Abstract
Speciation with gene flow is expected to generate a heterogeneous pattern of genomic differentiation. The few genes under or physically linked to loci experiencing strong disruptive selection can diverge, whereas gene flow will homogenize the remainder of the genome, resulting in isolated “genomic islands of speciation.” We conducted an experimental test of this hypothesis in Rhagoletis pomonella, a model for sympatric ecological speciation. Contrary to expectations, we found widespread divergence throughout the Rhagoletis genome, with the majority of loci displaying host differences, latitudinal clines, associations with adult eclosion time, and within-generation responses to selection in a manipulative overwintering experiment. The latter two results, coupled with linkage disequilibrium analyses, provide experimental evidence that divergence was driven by selection on numerous independent genomic regions rather than by genome-wide genetic drift. “Continents” of multiple differentiated loci, rather than isolated islands of divergence, may characterize even the early stages of speciation. Our results also illustrate how these continents can exhibit variable topography, depending on selection strength, availability of preexisting genetic variation, linkage relationships, and genomic features that reduce recombination. For example, the divergence observed throughout the Rhagoletis genome was clearly accentuated in some regions, such as those harboring chromosomal inversions. These results highlight how the individual genes driving speciation can be embedded within an actively diverging genome.
Keywords: host race, inversion, island of speciation, latitudinal cline, Rhagoletis pomonella
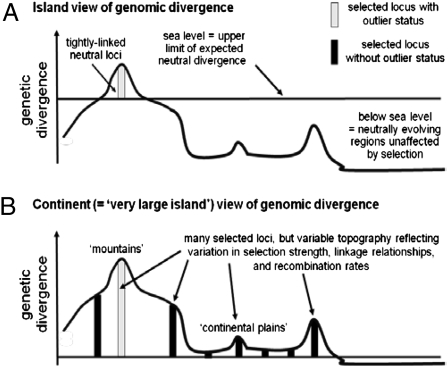
A seminal question in evolutionary biology is the role of genome structure in speciation, especially for taxa diverging with gene flow (e.g., in parapatry or sympatry). The emerging field of population genomics has recently focused attention on an “islands” metaphor, in which speciation is initiated via divergent selection on only a handful of genes (e.g., two or three) that reside in just a few isolated chromosomal regions (Fig. 1A). These “genomic islands of speciation” show elevated differentiation between taxa compared with the remainder of the genome, which is homogenized by gene flow and thus relatively undifferentiated (1–5) (Fig. 1A). Genomic islands have been hypothesized to promote speciation, because reduced effective gene flow in regions surrounding loci under selection could facilitate further differentiation through a process of divergence hitchhiking (3, 6, 7) (Fig. 1A).
Fig. 1.
Schematic representation of the (A) island versus (B) continent view of genomic divergence. These views represent ends of a continuum, rather than being mutually exclusive. For example, “continents” of divergence can be conceptualized as very large islands with variable topography.
Alternatively, selection acting on many loci distributed through-out the genome also could drive speciation with gene flow (8, 9) (Fig. 1B). This process also can produce a variable pattern of genomic divergence due to differences in selection intensities, linkage relationships, and recombination rates among loci (Fig. 1B). For example, loci residing in or near regions of reduced recombination, such as chromosomal inversions or centromeres, might exhibit increased differentiation (5, 10–13). However, in the case of selection on many loci, genomic regions displaying lower levels of differentiation might not represent neutrally evolving regions, but rather might reflect more weakly selected loci in regions of high recombination (Fig. 1B). Thus, even in early stages of speciation, many loci may be differentiated above neutral, “sea-level” expectations, such that the genomes of taxa differ by many “archipelagoes” or even whole “continents of divergence.” We stress that the island versus continent views of genomic divergence represent ends of a continuum, rather than mutually exclusive hypotheses; for example, continents of divergence can be conceptualized as very large islands with variable topography, such as high mountain tops and lowland continental plains, all above neutral sea level (Fig. 1B).
Experimental tests of the island versus continent scenarios are lacking. Indirect support for the island hypothesis has come from genome scans of populations, which test for “outlier loci” whose differentiation exceeds neutral expectations, implying divergent natural selection (11, 14). Outlier loci typically compose only a small proportion of the genome (roughly 5–10%; range, 0.4–24.5%; mean, 8.5%, n = 18 studies) (11, 14). A few studies have mapped the location of outlier loci in the genome (3, 5, 12, 13, 15, 16). In some cases, outlier loci appear to be clustered within specific and isolated genomic regions (3, 5, 15).
In contrast, evidence for genomically widespread divergence is rare. This scarcity might stem from the limitations inherent in relying on genome scans alone for detecting selection. Genome scans conducted without complementary selection experiments and mapping studies can predestine an island view, because only the most diverged regions will be identified as statistical outliers. Other loci affected by selection, but more weakly so, will go unnoticed and be considered part of the mostly “undifferentiated” genome (14). Until appropriate tests are conducted, it will not be possible to resolve the extent to which speciation is driven via islands of divergence in a few genomic regions versus divergence spread across the genome.
Here we report a direct test of the island hypothesis versus the continent hypothesis for Rhagoletis pomonella using a combination of manipulative experiments, genetic mapping/linkage disequilibrium analyses, and field data on genomic divergence. The known geographic context (sympatric with gene flow) and historical time frame (~150 years ago) of the host shift of R. pomonella from hawthorn (Crataegus sp.) to apple (Malus pumila) provide the necessary preconditions for a strong test of genomic differentiation underlying speciation with gene flow (17–19). In addition, the existence of known chromosomal inversions in several regions of the genome allows tests of the idea that regions of reduced recombination will exhibit particularly strong genetic divergence (20).
To quantify genome differentiation, we surveyed apple and hawthorn flies along a latitudinal transect in the United States (SI Appendix, Table S1 and Fig. S1) for 33 microsatellites and 6 allozymes distributed relatively evenly throughout the genome (Fig. S2) (Materials and Methods). The population survey was coupled with a manipulative selection experiment in which hawthorn flies were exposed to nondiapause versus diapause rearing conditions as pupae, along with an analysis of adult eclosion time (Materials and Methods). These treatments emulated important environmental differences experienced by the races in the wild due to the 3- to 4-week earlier fruiting time of apples compared with hawthorns (17–19). The selection experiment and eclosion study allowed for clear interpretation of geographic and host-related variations for the host races in terms of selection acting on the marker loci or linked genes. Linkage disequilibrium analysis allowed estimation of the number of independent genomic regions exhibiting divergence between the races in nature and responses to selection in the rearing experiment.
Results and Discussion
Population Genomic Analyses.
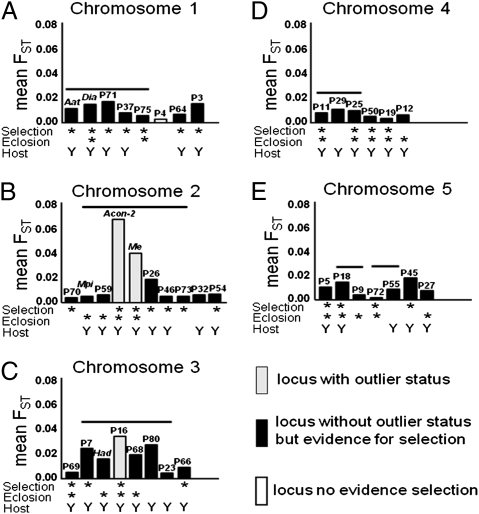
Results of standard outlier analyses of the microsatellites and allozymes were consistent with the genomic island hypothesis. Three significant outlier loci were detected between the host races: microsatellite P16, mapping to chromosome 3, and the allozymes Acon-2 and Me, linked on chromosome 2 (Fig. 2 and SI Appendix, Fig. S3). Thus, only two independent genomic regions exhibited outlier status. Previous allozyme studies indicated that a third region on chromosome 1 involving Aat-2 and Dia-2 also displays host-related differentiation (20–23).
Fig. 2.
Mean FST for loci on chromosomes 1–5 between apple host race populations. Asterisks below graphs denote loci responding significantly in the selection experiment or adult eclosion study (see SI Appendix, Table S2, for exact significance levels). Y (yes) and N (no) denote whether or not a locus displayed significant host-related differentiation, as determined by Fisher's exact test. Horizontal lines above bars represent groups of loci found to be in linkage disequilibrium with one another, as determined by the composite method of Weir (35).
Geographic Variation Among Populations.
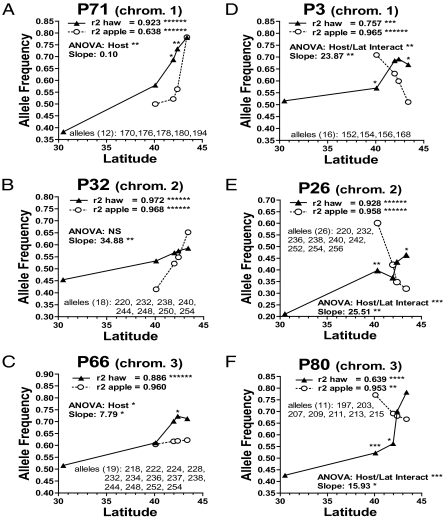
In sharp contrast to the outlier results, Monte Carlo bootstrapping analysis of the population data, patterns of linkage disequilibrium, and results of the manipulative selection experiment and adult eclosion study revealed widespread genomic differentiation. All 33 microsatellites and all 6 allozymes displayed significant clinal variations among hawthorn fly populations (Fig. 3 and SI Appendix, Figs. S4–S8 and Table S2). Geographic variation also was pronounced in the apple fly race, with 21 of the 33 microsatellites and the allozyme Had exhibiting significant latitudinal clines (Fig. 3 and SI Appendix, Figs. S4–S8 and Table S2). For roughly half of the loci (n = 22), the sign of allele frequency change with latitude was in the same direction in both host races, with the slopes of the clines being similar (n = 11), steeper (n = 2), or shallower (n = 9) for apple versus hawthorn populations; however, for the other half of the genes (n = 17), latitudinal clines among apple populations were in the opposite direction of those for the hawthorn race, significantly so for 15 loci (Fig. 3 and SI Appendix, Figs. S4–S8 and Table S2). As a result, 26 of the 33 microsatellites and all 6 allozymes displayed significant variation between apple and hawthorn flies (Fig. 2 and SI Appendix, Table S2). Linkage disequilibrium analyses confirmed that these 26 microsatellites were not just limited to the three previously identified rearranged regions showing divergence on chromosomes 1–3 (20–23), but rather that patterns of linkage disequilibrium implied that loci displaying host-associated differentiation were dispersed throughout the genome, representing a minimum of 17 different regions/genes (Fig. 2 and SI Appendix, Fig. S2 and Tables S2 and S3). The clinal variation generated a complex geographic mosaic of host-related divergence, with different loci being significant at different sites and alleles often reversing with respect to which host race in which they were more common, depending on latitude. Thus, although the races exhibited significant differences at individual sites, it is the pattern of change across the landscape that demonstrates that host-associated selection pressures or gene-by-environment interactions change in different ways with latitude, in turn affecting patterns of differentiation throughout the genome.
Fig. 3.
Representative microsatellites on chromosomes 1–3 where clines for the apple race are in the same (A–C) or opposite direction (D–F) as hawthorn race (see SI Appendix, Figs. S4–S8, for all 33 microsatellites scored on chromosomes 1–5). R2 represents the explained variation for regression between allele frequencies and latitude. ANOVA, host or host-by-latitude interaction, indicates that a locus displayed significant host-related differentiation in Monte Carlo ANOVA analysis. Slope indicates the F-test for heterogeneity in slopes between regressions for host races. Asterisks above hawthorn triangles indicate significant results of Fisher’ exact test for host-related frequency difference at sympatric sites. Alleles refer to the total number of variants segregating for a locus (in parentheses) and sizes (in base pairs) of different alleles in pooled allele class. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001; ******P ≤ 0.000001.
Manipulative Selection Experiment.
Results of the selection experiment indicate that the genomic divergence observed in R. pomonella was caused by natural selection. A total of 26 of the 39 loci tested (22 microsatellites and 4 allozymes) displayed significant allele frequency responses to rearing conditions in the selection experiment, as determined by Fisher's exact test (Fig. 2 and SI Appendix, Fig. S9 and Table S2). Patterns of linkage dis-equilibrium imply that these 26 loci represent a minimum of 16 different loci or genomic regions responding significantly and independently in the selection experiment (Fig. 2 and SI Appendix). The markers composing each of these 16 regions were in linkage disequilibrium with one another (when multiple loci demarcated a region), but in linkage equilibrium with all other markers in the selection experiment and in natural fly populations (SI Appendix, Table S3). There was therefore no significant genetic (allelic) correlation between a locus responding in one of these 16 regions and a second locus responding in a different region in the selection experiment. Moreover, map distances of at least 7 cM (and usually more) separated these regions when they resided on the same chromosome in our mating crosses (SI Appendix, Fig. S2). These findings imply that the observed response in the selection experiment was not due to a single selected gene on each chromosome. The large majority of significant loci (21 of 26; 80.8%) responded in the predicted direction in the selection experiment (χ2 = 9.85, P = 0.0017, 1 df for significant deviation from the 50:50 null hypothesis), with rearing conditions emulating the earlier fruiting apple favoring apple race alleles at the Grant, Michigan site in diapausing flies (SI Appendix, Fig. S9 and Table S2). Selection coefficients (s) estimated for the 26 significant loci in the diapause experiment ranged from 0.061 to 0.606 (mean, 0.180 ± 0.003; SI Appendix, Table S2).
Adult Eclosion Analysis.
Results of the eclosion experiment provide further evidence of widespread divergent selection. A total of 15 of the 33 microsatellites and 5 of the 6 allozymes showed either a significant main effect of genotype or a significant host-by-genotype interaction with eclosion time (SI Appendix, Tables S2 and S4). Based on patterns of linkage disequilibrium, the 20 loci significantly related to eclosion time represented a minimum of 12 different, independent genes/genomic regions. Ten of these 12 regions also significantly responded in the selection experiment (SI Appendix, Table S2). Forward and backward linear regressions considering only sets of loci in linkage equilibrium among the 12 regions displaying significant eclosion time responses resulted in an R2 value of 0.466 for apple flies (six loci—Mpi, Had, P69, P12, P5, and P9—included in the regression equation) and an R2 value of 0.750 for hawthorn flies (eight loci—P75, Mpi, Had, P19, P50, P18, P27, and P72—included in the regression equation).
Evidence That Selection Drives Genomic Divergence.
Our collective results, coupled with the results of previous mark-recapture field studies (24), discount genetic drift or isolation by distance as causes for the latitudinal clines and host-related divergence in R. pomonella. Of the 17 genomic regions identified in the population survey as displaying host differences, a total of 16 contained loci that responded significantly in the selection experiment and/or were significantly related to adult eclosion time (SI Appendix, Fig. S9 and Tables S2 and S4). Only the region demarcated by microsatellite P32 on chromosome 2 did not do so. Indeed, the magnitude of the response of loci in the selection experiment (as quantified by marginal fitness values) was significantly related to the degree of host-associated genetic differentiation (mean FST across sites) observed between natural populations (Spearman's rank correlation, 0.346 ± 0.027; P ≤ 0.033, n = 39 loci; SI Appendix, Fig. S10). This correlation did not arise due to only one locus or a few loci with particularly high FST (range of rank correlation coefficients under a jackknife analysis of 0.296–0.425), arguing against an island view of genomic divergence.
In addition, previous mark-recapture studies have estimated interhost migration and mating occurring at a rate of 4–6% per generation between sympatric hawthorn and apple fly populations at the Grant, Michigan site (24). Given this level of local genetic exchange, genetic drift alone is not expected to generate clinal- and host-related differences between apple and hawthorn races for marker loci; instead, natural selection is required. The logic behind this is that gene flow between races would move alleles between populations and obliterate local allele frequency differences. In that case, isolation by distance and a lack of selection would (at best) generate similar clines in both races. Thus, the observation of parallel, crossing, and even opposite, rather than overlapping, clines between the races is best explained by selection acting strongly along environmental gradients in different ways in the two races.
In summary, analysis of a rapidly evolving class of genetic markers across multiple sites in nature, a selection experiment, field and eclosion studies, mapping, and linkage disequilibrium analyses have revealed that large “continents” of divergence, rather than isolated islands, characterize genomic differentiation in R. pomonella. The observed latitudinal clines and host-related allele frequency differences between apple and hawthorn flies are thus not limited to a few genes.
Variable Topology of Genomic Divergence.
Even when divergence is genomically widespread, a flat, level topology for genetic differentiation is not predicted (Fig. 1B). For example, even if selection is strong, as estimated in the current study, some loci will exhibit greater divergence (i.e., less homogenization by gene flow) in nature than others, depending on linkage relationships and recombination rates.
The data from R. pomonella illustrate this point (Fig. 2). Two regions were identified as highly diverged statistical outliers, and several other loci also were more differentiated than most regions (Fig. 2 and SI Appendix, Fig. S3). There are likely and complementary explanations for this variable topography. First, some regions will experience stronger selection than others. Second, some markers will be in tighter linkage with selected loci than others, and thus will exhibit stronger differentiation even for an equal strength of selection on the selected locus itself (10). Third, loci in regions of reduced recombination might exhibit strong differentiation. In this regard, many of the most well-differentiated markers in Rhagoletis were in seen regions where loci were in linkage disequilibrium; such regions are known to or likely to harbor inversions. Indeed, the mean FST between apple and hawthorn host races for the 25 loci displaying linkage disequilibrium (i.e., loci putatively in rearrangements) was significantly higher than that for the 14 loci in linkage equilibrium (0.0141 ± 0.00297 vs. 0.0066 ± 0.00116; P = 0.0233, one-tailed Mann-Whitney U test). This was also true in an analysis that considered the fact that the 25 loci displaying linkage disequilibrium were not completely independent. That analysis compared the mean FST value within each independent region (n = 6 regions) harboring loci in linkage disequilibrium to the loci displaying linkage equilibrium (n = 14 loci, as above), and once again found greater differentiation in regions of disequilibrium (FST = 0.0142 ± 0.00299; P = 0.0197).
Inversions alone are unlikely to explain our overall results, however. Many of the 14 loci outside the 6 regions of linkage disequilibrium still showed significant differentiation between the host races (11/14), responded significantly in the selection experiment (10/14), and were significantly associated with adult eclosion time (6/14). Finally, even if these regions were inside inversions, numerous independent genomic regions (i.e., numerous independent inverted regions) would nonetheless exhibit divergence.
Thus, we are not proposing that all loci in the genome are under selection. Rather, our results suggest that selection is widespread and strong enough, and recombination is low enough, to allow detection of widespread divergence in presumably neutral markers throughout the genome. In this respect, we tested for a response to only one aspect of host ecology (diapause life history) in the current study. Assays for other components of host adaptation (e.g., host preference) could expand the scope of genomic differentiation. Regardless, our findings indicate that low baseline levels of genomic differentiation do not necessarily represent regions of neutral differentiation, but instead might reflect limitations in the ability to statistically distinguish moderate- to low-level selection from background neutral expectations.
Conclusion.
Further comprehensive tests for widespread genomic divergence are needed to determine whether the data for R. pomonella are the exception or the rule. Theory suggests that it is possible for speciation to proceed in the face of gene flow when selection acts on dispersed loci across the genome (25), but that the likelihood of this depends on such factors as the strength of selection, availability of standing genetic variation (26), and structural features of the genome that reduce recombination (27). These factors are important in accentuating the effectiveness of disruptive selection in generating divergence. Thus, whereas inversion polymorphism may enhance the scope of differentiation in R. pomonella, this does not alter the finding that widespread selection affects multiple regions across the fly's genome. Continents, rather than islands, of differentiation may occur, even in the early stages of speciation with gene flow. In this regard, sequence analysis of anonymous cDNA loci has implied that clinal variation in R. pomonella is of secondary origin, due to past episodes of introgression between Mexican and U.S. hawthorn fly populations (28, 29). Thus, the proximate ecological host shifts leading to the recent sympatric formation of the apple race, as well as other sibling species in the R. pomonella group, were facilitated by preexisting genetic variation. Therefore, the ancestral hawthorn race in the United States could be considered a hybrid zone writ large through space and time involving much of the genome, raising the possibility that many cases of speciation with gene flow may have genetic underpinnings similar to hybrid speciation (30, 31).
Decreasing costs of high throughput sequencing will make it increasingly possible to realistically analyze even greater numbers of genes and populations. As a result, the focus of speciation research likely will shift from searching for individual reproductive isolation “speciation genes” (2, 4) to questioning the genomic architecture of divergence. Attention to genome-wide patterns of differentiation will modify our view of the genetics of speciation by allowing the study of individual “speciation genes” as part of a collective and evolving genome.
Materials and Methods
Genetic Scoring of Flies.
Collection protocols of flies for the population survey are described in SI Appendix. DNA was isolated and purified from head or whole body fly tissue using Puregene extraction kits (Gentra Systems). Purified DNAs were transferred to 96-well plates for microsatellite PCR amplification and genotyping of 33 loci characterized from an enriched GT-dinucleotide repeat R. pomonella library (33) (GenBank accession numbers for Rhagoletis microsatellites are AY734885–AY734965.) Microsatellite loci are designated with the prefix “P,” followed by a suffix number indicating the order in which they were originally characterized. SI Appendix, Table S5, provides a complete list and PCR primer pairs for all currently characterized microsatellite loci in R. pomonella, including those scored in the current study. The 33 microsatellites analyzed were chosen because they displayed no systematic evidence for heterozygote deficiency from Hardy-Weinberg equilibrium due to null alleles, as determined using Micro-Checker (34). Total genomic DNA was PCR-amplified using locus-specific primers for 38 cycles of 94 °C for 20 s, 55 °C for 15 s, and 72 °C for 30 s, followed by a final incubation for 10 min at 72 °C. Genotyping was performed on a Beckman-Coulter CEQ8000 genetic analysis system. Microsatellite alleles were sized using the Fragment Analysis program (Beckman-Coulter). Data for the allozymes were assembled from previous genetic surveys of the study sites (18–23). Total sample sizes are given in SI Appendix, Table S1.
Mapping of Microsatellite Loci.
Linkage relationships of microsatellite loci were determined from seven single-pair crosses constructed previously using R. pomonella flies reared to adulthood from larval-infested apple fruit collected at the Grant, Michigan site in 1995 (20). Parental adults and F1 offspring were genotyped for microsatellites as described above. Recombination does not occur in R. pomonella males, allowing rapid determination of linkage relationships in F1 offspring, because they inherit whole chromosomes from male parents. Recombination rates and gene orders can then be estimated for maternal chromosomes by eliminating one or the other of the sets of paternally inherited alleles for a chromosome in an F1 offspring. Previous analysis has shown that there is no single linear gene order for chromosomes 1–3 (20). In the present study, significant heterogeneity in recombination rates between microsatellite loci (i.e., proportions of observed exchange events uncorrected for multiple exchanges) was also observed among the seven test crosses for chromosomes 1–3, as well as for chromosomes 4 and 5. No recombination was observed in at least one test cross for each linkage group except chromosome 4 (SI Appendix, Fig. S2), whereas these same loci demonstrated free recombination (map distances of ~50 cM) in other crosses. Furthermore, at least two loci in each chromosome displayed significant levels of linkage disequilibrium within apple and hawthorn populations (SI Appendix, Fig. S2 and Table S3), as determined by the composite disequilibrium value of Weir (35). Thus, recombination distances between microsatellites should be viewed in terms of evolutionary map distances of average exchange between markers. Although no single universal order may exist for chromosomes, we arranged evolutionary map distances into networks depicting how often exchange between microsatellites can be expected (SI Appendix, Fig. S2). Our results demonstrate that the microsatellites were widely dispersed through the genome.
Statistical Analysis of Population Survey Data.
We used two general approaches to test the microsatellites and allozymes for host-related and geographic allele frequency differentiation: standard population genomics FST outlier approaches (36–40) and a Monte Carlo approach using nonparametric bootstrapping.
Outlier Analyses.
We tested for FST outliers in two different ways, using the methods of Beaumont and Nichols (36) and Foll and Gaggiotti (39). For both methods, we conducted separate analyses for each of the four sympatric sites and pooled variants segregating at each microsatellite and allozyme locus into two major allele classes, as described below for the Monte Carlo bootstrapping analysis. We calculated FST values in apple and hawthorn populations using a MATLAB computer program written by J.F. based on the standard formula of Wright (41). The two different outlier methods yielded congruent results (Results). See SI Appendix for more details.
Monte Carlo Bootstrapping Analysis.
Our second approach to analyzing the population data involved nonparametric Monte Carlo bootstrapping. We examined every possible combination of alleles at a microsatellite or allozyme locus to determine the combination (i) that explained the largest amount of latitudinal allele frequency variation among apple and hawthorn fly populations across sites, and (ii) produced the highest levels of linkage disequilibrium with other markers mapping to the same chromosome. Our metric for assessing geographic variation was the product, calculated separately for apple and hawthorn races, of the variance explained by the linear regression of allele frequencies on latitude (R2 value), multiplied by the absolute value of the slope (b) of the regression line (i.e., R2 b). We used the standardized composite linkage disequilibrium coefficient of Weir (34) to quantify nonrandom associations of alleles between pairs of loci within apple and hawthorn fly populations at sites. A total composite disequilibrium coefficient for apple and hawthorn host races was calculated as the mean coefficient across sites for linked loci. This total composite value was then multiplied by the R2 b value for a locus to assess combinations of alleles producing the highest levels of both geographic variation and linkage disequilibrium. To test whether the R2 b value for a locus indicated significant latitudinal variation, we pooled microsatellite and allozyme allele frequencies for the locus separately across apple and hawthorn sites, and constructed random apple and hawthorn fly data sets by resampling alleles with replacement from their respective host race gene pools. We then tested every possible allele combination for a simulated data set to determine whether a combination existed that had a higher R2 b value than the actual value. We determined statistical significance by assessing the proportion of 100,000 simulation runs for apple and hawthorn populations that generated a greater R2 b value than the actual value.
We tested for host-related genetic differentiation in three different ways using the combinations of alleles at loci determined above, by calculating (i) the significance level for the heterogeneity in slopes (b) between the linear regressions of allele frequencies on latitude among sympatric apple versus hawthorn fly populations, as determined by F-tests (42); (ii) F-ratios for the host and host-by-latitude interaction effects generated from a two-way ANOVA of the variables host and latitude on allele frequencies across sympatric apple and hawthorn fly populations (statistically testing the F-ratios by Monte Carlo bootstrapping in a similar manner as discussed above for R2 b values, excluding Brazos Bend, Texas site); and (iii) the overall significance level for Fisher's exact tests for allele frequency differences between hawthorn and apple populations at individual sympatric sites.
Selection Experiment.
The rationale behind the selection experiment was to expose pupae to dichotomous rearing conditions inducing diapause versus direct nondiapause development to test for a genetic response at the microsatellite and allozyme loci. The flies analyzed in the current study formed part of a larger selection experiment on allozymes performed on 6,460 wild-collected hawthorn flies sampled as larvae from ~12,000 infested fruits collected from the Grant, Michigan site on Sept. 15, 1989 (32). Fly pupae were exposed to either 7 days (diapausing) or 35 days (nondiapausing) rearing conditions under a 15-/9-h light/dark cycle in a constant-temperature (26 °C) room. We used only flies that pupated within a 3-day collection period in the lab, to help standardize prediapause rearing conditions before pupation. After 7 days, pupae in the diapause treatment were transferred to a 4 °C refrigerator for 5 months to simulate winter. After this time, pupae in the diapause treatment were removed from the cold and placed in a 21 °C incubator with a 14-/10-h light/dark cycle. Newly eclosing (i.e., emerging) adults were collected on a daily basis and stored at −80 °C. In contrast, pupae in the nondiapause treatment remained exposed to 26 °C, 15-/9-h light/dark conditions for 35 days and were not overwintered. R. pomonella has a facultative pupal diapause. If exposed to warm prewinter conditions for an extended period, flies will forgo an extended diapause, directly develop into adults, and eclose. The 28-day difference in the treatments emulates the approximate 3- to 4-week difference in the mean fruiting times of hawthorn versus apple in the field. Thus, adults eclosing in the 7-day treatment following the 5-month chilling period represent flies developing under conditions akin to those experienced by the hawthorn race. In comparison, nondiapausing hawthorn flies that eclosed in the 35-day treatment without chilling would represent individuals selected against if the hawthorn race were to shift to the earlier fruiting apple; these nondiapausing flies would emerge in the late fall at times when suitable host fruit were not available for mating and oviposition. Nondiapausing adults that eclosed ≤35 days postpuparium formation were collected on a daily basis as they emerged in the 35-day treatment and stored at –80 °C. DNA isolated from the heads of adults were genotyped for microsatellites, as described above, whereas the thorax and abdomen were used to score the same flies for allozymes. The total numbers of hawthorn flies genotyped in the selection experiment were n = 90 for the 7-day diapause treatment and n = 146 for the 35-day nondiapause treatment. Allele frequency differences between flies eclosing in the diapause versus nondiapause rearing treatments were tested for significance by Fisher's exact test. For the selection experiment analysis, all of the alleles segregating at a microsatellite or allozyme locus were pooled into two major classes, as described above for the population survey. More details are provided in SI Appendix.
Eclosion Study.
The eclosion study tested for genetic relationships of loci with adult emergence time. The hawthorn flies analyzed in the study represented the same 90 individuals genotyped for the 7-day treatment in the selection experiment. The apple flies analyzed in the eclosion study (n = 96 genotyped) came from a parallel 7-day prewinter, 5-month overwinter treatment performed on wild-collected apple flies sampled in infested fruit from the Grant, Michigan site on August 15, 1989 (32). Microsatellite and allozyme loci were tested for significance with eclosion time in two-way ANOVA analyses, with host and genotype as main effects.
Supplementary Material
Acknowledgments
We thank P. Abbot, A. Forbes, D. Funk, J. Mallet, A. Meyer, D. Schluter, F. Ubeda de Torres, and two anonymous reviewers for useful discussions. S. Velez and A. Forbes were involved in initial microsatellite development. P.N. and J.F. were fellows of the Wissenschaftskolleg zu Berlin during manuscript preparation. This work was supported by grants from the National Science Foundation and US Department of Agriculture (to J.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000939107/-/DCSupplemental.
References
- 1.Feder JL. In: Endless Forms. Howard DJ, Berlocher SH, editors. Oxford: Oxford Univ. Press; 1998. pp. 130–144. [Google Scholar]
- 2.Wu C-I. The genic view of the process of speciation. J Evol Biol. 2001;14:851–865. [Google Scholar]
- 3.Via S, West J. The genetic mosaic suggests a new role for hitchhiking in ecological speciation. Mol Ecol. 2008;17:4334–4345. doi: 10.1111/j.1365-294X.2008.03921.x. [DOI] [PubMed] [Google Scholar]
- 4.Noor MAF, Feder JL. Speciation genetics: Evolving approaches. Nat Rev Genet. 2006;7:851–861. doi: 10.1038/nrg1968. [DOI] [PubMed] [Google Scholar]
- 5.Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3:e285. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smadja C, Galindo J, Butlin RK. Hitching a lift on the road to speciation. Mol Ecol. 2008;17:4177–4180. doi: 10.1111/j.1365-294x.2008.03917.x. [DOI] [PubMed] [Google Scholar]
- 7.Via S. Natural selection in action during speciation. Proc Natl Acad Sci USA. 2009;106(Suppl 1):9939–9946. doi: 10.1073/pnas.0901397106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rice WR, Hostert EE. Laboratory experiments on speciation: What have we learned in forty years? Evolution. 1993;47:1637–1653. doi: 10.1111/j.1558-5646.1993.tb01257.x. [DOI] [PubMed] [Google Scholar]
- 9.Nosil P, Harmon LJ, Seehausen O. Ecological explanations for (incomplete) speciation. Trends Ecol Evol. 2009;24:145–156. doi: 10.1016/j.tree.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Feder JL, Nosil P. The efficacy of divergence hitchhiking in generating genomic islands during ecological speciation. Evolution. 2010 doi: 10.1111/j.1558-5646.2010.00943.x. doi:10.1111/j.1558-5646.2010.00943.x. [DOI] [PubMed] [Google Scholar]
- 11.Nosil P, Funk DJ, Ortiz-Barrientos D. Divergent selection and heterogeneous genomic divergence. Mol Ecol. 2009;18:375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- 12.Noor MAF, Garfield DA, Schaeffer SW, Machado CA. Divergence between the Drosophila pseudoobscura and D. persimilis genome sequences in relation to chromosomal inversions. Genetics. 2007;177:1417–1428. doi: 10.1534/genetics.107.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yatabe Y, Kane NC, Scotti-Saintagne C, Rieseberg LH. Rampant gene exchange across a strong reproductive barrier between the annual sunflowers, Helianthus annuus and H. petiolaris. Genetics. 2007;175:1883–1893. doi: 10.1534/genetics.106.064469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butlin RK. Population genomics and speciation. Genetica. 2008;138:409–418. doi: 10.1007/s10709-008-9321-3. [DOI] [PubMed] [Google Scholar]
- 15.Emelianov I, Marec F, Mallet J. Genomic evidence for divergence with gene flow in host races of the larch budmoth. Proc Biol Sci. 2004;271:97–105. doi: 10.1098/rspb.2003.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harr B. Genomic islands of differentiation between house mouse subspecies. Genome Res. 2006;16:730–737. doi: 10.1101/gr.5045006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bush GL. The taxonomy, cytology, and evolution of the genus Rhagoletis in north America (Diptera: Tephritidae) Bull Mus Comp Zool. 1966;134:431–562. [Google Scholar]
- 18.Feder JL, Chilcote CA, Bush GL. Genetic differentiation between sympatric host races of the apple maggot fly Rhagoletis pomonella. Nature. 1988;336:61–64. [Google Scholar]
- 19.McPheron BA, Smith DC, Berlocher SH. Genetic differences between host races of Rhagoletis pomonella. Nature. 1988;336:64–66. [Google Scholar]
- 20.Feder JL, Roethele JB, Filchak K, Niedbalski J, Romero-Severson J. Evidence for inversion polymorphism related to sympatric host race formation in the apple maggot fly, Rhagoletis pomonella. Genetics. 2003;163:939–953. doi: 10.1093/genetics/163.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feder JL, Bush GL. Gene frequency clines for host races of Rhagoletis pomonella in the midwestern United States. Heredity. 1989;63:245–266. [Google Scholar]
- 22.Feder JL, Chilcote CA, Bush GL. The geographic pattern of genetic differentiation between host-associated populations of Rhagoletis pomonella (Diptera: Tephritidae) in the eastern United States and Canada. Evolution. 1990;44:570–594. doi: 10.1111/j.1558-5646.1990.tb05939.x. [DOI] [PubMed] [Google Scholar]
- 23.Berlocher SH. Radiation and divergence in the Rhagoletis pomonella species group: Inferences from allozymes. Evolution. 2000;54:543–557. doi: 10.1111/j.0014-3820.2000.tb00057.x. [DOI] [PubMed] [Google Scholar]
- 24.Feder JL, et al. Host fidelity is an effective premating barrier between sympatric races of the apple maggot fly. Proc Natl Acad Sci USA. 1994;91:7990–7994. doi: 10.1073/pnas.91.17.7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gavrilets S. Fitness Landscapes and the Origin of Species. Princeton: Princeton Univ. Press; 2004. [Google Scholar]
- 26.Barrett RD, Schluter D. Adaptation from standing genetic variation. Trends Ecol Evol. 2008;23:38–44. doi: 10.1016/j.tree.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 27.Feder JL, Nosil P. Chromosomal inversions and species differences: When are genes affecting adaptive divergence and reproductive isolation expected to reside within inversions? Evolution. 2009;63:3061–3075. doi: 10.1111/j.1558-5646.2009.00786.x. [DOI] [PubMed] [Google Scholar]
- 28.Feder JL, et al. Allopatric genetic origins for sympatric host-plant shifts and race formation in Rhagoletis. Proc Natl Acad Sci USA. 2003;100:10314–10319. doi: 10.1073/pnas.1730757100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feder JL, et al. Mayr, Dobzhansky, and Bush and the complexities of sympatric speciation in Rhagoletis. Proc Natl Acad Sci USA. 2005;102(Suppl 1):6573–6580. doi: 10.1073/pnas.0502099102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seehausen O. Hybridization and adaptive radiation. Trends Ecol Evol. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Mallet J. Hybrid speciation. Nature. 2007;446:279–283. doi: 10.1038/nature05706. [DOI] [PubMed] [Google Scholar]
- 32.Feder JL, Roethele JB, Wlazlo B, Berlocher SH. Selective maintenance of allozyme differences among sympatric host races of the apple maggot fly. Proc Natl Acad Sci USA. 1997;94:11417–11421. doi: 10.1073/pnas.94.21.11417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Velez S, Taylor MS, Noor MAF, Lobo NF, Feder JL. Isolation and characterization of microsatellite loci from the apple maggot fly, Rhagoletis pomonella (Diptera: Tephritidae) Mol Ecol Notes. 2006;6:90–92. [Google Scholar]
- 34.Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. Micro-Checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. [Google Scholar]
- 35.Weir BS. Inferences about linkage disequilibrium. Biometrics. 1979;35:235–254. [PubMed] [Google Scholar]
- 36.Beaumont MA, Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proc Biol Sci. 1996;263:1619–1626. [Google Scholar]
- 37.Beaumont MA, Balding DJ. Identifying adaptive genetic divergence among populations from genome scans. Mol Ecol. 2004;13:969–980. doi: 10.1111/j.1365-294x.2004.02125.x. [DOI] [PubMed] [Google Scholar]
- 38.Beaumont MA. Adaptation and speciation: What can F(st) tell us? Trends Ecol Evol. 2005;20:435–440. doi: 10.1016/j.tree.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 39.Foll M, Gaggiotti OA. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: A Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G. LOSITAN: A workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinformatics. 2008;9:323. doi: 10.1186/1471-2105-9-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sokal RR, Rolf FJ. Biometry. 2nd Ed. San Francisco: Freeman; 1981. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.