins (RhD and RhCE). Erythroid RhD is recognized for its key role in blood group incompatibility (Rh+, Rh-) (1, 2) whereby Rh- individuals (lacking RhD antigen) can develop an immune response when exposed to Rh+ blood. Along with RhD, RhAG and RhCE are specific components of the erythrocyte Rh complex with incompletely defined stoichiometry (3, 4). RhBG and RhCG are found in a variety of tissues including brain, liver, gut, and kidney. Proteins of the Rh family have 20–27% identity to the “methyl-ammonia permeases” (MEPs) in yeast and to the “ammonia transporters” (Amts) in prokaryotes, plants, and some invertebrates (5). RhAG, RhBG, and RhCG have been shown to transport ammonia (6, 7). RhCG is responsible for ammonia secretion across the membranes of the epithelial cells of the renal collecting duct (8, 9). RhCG knockout mice show abnormal acidification of the blood due to impaired urinary removal of 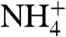 emphasizing the importance of renal transport for human physiology.
emphasizing the importance of renal transport for human physiology.
Aqueous ammonia (NH3) and ammonium ion ( ) are in equilibrium governed by a pKa of 9.25. Thus at physiological pH, the charged
) are in equilibrium governed by a pKa of 9.25. Thus at physiological pH, the charged 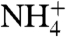 form predominates, and the cell membrane is impervious to the charged ionic form. Prior to their structure, the preponderance of evidence suggested that the Amts and their homologs transport the charged species,
form predominates, and the cell membrane is impervious to the charged ionic form. Prior to their structure, the preponderance of evidence suggested that the Amts and their homologs transport the charged species,  , however, the atomic structure of AmtB and conductivity measurements (10, 11) showed that the prokaryotic Amts serve as channels for passage of uncharged NH3 across the membrane, later confirmed by subsequent transport measurements (12, 13), as had been postulated previously (14).
, however, the atomic structure of AmtB and conductivity measurements (10, 11) showed that the prokaryotic Amts serve as channels for passage of uncharged NH3 across the membrane, later confirmed by subsequent transport measurements (12, 13), as had been postulated previously (14).  is formed by reprotonation of NH3 in solution.
is formed by reprotonation of NH3 in solution.
Here, we report the atomic resolution structure of a eukaryotic Rh glycoprotein, human RhCG, expressed in modified HEK293s cells (15). The structural differs from its prokaryotic homologs in having an additional transmembrane helix and in other Rh-specific structural features that further elaborate the NH3 transport mechanism vital to human renal function. The high sequence similarity of RhCG to the erythroid Rh proteins allows construction of reliable models of their structures that inform the stoichiometry and assembly of the erythrocyte complex.
Results and Discussion
RhCG Forms Trimers of 12 Crossing Transmembrane Channels.
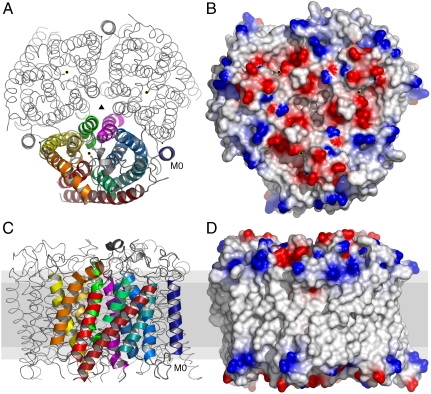
The atomic structure of human RhCG at 2.1 Å resolution contains an additional N-terminal transmembrane helix, termed M0 relative to Amts (10–12) or the prokaryotic NeRh (16, 17). The 12 transmembrane α-helices (M0–M11) form a right-handed helical bundle that surrounds an apparently empty channel (Fig. 1). As in bacterial Amt proteins, helices M1–M5 and M6–M10 are related by an in-plane quasi-2-fold symmetry with respect to the membrane plane. Helices M0 and M11 break this quasi-symmetry and are linked by loops to the 10-helix core. These loops vary in sequence and length between the human Rh family (Fig. S1). In RhD and RhCE, the loops are relatively short and are resistant to proteolytic cleavage. In the human Rh glycoproteins, RhAG, RhBG, and RhCG, the loops (with the exception of M10–M11 in RhAG) are longer in sequence and are susceptible to proteolytic cleavage (4). These differences suggest that the loops are functionally differentiated.
Fig. 1.
Molecular structure of RhCG. (A) Cytoplasmic view of the RhCG with monomer in cartoon representation and complete symmetry-generated biological molecule as ribbon. Threefold symmetry axis indicated by ▴, and • indicates the channel position of each monomer. Transmembrane helices are progressively colored blue to red, indicating succession from N to C terminus. (B) Cytoplasmic molecular surface of RhCG colored by electrostatic potential (19) reveals channel apertures surrounded by negative charge (red) with positive charge (blue) at the periphery. (C and D) Representation of A and B, respectively, rotated 90° in membrane (gray) cross-section with cytoplasmic surface up.
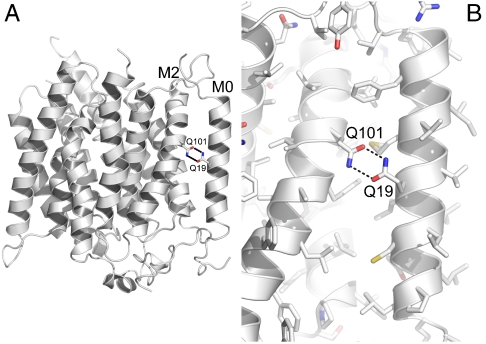
The RhCG M0 region is conserved across higher eukaryotes (Fig. S2). However, in the prokaryotic Amts (10–12) and NeRh (16, 17), a shorter stretch of residues constitute a cleaved signal sequence. M0 lies against the lipid-accessible surface of the trimer and at the interface between subunits. A single glutamine (Q19), halfway along the M0 helix in the center of the lipid bilayer, is hydrogen bonded to glutamine (Q101) on M2 (Fig. 2). In all members of the Rh family that have M0, a glutamine or glutamate are conserved at positions equivalent to 19 and 101 (Fig. S2). These polar residues in the center of a transmembrane region would otherwise be exposed to lipid. Therefore, M0 is pinned to M2 by this conserved midmembrane interaction.
Fig. 2.
The “M0 lock” in RhCG. (A) Cartoon representation of RhCG rendered in PyMol (20). The side chains of Q19 on M0 and Q101 on M2 are shown as stick representation colored by atom type. The hydrogen bonding between the side chains are indicted by dashed lines. (B) Closer view of the M0 lock with all side chains displayed as sticks and colored by atom type (C: white; O: red; N: blue; S: yellow).
Each of the human Rh glycoproteins contains a single conserved N-glycosylation site (N48 in RhCG) on the loop between M0 and M1. For our construct, expressed in a human endothelial kidney (HEK) cell line engineered to produce a short homogeneous core oligosaccharide (Man5GlcNAc2), termed GnTl- (15) (SI Methods), N48 would be homogeneously glycosylated. The mass of 55.1 ± 0.1 kDa measured by MALDI mass spectroscopy is consistent with this predicted mass of 55.163 kDa for the singly glycosylated species.
In RhCG, the M0–M1 loop (∼25 amino acids) contains several hydrophobic residues (A36, A38, W40, W41, and L49) that would be shielded from solvent by the normal branched polysaccharide. Similar hydrophobic character is seen in the M0–M1 loop of the other human Rh glycoproteins. The nonglycosylated RhD/CE have a shorter (∼11 amino acids) M0–M1 loop. The glycosylation may serve to cover the hydrophobic sequence and protect the longer loops from proteolytic degradation.
Mechanism of NH3 Permeation.
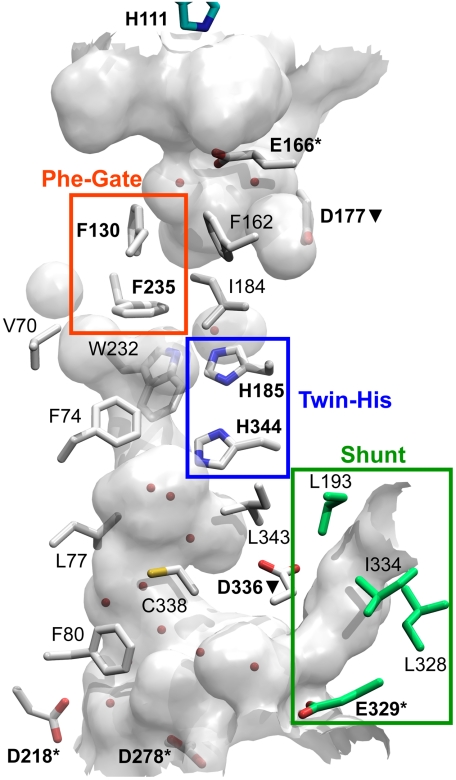
Comparison of RhCG to the prokaryotic Amt (10–12) and NeRh (16, 17) structures confirms distinct conserved family specific features including acidic residues that line the apertures, an external aperture gated by phenylalanines, a largely hydrophobic lumen of the channel, and twin coplanar histidines in the center of the channel proposed to facilitate passage of the substrate. Because of these conserved features, the RhCG channel aperture is expected to attract  , whereas the lumen excludes
, whereas the lumen excludes  and facilitates passage of neutral NH3. The central pore of the channel is hydrophobic (Fig. 3), and most of residues are conserved among the human Rh glycoproteins (Fig. S1) favoring passage of uncharged NH3 as in AmtB (10). This implies deprotonation of
and facilitates passage of neutral NH3. The central pore of the channel is hydrophobic (Fig. 3), and most of residues are conserved among the human Rh glycoproteins (Fig. S1) favoring passage of uncharged NH3 as in AmtB (10). This implies deprotonation of  within the vestibule.
within the vestibule.
Fig. 3.
RhCG NH3 channel and shunt. Partially transparent surface generated by HOLLOW (22) of the channel, voids, and shunt. Select side chains that compose this surface are displayed as sticks. Water molecules within the channel surface displayed as red spheres. All atoms colored by atom type with carbons in white except for the shunt with carbons in green.
Conserved twin histidines (H185 and H344), required for optimal NH3 permeation (21), are hydrogen bonded to each other via their NδH–Nδ and intrude into the center of the channel (Fig. 3, blue box). These coplanar and antiparallel histidines are conserved underlying their importance in the mechanism of transport. Despite these histidines, the hydrophobic lumen contains no ordered water molecules. The absence of any ordered water further supports the exclusion of  , which would require a replacement for its hydration shell, from the channel.
, which would require a replacement for its hydration shell, from the channel.
At the extracellular vestibule, Amt channels have a characteristic tryptophan proposed to aid in recruiting  through a π-cation interaction (10). RhCG and NeRh (16, 17) lack this tryptophan, however acidic residues E166 in the extracellular vestibule and D218, D278, and E329 in the intracellular vestibule may serve to attract
through a π-cation interaction (10). RhCG and NeRh (16, 17) lack this tryptophan, however acidic residues E166 in the extracellular vestibule and D218, D278, and E329 in the intracellular vestibule may serve to attract  (Fig. 3, labeled with *). Transport of NH3 by the Rh glycoproteins has been shown to be pH dependent (7, 23). Because the hydrophobicity of the channel lumen is phylogenetically well conserved, pH dependence may be conferred at the vestibules. In the kidney, the environment of the extracellular vestibule in RhCG differs between the apical and basal surface of the epithelium of the renal intercalated cell. The basal surface exposes the extracellular vestibule of RhCG to the kidney interstitium with a pH of ∼7.3, thereby resulting in a net extracellular charge of -2.6 (-2.3 in the vestibule) for each RhCG monomer, assuming unaltered side chain pKas. The apical surface exposes the extracellular vestibule to the urinary space with a pH of ∼5.5, suggesting protonation of five histidines (39, 46, 111, 247, and 374) for a net extracellular charge of +3.1 (+0.4 in the vestibule) assuming unaltered side chain pKas. Thus the extracellular vestibule in the urinary space has reduced affinity for
(Fig. 3, labeled with *). Transport of NH3 by the Rh glycoproteins has been shown to be pH dependent (7, 23). Because the hydrophobicity of the channel lumen is phylogenetically well conserved, pH dependence may be conferred at the vestibules. In the kidney, the environment of the extracellular vestibule in RhCG differs between the apical and basal surface of the epithelium of the renal intercalated cell. The basal surface exposes the extracellular vestibule of RhCG to the kidney interstitium with a pH of ∼7.3, thereby resulting in a net extracellular charge of -2.6 (-2.3 in the vestibule) for each RhCG monomer, assuming unaltered side chain pKas. The apical surface exposes the extracellular vestibule to the urinary space with a pH of ∼5.5, suggesting protonation of five histidines (39, 46, 111, 247, and 374) for a net extracellular charge of +3.1 (+0.4 in the vestibule) assuming unaltered side chain pKas. Thus the extracellular vestibule in the urinary space has reduced affinity for 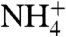 , perhaps accounting for the pH dependence of the delivery of NH3 to the urinary space via RhCG (24).
, perhaps accounting for the pH dependence of the delivery of NH3 to the urinary space via RhCG (24).
Another acidic residue (D177) near the vestibule has a key structural role rather than substrate recruitment (Fig. 3 D177▾). RhCG D177 is hydrogen bonded to the backbone amides of residues 180–183 in the extracellular vestibule. These interactions satisfy the available bonds and occlude the acidic side chain from solvent. The structures of Amt, NeRh, and RhCG show that this position plays a key structural role in capping the M5 helix (10). Similarly, at the intracellular vestibule, quasi-symmetric D336 acts as an N-terminal cap to the M10 helix (Fig. 3 D336▾). The D160A mutation in AmtB (25), and the homologous D177N mutation in RhCG (26) result in loss of transport of the orthologous radio-labeled substrate, C14-methyl ammonia, most probably, we argue, due to structural disruption.
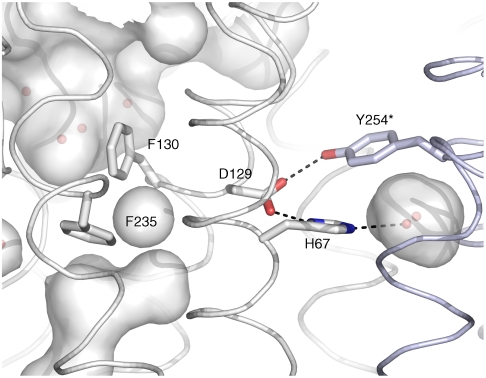
The extracellular pore in the Amt subfamily is gated by two phenylalanines, F235 and F130 (10). In contrast to Amt, the outer phenylalanine (F130) does not obstruct the pore of RhCG or NeRh (Fig. 3, orange box) (16, 17). This conformation may be favored due to the nearby D129, a buried, charged residue that is hydrogen bonded to the hydroxyl of Y254 of an adjacent monomer within the trimeric assembly and the HNδ of H67. Each of these residues is conserved in the human Rh glycoproteins. The limited mobility of D129 may restrict F130, thereby locking the gate in the open conformation. Small voids on each side of the F235 side chain allow for freedom of motion necessary to open the “Phe gate.” Mutation of F235 to valine reduced NH3 transport, supporting a role in passage (27).
An RhCG feature not present in Amts is a pocket that extends from the cytosolic aperture to the lateral exterior surface in contact with the lipid hydrocarbon chains that we term the “shunt” (Fig. 3, green box). Amino acids that line the shunt are similar among human Rh glycoproteins but conserved in RhD and RhCE, which do not function as NH3 channels. The equivalent region in the Amt structures contained a large void but no opening to the cytosol or membrane. The shunt is also seen in the structure of NeRh, suggesting that it is common to the Rh subfamily. The cytosolic aperture of this shunt has an acidic residue E329, which may play a role in recruiting charged 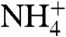 . The vestibule is polar and hydrated, but as the shunt approaches the lipidic region, the residues are largely hydrophobic and without any discrete water. Therefore, the shunt could be an alternative path for
. The vestibule is polar and hydrated, but as the shunt approaches the lipidic region, the residues are largely hydrophobic and without any discrete water. Therefore, the shunt could be an alternative path for  entry, and NH3 delivery into the hydrophobic portion of the lipid bilayer, bypassing the polar headgroups of the cytoplasmic leaflet.
entry, and NH3 delivery into the hydrophobic portion of the lipid bilayer, bypassing the polar headgroups of the cytoplasmic leaflet.
RhCG Transports Ammonia but Not Water.
The permeation of NH3 was measured using reconstituted RhCG-containing proteoliposomes, where the influx of NH3, but not 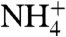 , induces a rise in internal pH assayed by the pH-sensitive fluorescence of 5-carboxy-fluorescein (CF) located inside. (Fig. 4A) (10, 28, 29). (See SI Materials). Relative rates were assayed by stopped-flow external addition of 5 mM NH4Cl. In the absence of protein, liposomes alone exhibited a rate of 11.1 ± 0.6 s-1 (n = 6). With RhCG, the rate was 14.3 ± 1.9 s-1 (n = 6). Doubling the RhCG increased the rate to 23.4 ± 2.4 s-1, demonstrating that RhCG transports NH3. Substitution of 5 mM NaCl for 5 mM NH4Cl made no detectable change in fluorescence, demonstrating that changes in CF fluorescence were due to NH3 influx rather than to water efflux resulting from any change in osmolarity. When the Escherichia coli acetate transporter, YaaH, was reconstituted with liposomes (as control for the reconstitution procedure), no significant change in the control rate at 10.4 ± 1.7 s-1 (n = 6) was observed, consistent with a lack of nonspecific protein-mediated permeability and no leakage of NH3 due to the reconstitution procedure. These results indicate that RhCG is an efficient channel for NH3.
, induces a rise in internal pH assayed by the pH-sensitive fluorescence of 5-carboxy-fluorescein (CF) located inside. (Fig. 4A) (10, 28, 29). (See SI Materials). Relative rates were assayed by stopped-flow external addition of 5 mM NH4Cl. In the absence of protein, liposomes alone exhibited a rate of 11.1 ± 0.6 s-1 (n = 6). With RhCG, the rate was 14.3 ± 1.9 s-1 (n = 6). Doubling the RhCG increased the rate to 23.4 ± 2.4 s-1, demonstrating that RhCG transports NH3. Substitution of 5 mM NaCl for 5 mM NH4Cl made no detectable change in fluorescence, demonstrating that changes in CF fluorescence were due to NH3 influx rather than to water efflux resulting from any change in osmolarity. When the Escherichia coli acetate transporter, YaaH, was reconstituted with liposomes (as control for the reconstitution procedure), no significant change in the control rate at 10.4 ± 1.7 s-1 (n = 6) was observed, consistent with a lack of nonspecific protein-mediated permeability and no leakage of NH3 due to the reconstitution procedure. These results indicate that RhCG is an efficient channel for NH3. 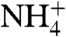 conductance is not precluded by the liposome assays, nor the different indications from electrophysiological recordings (6, 7, 30) resolved, however the structure of RhCG seems to imply exclusion of
conductance is not precluded by the liposome assays, nor the different indications from electrophysiological recordings (6, 7, 30) resolved, however the structure of RhCG seems to imply exclusion of 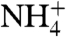 from the hydrophobic lumen.
from the hydrophobic lumen.
Fig. 4.
Channel NH3 transport in proteoliposomes. (A) Relative vesicular pH change indicated by CF fluorescence for RhCG at protein to lipid ratio 1∶90 by weight (open blue triangles) and 1∶45 (closed blue triangles) demonstrated protein dependent NH3 permeability relative to empty liposomes (black squares) and a control protein YaaH (red diamonds). (B) Proteoliposomes from the same sample preparations after addition of 275 mM sucrose at t = 0 were monitored for transport of water resulting in decreased vesicular volume, increased CF concentration, and subsequent self-quenching. Data displayed for the exponential fluorescence decay after lag time to reach self-quenching concentration and exponential fit (black lines).
Water permeability of RhCG was assessed by measuring contraction of proteoliposomes upon osmotic challenge, as assayed by concentration-dependent self-quenching of CF (29) (Fig. 4B). For protein-free liposomes, the fluorescence change on stopped-flow mixing with 275 mM sucrose indicated that water efflux was 4.0 ± 0.1 s-1 (n = 6) versus 3.9 ± 0.1 s-1 (n = 6) for RhCG proteoliposomes, indicating no additional water permeability in RhCG proteoliposomes. Doubling the protein concentration resulted in an increase to 4.6 ± 0.1 s-1 and YaaH gave 5.7 ± 0.1 s-1. This can be compared with reconstitutions of the water channel AqpZ into proteoliposomes under similar conditions which gave ∼150 s-1 (10). Thus, RhCG does not transport water and the liposomes were well sealed. This conclusion is critical to kidney physiology, in which the high solute concentration of the urinary space must be maintained.
Modeling the Erythrocyte Rh Complex.
Early identification of the erthrocyte Rh complex (3) suggested a composition of RhAG∶RhD∶RhCE with ∼2∶1∶1 stoichiometry, in tetrameric assemblies (4). The specific arrangement of proteins can now be addressed by protein modeling. Among the human Rh glycoproteins, 35% of residues and 46% of transmembrane positions are conserved (Fig. S1). Thus, the homotrimeric RhCG currently provides a better template for modeling the erythroid complex and the binary interfaces between them than any other known structure.
To probe the stoichiometry and configuration of the subunits in the erythrocyte complex, models of all possible trimeric arrangements of the RhAG, RhD, and RhCE subunits were built based on the RhCG trimer template (31). In the RhCG trimer, four exposed polar regions of each subunit are buried by a complementary region on the opposing subunit: P1(Y32, N252), P2(Y60, Q64, D65, S234), P3(H67, D129, Y254) (Fig. 5), and P4(S87, D432,W436). The polarity of these amino acid residues is preserved in the erythrocytic proteins (Fig. S1), and is distinct from the Amt and MEP proteins.
Fig. 5.
Phenylalanine gate and polar cluster P3. The partially transparent channel surface and nearby void surfaces are displayed to reveal the waters with observed electron density in the molecular structure. The side chains of Phe gate and P3 residues are shown as sticks colored by atom type. The P3 cluster for each monomer (structure model in white) involves Y254 from the adjacent monomer (labeled with * and in blue).
The modeled assemblies were used to test the various possible homo and heteroligomeric arrangements in the erthrocyte Rh complex. Conservation of the subunit interfaces argues that the human erythrocyte Rh complex is trimeric, just as in the RhCG trimer. Computational assessment of all homomeric and heteromeric trimer models shows various scores that are less favorable than in the parent RhCG structure (Table S1). However, the scores among the models are most favorable for homotrimers of RhAG, followed by trimers that replace one RhAG by RhD, or RhCE. They are least favorable for homotrimers of RhD, or RhCE, suggesting that RhAG is more compatible with the template trimeric configuration than RhD and RhCE (Table S1). Although the assessment scores cannot be used to distinguish between existing and nonexisting configurations, they suggest a stochastic subunit arrangement within trimers with the overall experimentally observed ratio of ∼2∶1∶1 for RhAG∶RhD∶RhCE (4).
Functional Insights Derived from Models.
RhD and RhCE do not transport NH3. However, NH3 permeation cannot be measured directly because these two proteins can only be expressed in the presence of RhAG and NH3 passage measured in two different experimental methods can be attributed to RhAG alone (24, 32, 33). In the RhD/CE protein models, the positions occupied by conserved histidines within the central region of the channel are replaced. The outer histidine (H185) found in the Rh glycoproteins is replaced by tyrosine and the inner histidine (H344) by phenylalanine. These substitutions do not fully obstruct the lumen of the channel, however the histidines (in AmtB) have been shown to be essential; mutants in which the histidines were replaced by phenylalanine and alanine lost NH3 conduction (21). The histidine C-H hydrogen-bonding potential may be a key to freezing the tautomeric form of NH3 as suggested based on electron density difference maps between high and low NH3 concentration in crystal structures (10).
Debate continues as to a possible role for the Rh erythroid complex as the long-sought-after conductor of CO2 in the red blood cell. Such a role was suggested based on reduced growth of green algae following sRNAi down regulation of a native Rh homolog (34, 35). Support for this proposal was augmented by a feature of the prokaryotic NeRh structure (16), where a CO2 binding site was identified in a cleft near the cytosolic aperture of the channel. However, RhCG shows that this site is not present in the human Rh glycoproteins. RhCG Q283 replaces an alanine at the equivalent position in NeRh. This glutamate side chain occupies the proposed CO2 site and it is conserved in all the human Rh glycoproteins. Therefore this site is not available for CO2 transport in human Rh.
Nevertheless, RhAG and the erythrocyte aquaporin AQP1 do allow passage of both NH3 and the less hydrophilic CO2 measured based on changes in surface pH in Xenopus oocytes (36). However, RhAG demonstrated 4-fold greater preference for passage of NH3 versus CO2 than seen in AQP1. Furthermore, AQP1 is far more abundant than RhAG in the erythrocyte, making it the more probable primary pathway for CO2. However, the potential remains for some Rh mediated CO2 passage through the central channel in the same manner proposed for NH3.
Renal Rh Glycoprotein Ammonia Transport.
 secretion in the kidneys is critical to systemic acid-base homeostasis (37–39). In humans, inability to appropriately dispose of excess H+ in urine results in distal renal tubular acidosis that depresses arterial blood pH (< 7.35), and in extreme cases results in kidney stones, rickets, and renal failure (40, 41). Amongst many epithelial cell types, RhCG and RhBG are found in the cells of the collecting tubule and collecting duct acid-secreting intercalated cells as well as in the majority of the cells of the distal collecting tubule (7, 8, 42–46).
secretion in the kidneys is critical to systemic acid-base homeostasis (37–39). In humans, inability to appropriately dispose of excess H+ in urine results in distal renal tubular acidosis that depresses arterial blood pH (< 7.35), and in extreme cases results in kidney stones, rickets, and renal failure (40, 41). Amongst many epithelial cell types, RhCG and RhBG are found in the cells of the collecting tubule and collecting duct acid-secreting intercalated cells as well as in the majority of the cells of the distal collecting tubule (7, 8, 42–46).
Growing consensus supports maintenance of acid/base equilibrium and  excretion by the following model (Fig. 6).
excretion by the following model (Fig. 6).
Fig. 6.
Proposed mechanism of acid/ammonium balance in the epithelium of the acid-secreting renal intercalated cells. Channels and pumps are indicated as tubes through the membrane. Substrates are indicated as atomic models. The net flow of respective substrates and diffusion through the membrane are indicated by arrows. Figure is based on the proposed epithelial model for acid-secreting intercalated cells (7, 47, 48).
Whereas renal RhBG is found predominately on the basal epithelial surface (in contact with the kidney interstitium), RhCG is abundant in both the basal and apical surfaces (in contact with the urinary space). Based on studies of the RhCG knockout mouse, NH3 can pass through the epithelial cell membrane, however RhCG dramatically increases the permeability as evidenced by recovery rates under acid load (8, 9). The diffusion of NH3 through RhCG in the distal portions of the nephron epithelium appears to be driven from the interstitium, by progressively lowered pH to the epithelial cell cytosol, and into the lumen.
The pH balance is maintained by the basolateral  exchanger (49–51), the luminal V-type H+ ATPase, and luminal H+/K+-ATPase. The secretion of protons, mostly from H+-ATPases, and to a lesser extent by H+/K+-ATPase acidifies urine (48, 52). The more acidic environment of the urine shifts the equilibrium
exchanger (49–51), the luminal V-type H+ ATPase, and luminal H+/K+-ATPase. The secretion of protons, mostly from H+-ATPases, and to a lesser extent by H+/K+-ATPase acidifies urine (48, 52). The more acidic environment of the urine shifts the equilibrium  ∼100∶1 at pH 7.25 toward 104∶1 at pH 5.25 in urine. This equilibrium favors concentration of
∼100∶1 at pH 7.25 toward 104∶1 at pH 5.25 in urine. This equilibrium favors concentration of 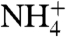 in the urinary space, thereby lowering the [NH3] and so providing the NH3 gradient to drive concentration. This also allows for the accumulation of high
in the urinary space, thereby lowering the [NH3] and so providing the NH3 gradient to drive concentration. This also allows for the accumulation of high  , resulting in net excretion of
, resulting in net excretion of  . Urinary concentrations of
. Urinary concentrations of 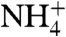 of greater than 200 mM can exist, while across the cell membrane
of greater than 200 mM can exist, while across the cell membrane 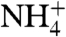 remains ∼8 mM (7, 38). Consequently, long-term maintenance of blood pH is dependent on NH3 passage by RhCG in the kidney.
remains ∼8 mM (7, 38). Consequently, long-term maintenance of blood pH is dependent on NH3 passage by RhCG in the kidney.
Methods
RhCG was expressed in HEK293S cells (15) and purified using FLAG affinity purification and size-exclusion chromatography. Crystals were grown in vapor diffusion and diffraction data were collected from cryoprotected crystals at beamline 8.3.1 of the Advanced Light Source (Lawrence Berkeley National Laboratories). The structure was determined by molecular replacement in Phaser (53) using NeRh (3B9W) as search model. Model building and refinement were calculated in CCP4 (54) and REFMAC (55), ARP/wARP (56), and Coot (57) (Table 1). Proteoliposome permeability of NH3 was indicated by pH-dependent response of CF by stopped-flow cytometry. Comparative models were generated using MODELLER-9v6 (31) and assessed using Z-DOPE (58), a normalized atomic distance-dependent statistical potential based on known protein structures. Detailed methods are provided in SI Text.
Table 1.
Crystallographic statistics for RhCG
| Data | |
| Unit cell | |
| a = b, c, Å | 98.83, 101.08 |
| α = β, γ, ° | 90, 120 |
| Space group | P63 |
| Wavelength, Å | 1.1159 |
| Resolution, Å | 2.10 |
| Completeness, % | 99.7 (95.8) |
| Multiplicity | 10.6 (8.6) |
| I/σ | 39.7 (3.4) |
| Rsym, % | 8.9 (63.7) |
| Refinement | |
| Rcryst, % | 16.7 (17.5) |
| Rfree, % | 20.3 (21.2) |
| Mean B factor, Å2 | 25.1 |
| rmsd bond length, Å | 0.012 |
| rmsd bond angle, ° | 1.274 |
Numbers in parentheses correspond to the high-resolution shell for data refinement (2.18–2.10 Å).
Supplementary Material
Acknowledgments.
We thank Witek Kwiatkowski (Salk Institute) for MALDI-MS analysis, James Holton and George Meigs who provided beamline support, Melissa del Rosario and Xiaojin Li (Holland Laboratory, American Red Cross) who gave molecular biology assistance, and Zach Newby who implemented HEK expression. We thank Albert F. Gruswitz (Boundless Creativity, LLC) for providing graphic assistance with Fig. 6 and Patricia Greene for assistance in editing the manuscript. This research was supported by National Institutes of Health Grants R01 GM24485, GM73210, and GM74929.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003587107/-/DCSupplemental.
Data deposition: Atomic coordinates and structure factors for the human RhCG crystal structure are deposited in the Protein Data Bank under accession code 3HD6. Comparative models deposited as the “HumanRhesus” model set into MODBASE (18). Correspondence and requests for materials should be addressed to stroud@msg.ucsf.edu.
References
- 1.Levine P, Stetson RE. An unusual case of intra-group agglutination. J Am Med Assoc. 1939;113:126–127. doi: 10.1001/jama.251.10.1316. [DOI] [PubMed] [Google Scholar]
- 2.Colin Y, et al. Genetic basis of the RhD-positive and RhD-negative blood group polymorphism as determined by Southern analysis. Blood. 1991;78:2747–2752. [PubMed] [Google Scholar]
- 3.Hartel-Schenk S, Agre P. Mammalian red cell membrane Rh polypeptides are selectively palmitoylated subunits of a macromolecular complex. J Biol Chem. 1992;267:5569–5574. [PubMed] [Google Scholar]
- 4.Eyers SA, Ridgwell K, Mawby WJ, Tanner MJ. Topology and organization of human Rh (rhesus) blood group-related polypeptides. J Biol Chem. 1994;269:6417–6423. [PubMed] [Google Scholar]
- 5.Marini AM, Urrestarazu A, Beauwens R, Andre B. The Rh (rhesus) blood group polypeptides are related to NH4+ transporters. Trends Biochem Sci. 1997;22:460–461. doi: 10.1016/s0968-0004(97)01132-8. [DOI] [PubMed] [Google Scholar]
- 6.Bakouh N, et al. NH3 is involved in the NH4+ transport induced by the functional expression of the human Rh C glycoprotein. J Biol Chem. 2004;279:15975–15983. doi: 10.1074/jbc.M308528200. [DOI] [PubMed] [Google Scholar]
- 7.Mak DO, Dang B, Weiner ID, Foskett JK, Westhoff CM. Characterization of ammonia transport by the kidney Rh glycoproteins RhBG and RhCG. Am J Physiol. 2006;290:297–305. doi: 10.1152/ajprenal.00147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biver S, et al. A role for Rhesus factor Rhcg in renal ammonium excretion and male fertility. Nature. 2008;456:339–343. doi: 10.1038/nature07518. [DOI] [PubMed] [Google Scholar]
- 9.Lee HW, et al. Collecting duct-specific Rh C glycoprotein deletion alters basal and acidosis-stimulated renal ammonia excretion. Am J Physiol. 2009;296:1364–1375. doi: 10.1152/ajprenal.90667.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khademi S, et al. Mechanism of ammonia transport by Amt/MEP/Rh: Structure of AmtB at 1.35 A. Science. 2004;305:1587–1594. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- 11.Zheng L, Kostrewa D, Berneche S, Winkler FK, Li XD. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc Natl Acad Sci USA. 2004;101:17090–17095. doi: 10.1073/pnas.0406475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrade SL, Dickmanns A, Ficner R, Einsle O. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc Natl Acad Sci USA. 2005;102:14994–14999. doi: 10.1073/pnas.0506254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Javelle A, Thomas G, Marini AM, Kramer R, Merrick M. In vivo functional characterization of the Escherichia coli ammonium channel AmtB: Evidence for metabolic coupling of AmtB to glutamine synthetase. Biochem J. 2005;390:215–222. doi: 10.1042/BJ20042094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soupene E, Lee H, Kustu S. Ammonium/methylammonium transport (Amt) proteins facilitate diffusion of NH3 bidirectionally. Proc Natl Acad Sci USA. 2002;99:3926–3931. doi: 10.1073/pnas.062043799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reeves PJ, Kim JM, Khorana HG. Structure and function in rhodopsin: A tetracycline-inducible system in stable mammalian cell lines for high-level expression of opsin mutants. Proc Natl Acad Sci USA. 2002;99:13413–13418. doi: 10.1073/pnas.212519199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, Jayachandran S, Nguyen HH, Chan MK. Structure of the Nitrosomonas europaea Rh protein. Proc Natl Acad Sci USA. 2007;104:19279–19284. doi: 10.1073/pnas.0709710104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lupo D, et al. The 1.3-A resolution structure of Nitrosomonas europaea Rh50 and mechanistic implications for NH3 transport by Rhesus family proteins. Proc Natl Acad Sci USA. 2007;104:19303–19308. doi: 10.1073/pnas.0706563104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pieper U, Eswar N, Stuart AC, Ilyin VA, Sali A. MODBASE, a database of annotated comparative protein structure models. Nucleic Acids Res. 30:255–259. doi: 10.1093/nar/30.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeLano WL. Manheim, Germany: Schrödinger, LLC; 2008. The PyMOL Molecular Graphics System, Ver. 1.2r3pre. [Google Scholar]
- 21.Javelle A, et al. An unusual twin-His arrangement in the pore of ammonia channels is essential for substrate conductance. J Biol Chem. 2006;281:39492–39498. doi: 10.1074/jbc.M608325200. [DOI] [PubMed] [Google Scholar]
- 22.Ho BK, Gruswitz F. HOLLOW: Generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct Biol. 2008;8:49. doi: 10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westhoff CM, Ferreri-Jacobia M, Mak DO, Foskett JK. Identification of the erythrocyte Rh blood group glycoprotein as a mammalian ammonium transporter. J Biol Chem. 2002;277:12499–12502. doi: 10.1074/jbc.C200060200. [DOI] [PubMed] [Google Scholar]
- 24.Westhoff CM, Wylie DE. Transport characteristics of mammalian Rh and Rh glycoproteins expressed in heterologous systems. Transfus Clin Biol. 2006;13:132–138. doi: 10.1016/j.tracli.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 25.Javelle A, Severi E, Thornton J, Merrick M. Ammonium sensing in Escherichia coli. Role of the ammonium transporter AmtB and AmtB-GlnK complex formation. J Biol Chem. 2004;279:8530–8538. doi: 10.1074/jbc.M312399200. [DOI] [PubMed] [Google Scholar]
- 26.Marini AM, Boeckstaens M, Benjelloun F, Cherif-Zahar B, Andre B. Structural involvement in substrate recognition of an essential aspartate residue conserved in Mep/Amt and Rh-type ammonium transporters. Curr Genet. 2006;49:364–374. doi: 10.1007/s00294-006-0062-5. [DOI] [PubMed] [Google Scholar]
- 27.Zidi-Yahiaoui N, et al. Ammonium transport properties of HEK293 cells expressing RhCG mutants: preliminary analysis of structure/function by site-directed mutagenesis. Transfus Clin Biol. 2006;13:128–131. doi: 10.1016/j.tracli.2006.02.025. [DOI] [PubMed] [Google Scholar]
- 28.Roos A, Boron WF. Intracellular pH. Physiol Rev. 1981;61:296–434. doi: 10.1152/physrev.1981.61.2.296. [DOI] [PubMed] [Google Scholar]
- 29.Priver NA, Rabon EC, Zeidel ML. Apical membrane of the gastric parietal cell: Water, proton, and nonelectrolyte permeabilities. Biochemistry. 1993;32:2459–2468. doi: 10.1021/bi00061a002. [DOI] [PubMed] [Google Scholar]
- 30.Bakouh N, Benjelloun F, Cherif-Zahar B, Planelles G. The challenge of understanding ammonium homeostasis and the role of the Rh glycoproteins. Transfus Clin Biol. 2006;13:139–146. doi: 10.1016/j.tracli.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 31.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 32.Marini AM, et al. The human Rhesus-associated RhAG protein and a kidney homologue promote ammonium transport in yeast. Nat Genet. 2000;26:341–344. doi: 10.1038/81656. [DOI] [PubMed] [Google Scholar]
- 33.Ripoche P, et al. Human Rhesus-associated glycoprotein mediates facilitated transport of NH(3) into red blood cells. Proc Natl Acad Sci USA. 2004;101:17222–17227. doi: 10.1073/pnas.0403704101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soupene E, et al. Rhesus expression in a green alga is regulated by CO2. Proc Natl Acad Sci USA. 2002;99:7769–7773. doi: 10.1073/pnas.112225599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soupene E, Inwood W, Kustu S. Lack of the Rhesus protein Rh1 impairs growth of the green alga Chlamydomonas reinhardtii at high CO2. Proc Natl Acad Sci USA. 2004;101:7787–7792. doi: 10.1073/pnas.0401809101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Musa-Aziz R, Chen LM, Pelletier MF, Boron WF. Relative CO2/NH3 selectivities of AQP1, AQP4, AQP5, AmtB, and RhAG. Proc Natl Acad Sci USA. 2009;106:5406–5411. doi: 10.1073/pnas.0813231106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamm LL, Simon EE. Roles and mechanisms of urinary buffer excretion. Am J Physiol. 1987;253:F595–605. doi: 10.1152/ajprenal.1987.253.4.F595. [DOI] [PubMed] [Google Scholar]
- 38.Knepper MA, Packer R, Good DW. Ammonium transport in the kidney. Physiol Rev. 1989;69:179–249. doi: 10.1152/physrev.1989.69.1.179. [DOI] [PubMed] [Google Scholar]
- 39.DuBose TD, Jr, Good DW, Hamm LL, Wall SM. Ammonium transport in the kidney: New physiological concepts and their clinical implications. J Am Soc Nephrol. 1991;1:1193–1203. doi: 10.1681/ASN.V1111193. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez Soriano J. Renal tubular acidosis: The clinical entity. J Am Soc Nephrol. 2002;13:2160–2170. doi: 10.1097/01.asn.0000023430.92674.e5. [DOI] [PubMed] [Google Scholar]
- 41.Laing CM, Toye AM, Capasso G, Unwin RJ. Renal tubular acidosis: Developments in our understanding of the molecular basis. Int J Biochem Cell B. 2005;37:1151–1161. doi: 10.1016/j.biocel.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 42.Quentin F, et al. RhBG and RhCG, the putative ammonia transporters, are expressed in the same cells in the distal nephron. J Am Soc Nephrol. 2003;14:545–554. doi: 10.1097/01.asn.0000050413.43662.55. [DOI] [PubMed] [Google Scholar]
- 43.Verlander JW, et al. Localization of the ammonium transporter proteins RhBG and RhCG in mouse kidney. Am J Physiol. 2003;284:F323–337. doi: 10.1152/ajprenal.00050.2002. [DOI] [PubMed] [Google Scholar]
- 44.Han KH, et al. Expression of the ammonia transporter, rh C glycoprotein, in normal and neoplastic human kidney. J Am Soc Nephrol. 2006;17:2670–2679. doi: 10.1681/ASN.2006020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seshadri RM, et al. Renal expression of the ammonia transporters, Rhbg and Rhcg, in response to chronic metabolic acidosis. Am J Physiol. 2006;290:397–408. doi: 10.1152/ajprenal.00162.2005. [DOI] [PubMed] [Google Scholar]
- 46.Brown AC, et al. RhCG is the major putative ammonia transporter expressed in the human kidney, and RhBG is not expressed at detectable levels. Am J Physiol. 2009;296:1279–1290. doi: 10.1152/ajprenal.00013.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim HY, et al. Basolateral expression of the ammonia transporter familymember Rh C glycoprotein in the mouse kidney. Am J Physiol. 2009;296:543–555. doi: 10.1152/ajprenal.90637.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wagner CA, Devuyst O, Bourgeois S, Mohebbi N. Regulated acid-base transport in the collecting duct. Pflug Arch Eur J Phy. 2009;458:137–156. doi: 10.1007/s00424-009-0657-z. [DOI] [PubMed] [Google Scholar]
- 49.Weiner ID, Weill AE, New AR. Distribution of Cl-/HCO3- exchange and intercalated cells in rabbit cortical collecting duct. Am J Physiol. 1994;267:952–964. doi: 10.1152/ajprenal.1994.267.6.F952. [DOI] [PubMed] [Google Scholar]
- 50.Petrovic S, et al. SLC26A7: A basolateral Cl-/HCO3- exchanger specific to intercalated cells of the outer medullary collecting duct. Am J Physiol. 2004;286:161–169. doi: 10.1152/ajprenal.00219.2003. [DOI] [PubMed] [Google Scholar]
- 51.Stehberger PA, et al. Distal renal tubular acidosis in mice lacking the AE1 (band3) Cl-/HCO3- exchanger (slc4a1) J Am Soc Nephrol. 2007;18:1408–1418. doi: 10.1681/ASN.2006101072. [DOI] [PubMed] [Google Scholar]
- 52.Wagner CA, et al. Renal vacuolar H+-ATPase. Physiol Rev. 2004;84:1263–1314. doi: 10.1152/physrev.00045.2003. [DOI] [PubMed] [Google Scholar]
- 53.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 54.CCP4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 55.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 56.Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. ARP/wARP and molecular replacement. Acta Crystallogr D. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- 57.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 58.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.