Abstract
Sulforaphane [1-isothiocyanato-4-(methylsulfinyl)butane], a naturally occurring isothiocyanate derived from cruciferous vegetables, is a highly potent inducer of phase 2 cytoprotective enzymes and can protect against electrophiles including carcinogens, oxidative stress, and inflammation. The mechanism of action of sulforaphane is believed to involve modifications of critical cysteine residues of Keap1, which lead to stabilization of Nrf2 to activate the antioxidant response element of phase 2 enzymes. However, the dithiocarbamate functional group formed by a reversible reaction between isothiocyanate of sulforaphane and sulfhydryl nucleophiles of Keap1 is kinetically labile, and such modification in intact cells has not yet been demonstrated. Here we designed sulforaphane analogs with replacement of the reactive isothiocyanate by the more gentle electrophilic sulfoxythiocarbamate group that also selectively targets cysteine residues in proteins but forms stable thiocarbamate adducts. Twenty-four sulfoxythiocarbamate analogs were synthesized that retain the structural features important for high potency in sulforaphane analogs: the sulfoxide or keto group and its appropriate distance to electrophilic functional group. Evaluation in various cell lines including hepatoma cells, retinal pigment epithelial cells, and keratinocytes as well as in mouse skin shows that these analogs maintain high potency and efficacy for phase 2 enzyme induction as well as the inhibitory effect on lipopolysaccharide-induced nitric oxide formation like sulforaphane. We further show in living cells that a sulfoxythiocarbamate analog can label Keap1 on several key cysteine residues as well as other cellular proteins offering new insights into the mechanism of chemoprotection.
The increasing societal burden of cancer and other chronic degenerative diseases has led to an intense interest in the development of strategies designed to reduce the risk of these conditions. Chemoprotection with natural or synthetic agents offers an attractive approach to boost the body’s defenses to ward off environmental and endogenous insults (1, 2). The plant product sulforaphane [1-isothiocyanato-4-(methylsulfinyl)butane], derived from glucosinolates present in broccoli and other cruciferous vegetables (3), has served as a prototype for our understanding of chemoprotection by induction of phase 2 cytoprotective enzymes including NAD(P)H:quinone oxidoreductase (NQO1) (4). A broad series of animal and human studies has demonstrated the potential of sulforaphane to protect against the onset, or reduce the severity, of cancer (5–8), retinal disease (9), and skin damage (10–12) resulting from oxidative or electrophile damage (5, 6, 8), UV irradiation (10, 11), or genetic predisposition (9, 13).
Like other known phase 2 enzyme inducers (14), sulforaphane is an electrophilic compound that covalently modifies cysteine residues in proteins (15, 16). Such enzyme induction likely involves the Keap1-Nrf2-antioxidant response element pathway (15, 17). The prevailing hypothesis for sulforaphane’s cellular mechanism is that the natural product covalently modifies Keap1 on one or more of its 27 cysteine residues, altering the Keap1-Nrf2-Cullin-3 protein complex, and allowing Nrf2 to translocate to the nucleus where it activates gene expression (Fig. 1A). However, other protein targets for sulforaphane have also been proposed (4, 18), and mechanistic questions remain.
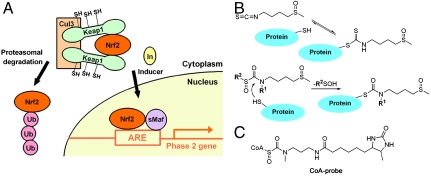
Fig. 1.
Proposed chemoprotective mechanism of sulforaphane for phase 2 gene activation involves covalent Cys modification of Keap1. (A) Mechanism of phase 2 enzyme induction. In the basal state, the transcription factor Nrf2 is efficiently ubiquitylated and targeted for proteasomal degradation by forming a complex with Keap1 and Cul3 E3 ligase. Inducers such as sulforaphane modify cysteines of Keap1, which leads to stabilization and translocation of Nrf2 to the nucleus, and its binding to ARE and stimulation of phase 2 gene transcription. (B) Reactions of sulforaphane and sulfoxythiocarbamate analog with a thiol. The isothiocyanate group of sulforaphane reacts with sulfhydryl groups on protein to form dithiocarbamate adduct that appear to be kinetically labile (Top), whereas sulfoxythiocarbamate analog forms a relatively stable thiocarbamate derivative (Bottom). (C) Structure of sulfoxythiocarbamate CoA probe developed previously (24).
Whereas sulforaphane has been shown to react directly with purified, recombinant Keap1 in solution (15, 19), it has not yet been established that sulforaphane modifies Keap1 in cells. Human Keap1 is a 70-kDa cysteine-rich protein (624 amino acids and 27 cysteines), comprising five domains: (i) N-terminal region (NTR), (ii) BTB (broad complex, tramtrack, or bric-a-brac), an evolutionarily conserved protein-protein interaction motif that often dimerizes with other BTB domains (20), (iii) intervening region (IVR), a cysteine-rich region, (iv) double glycine region (DGR), a domain that comprises six Kelch motifs and binds to the Neh2 domain of Nrf2 (21), and (v) C-terminal region (CTR). It is difficult to express soluble Keap1, which is highly prone to oxidation and oligomerizes easily. Sulforaphane contains the highly reactive isothiocyanate functionality that forms dithiocarbamate products with sulfhydryl nucleophiles both enzymatically catalyzed by glutathione transferase and nonenzymatically (Fig. 1B) (22, 23). Such dithiocarbamate adduct formation is a reversible process, which complicates their isolation and characterization (22).
In prior studies on coenzyme A (CoA)-utilizing enzymes, it was found that a sulfoxythiocarbamate-CoA analog (CoA-probe, Fig. 1C) was a relatively mild but selective electrophilic probe for acetyltransferases (24). Mass spectrometric analysis revealed that cysteine residues were selectively targeted by this CoA probe, and stable thiocarbamate adducts could be readily isolated. We considered the possibility that replacing the isothiocyanate moiety of sulforaphane with a sulfoxythiocarbamate group (Fig. 1B) might allow retention of the natural product’s chemoprotective properties and confer enhanced potential for mechanistic analysis. Below, we report the design and synthesis of a series of sulfoxythiocarbamate sulforaphane analogs and describe their chemopreventive and protein labeling properties.
Results
Design and Synthesis of Sulfoxythiocarbamate Sulforaphane Analogs.
The simple cellular bioassay of measuring NQO1 enzyme activities of murine hepatoma cells has accurately predicted and quantified the induction potency of many compounds. These potencies have been expressed as CD (Concentrations required to Double the NQO1 activity) values (25, 26). With this assay, several structural derivatives of sulforaphane have been compared for their potencies (3, 27). These findings highlight the importance of the sulfoxide group of sulforaphane in addition to the isothiocyanate group. Thus n-hexyl (CD 15 μM), n-cyclohexyl (CD 56 μM), n-phenyl (inactive), and n-benzyl isothiocyanate (2–3 μM) are much less potent inducers when compared to sulforaphane (CD = 0.22 μM) (27). This sulfoxide group could be replaced by a polar functional group, ketone, without losing its activity while the corresponding sulfone (CD 0.82 μM) and sulfide (CD 2.3 μM) showed somewhat lower potencies (27). In addition, the length of the methylene bridge between two important functional groups, isothiocyanate and sulfoxide, is an important determinant of potency since a shorter or longer bridge resulted in lower potencies (3). Interestingly, the structure-potency studies of potent triterpenoid Michael acceptor inducers also illustrated the critical importance of two electrophilic functional groups separated by a potentially similar distance to that found in sulforaphane (28). Considering these structural features, sulfoxythiocarbamate analogs were designed to retain the 4-methylsulfinylbutyl group moiety of sulforaphane with replacement of the isothiocyanate by a sulfoxythiocarbamate group (Fig 1B). In order to substitute the isothiocyanate functionality of sulforaphane with a sulfoxythiocarbamate group, two significant chemical constraints had to be overcome. First, sulfoxythiocarbamates, unlike isothiocyanates, require alkyl substitution on the sulfur atom (R2 group, Fig. 1B) to provide chemical stability. Second, the nitrogen atom in the sulfoxythiocarbamate must also be alkyl-substituted to reduce hydrolytic breakdown (R1 group, Fig. 1B). These chemical requirements led us to explore a range of derivatives (Fig. 2) whose substitution differences could influence potency, toxicity, and protein labeling. Since analogs of sulforaphane in which the sulfoxide is replaced by a carbonyl group show very similar inducer potencies (27), we also explored the effects of replacement of the sulfoxide moiety with a carbonyl functionality (8a–8f, Fig. 2). A key conserved feature of our analogs is a backbone containing an electrophilic carbonyl group attached via a butyl linker to the ketone or sulfoxide moiety. The distance between the sulfoxide that contributes to inducer potency and the electrophilic reactive carbon center of sulforaphane is thus preserved in sulfoxythiocarbamate analogs.
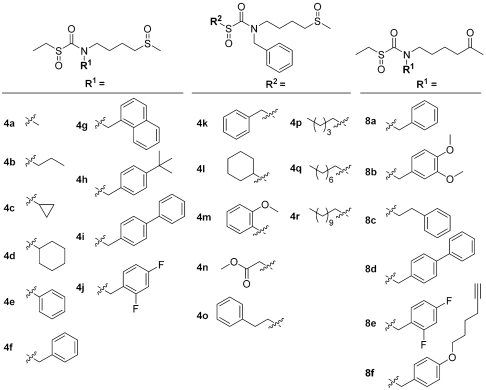
Fig. 2.
Structures of sulfoxythiocarbamate sulforaphane analogs. Two classes of analogs, one with sulfoxide group (4a–4r) and another with ketone-group (8a–8f), were generated with alkyl or aryl modifications on R1 and R2 positions.
The basic synthetic approach to the designed analogs is shown in two related sequences outlined in Scheme S1 in SI Text. Preparation of the sulfoxide-containing analogs, 4a–4r, began by controlled oxidation of 4-hydroxybutyl-methyl thioether. Mesylation followed by amine displacement, and thiocarbamylation gave the penultimate precursor that could be oxidized with 3-chloroperbenzoic acid or Oxone to generate the sulfoxythiocarbamate functionality. Synthesis of the keto analogs began with 6-chloro-2-hexanone that, after protection of the ketone, was derivatized in a similar fashion to provide the target compounds, 8a–8f.
Comparison of Reactivities of Sulforaphane and Sulfoxythiocarbamate Analogs with a Thiol.
To examine the relative electrophilicity of the sulfoxythiocarbamate and the isothiocyanate functionalities, we compared the reaction rates of β-mercaptoethanol with 4i, 8a, and sulforaphane. These reactions could be readily fit to a pseudofirst order kinetic model with the sulfoxythiocarbamates showing approximately 70-fold slower rate constants compared to sulforaphane (Figs. S1 and S2). This finding is consistent with the hypothesis that the sulfoxythiocarbamate is a significantly weaker electrophile toward thiols relative to the isothiocyanate group.
NQO1 Induction by Sulfoxythiocarbamate Analogs in Cells.
Next, our synthetic sulforaphane analogs were assayed in a murine hepatoma cell line (Hepa1c1c7) to quantify the induction of NQO1 activity as a marker of phase 2 enzyme induction (25, 26). Cellular toxicity of the sulfoxythiocarbamates was also determined by measuring cell survival based on protein concentration (29). All sulfoxythiocarbamate compounds tested were found to be inducers of NQO1, and dose-response curves were used to obtain CD values (Table 1) (26). As illustrated, benzyl substituents (4f–4j) in R1 have higher potencies when compared to an aliphatic alkyl chain or phenyl group (4a–4e). Interestingly, benzyl isothiocyanate is also a more potent inducer than hexyl or phenyl isothiocyanate (27). In addition, compounds with hydrophobic substitutions on sulfur, nitrogen, or both (4g–4i, 4o, 4r, and 8d) tended to give the lowest CD values (2–6 μM). However, larger alkyl and bulky benzyl substituents on sulfoxythiocarbamate analogs (4i, 4q, 4r, and 8d) tended to increase the cellular toxicities (LD50). We thus focused on a set of compounds (4f, 8a, 8f, and structures in Fig. S3D) that showed relatively high inducer potencies (low CD) and low toxicities (LD50). The chemoprotective index (CI = LD50/CD) (26, 30) of these three compounds in hepatoma cells was in the range of 8–20, within one order of sulforaphane (CI = 80), which while more potent, is also more toxic than some of the sulfoxythiocarbamates. NQO1 induction by 4f, 8a, and 8f was also analyzed in two other cell lines, human retinal pigment epithelial cells (ARPE-19) (31) and murine keratinocytes (PE) (32) (Table 1 and Fig. S3). We found that 8a (CD 1.5 μM) was essentially equipotent to sulforaphane (1.4 μM) in ARPE-19 cells, and this sulfoxythiocarbamate was at least fivefold less toxic than the natural product. Compounds 8f and 4f were just slightly less potent than 8a in this cell line. The results in the keratinocyte line showed that the CI values for 8a and sulforaphane were essentially identical. Furthermore, the maximal degree of NQO1 induction by 8a was greater than that of sulforaphane in keratinocytes (Fig. S3C).
Table 1.
NQO1 induction potency and toxicity of sulfoxythiocarbamate analogs in three cell lines including murine hepatoma cells (Hepa1c1c7), human retinal pigment epithelial cells (ARPE-19), and murine keratinocytes (PE)
| Hepa1c1c7 | ARPE-19 | PE | ||||||||||||
| Name |
CD, μM |
LD50, μM |
Name |
CD, μM |
LD50, μM |
Name |
CD, μM |
LD50, μM |
Name |
CD, μM |
LD50, μM |
Name |
CD, μM |
LD50, μM |
| 4a | 94 | > 200 | 4j | 7.3 | > 50 | 8a | 5.4 | 115 | 4f | 4.8 | > 200 | 4f | 13 | > 200 |
| 4b | 35 | > 200 | 4k | 9.8 | > 50 | 8b | 5.4 | 120 | 4i | 3.8 | 21 | 8a | 2.5 | 128* |
| 4c | 43 | > 200 | 4l | ND | — | 8c | 12 | > 50 | 4o | 1.7 | 29 | 8b | 2.8 | 131* |
| 4d | 64 | > 200 | 4m | 12.6 | > 50 | 8d | 2.1 | 12 | 8a | 1.5 | 120* | 8f | 3.8 | 50 |
| 4e | 43 | > 200 | 4n | ND | — | 8e | 7.1 | > 50 | 8b | 1.8 | 123* | SF | 0.25 | 13* |
| 4f | 13 | > 200 | 4o | 4.3 | 37 | 8f | 5.5 | 43 | 8f | 5 | 38 | |||
| 4g | 5.3 | > 50 | 4p | 8.2 | > 50 | SF | 0.23 | 18 | SF | 1.4 | 14* | |||
| 4h | 3.8 | 32 | 4q | 7 | 12.5 | |||||||||
| 4i | 2.8 | 16 | 4r | 2.6 | 10 | |||||||||
Cells were grown in 96-well plates for 24 h and incubated with serial dilutions of each compound for 48 h. NQO1 activity and total protein concentrations were determined in cell lysates by Prochaska method (26) and the BCA assay, respectively. The specific NQO1 activity or total protein concentrations were plotted vs. compound concentrations with values of mean ± SEM. (n = 8). The concentrations that give a twofold increase of NQO1 activity or 50% decrease of total protein concentrations when compared to control were determined as CD or LD50. Standard errors were < ± 20% for values shown. ND, not detemined; less than twofold induction.
*Values were determined by extrapolation.
Sulfoxythiocarbamate Analogs Work Through the Keap1/Nrf2/ARE Pathway.
To confirm that, like sulforaphane, the sulfoxythiocarbamates stimulate NQO1 via the antioxidant response element (ARE) transcriptional pathway, we examined eight sulfoxythiocarbamate analogs by using a previously established ARE-luciferase reporter stably transfected in MCF7 cells (AREc32) (33). Each of these compounds led to a significant increase in the luciferase signal, and the most powerful effects were caused by 8f and 4o as shown in Fig. S4A. A time course over three days of luciferase induction with 6.25 μM 8f and 4o revealed that the sulfoxythiocarbamates showed a 20-fold maximal induction and sustained increase in the level of reporter, comparing favorably with the effects of 2 μM sulforaphane (Fig. S4B). As expected, cellular depletion of glutathione with L-buthionine-sulfoximine enhanced luciferase induction by 8f and 4o in a fashion comparable to that of sulforaphane (33) (Fig. S4C). These results suggest that glutathione can antagonize sulfoxythiocarbamate action by chemical modification through thiol attack.
We further examined the specific importance of Nrf2 and Keap1 in NQO1 induction by bioassays with 8f in mouse embryonic fibroblasts (MEF) derived from wild-type, Nrf2-knockout, or Keap1/Nrf2-double knockout mice (Fig. S5). Like sulforaphane, compound 8f stimulates NQO1 activity in wild-type (WT) MEF in a dose-dependent manner (Fig. S5A). In sharp contrast, in Nrf2-knockout and in Keap1/Nrf2-double knockout MEF, the basal levels of NQO1 activity are much lower than in WT cells and are not affected by either sulforaphane or 8f. Furthermore, immunoblot analysis revealed that these differences in NQO1 enzyme activity between WT and knockout MEF correspond to differences in protein levels (Fig. S5 B and C). Taken together with the ARE-reporter assays, these experiments demonstrate that, in both human and murine cells, sulforaphane and its sulfoxythiocarbamate analogs induce the expression of cytoprotective (phase 2) genes by targeting the Keap1/Nrf2/ARE pathway.
Inhibition of Lipopolysaccharide-Induced Nitric Oxide Formation by Sulfoxythiocarbamate Analogs in Macrophage-Like Cells.
In addition to inducing phase 2 enzymes, sulforaphane has demonstrated antiinflammatory action including inhibition of the formation of reactive oxygen species in mouse peritoneal macrophage and murine macrophage-like cell line (RAW264.7) (34). To explore the effects of the sulfoxythiocarbamates on macrophages, we utilized a lipopolysaccharide macrophage activation assay and measured nitric oxide (NO) levels in the presence of 8a and 4f. These compounds blocked NO generation with IC50 values of 2.5 and 8 μM, respectively, modestly less potent than sulforaphane in this assay (IC50 0.5 μM) (Fig. S6).
Inducer Potency of Sulfoxythiocarbamate Analog 8a in Mouse Skin.
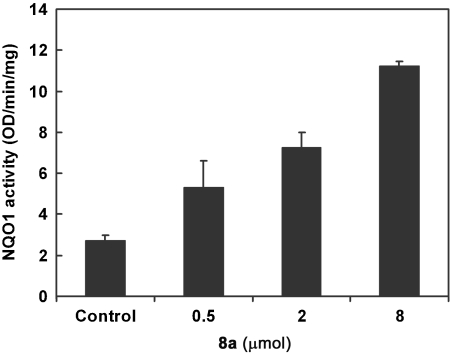
We next investigated the in vivo properties of sulfoxythiocarbamate 8a in a mouse epidermis NQO1 induction assay. Topical application of three doses of 8a at 24-h intervals led to fivefold induction of NQO1 in the dorsal skin (Fig. 3), with the level of induction at 0.5 μmol comparable to that of sulforaphane.
Fig. 3.
Evaluation of sulfoxythiocarbamate analog for NQO1 enzyme induction on mouse skin. The back of each SKH-1 hairless mouse (n = 3) was topically treated with three concentrations of 8a (indicated amounts in 40 μL of 80% aq. acetone) and solvent only over ca. 2.5 cm2 area for three doses at 24-h intervals. Mice were euthanized 24 h after the last dose, and dorsal skin was harvested. NQO1 specific activity was measured in supernatant fractions of homogenates of skin sections treated with 8a or solvent (control). Means ± SD are shown.
Labeling Keap1 by Sulfoxythiocarbamate Analog 8f in Cells.
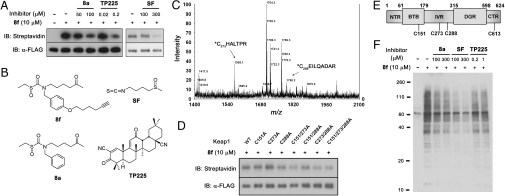
Since alkynyl sulfoxythiocarbamate analog 8f was shown to be a relatively potent, efficacious, and nontoxic chemopreventive agent, comparable to sulforaphane, we elected to use it as a mechanistic probe for cellular Keap1. Whereas sulforaphane covalently modifies specific cysteine residues of recombinant Keap1 in solution (15, 19), this has not been demonstrated in intact cells. We transiently transfected FLAG-tagged Keap1 in HEK293 cells and showed that it could be efficiently immunoprecipitated with immobilized anti-FLAG antibody, irrespective of exposure of cells to 8f (bottom blot, Fig. 4A). Taking advantage of the click reaction (35), we treated antibody-immobilized FLAG-Keap1 with biotin-azide (36) and then eluted it with FLAG peptide and subjected it to streptavidin blotting. As shown in Fig. 4A, cells treated with sulfoxythiocarbamate 8f produced a major and clean band at identical molecular weight to FLAG-Keap1, dependent on treatment with 8f. This provides evidence of covalent modification of transfected FLAG-Keap1 by 8f. To examine further the significance of this labeling, we attempted to compete the modification with the non-alkyne-containing inducer (8a), sulforaphane, and the extremely potent triterpenoid inducer (TP225) (Fig. 4B) (28). Treatment of FLAG-Keap1-transfected cells with each of the three inducers followed by incubation with 8f led to reduced labeling of FLAG-Keap1 compared with 8f by itself, as shown in Fig. 4A. It is noteworthy that TP225 blocked FLAG-Keap1 labeling approximately 1,000-fold more potently than either 8a or sulforaphane, consistent with the known potency of this powerful pentacyclic inducer (28).
Fig. 4.
Sulfoxythiocarbamate analog conjugates on Keap1 cysteines as well as potential target proteins in cells. (A) Labeling FLAG-Keap1 in cells. HEK293 cells transiently expressing FLAG-tagged Keap1 were treated with 8f for 0.5 h, or pretreated with 8a, sulforaphane, or TP225 for 0.5 h before incubation of 8f for 0.5 h. FLAG-Keap1 was immunoprecipitated from cell lysates with anti-FLAG antibody, subjected to reaction with biotin azide and eluted with FLAG peptide. Samples were immunoblotted with streptavidin (Top) and anti-FLAG antibody (Bottom). (B) Structure of compounds, 8f, 8a, sulforaphane, and TP225. (C) MALDI-MS spectrum of FLAG-Keap1 tryptic digests labeled by 8f. Eluted FLAG-Keap1 was digested by trypsin, followed by incubation with avidin-coated beads. Biotin-conjugated peptides were eluted with aq. acetonitrile and analyzed by MALDI mass spectrometry, which showed two peaks corresponding to modified Cys 273 and Cys 288 peptides. (D) Labeling FLAG-Keap1 cysteine to alanine mutants. Cells transiently expressing FLAG-tagged WT or mutant Keap1 were treated with 8f for 0.5 h. FLAG-Keap1 proteins, eluted after immunoprecipitation with anti-FLAG antibody and conjugation with biotin azide, were immunoblotted with streptavidin (Top) and anti-FLAG antibody (Bottom). (E) Schematic structure of Keap1 with five domains that include NTR, BTB, IVR, DGR, and CTR. Cys 288 and Cys 613 modified peptides were found by LTQ mass analysis. (F) Labeling potential target proteins in cells. After incubation of compounds as described in A, cell lysates were subjected to click reaction with biotin azide, and analyzed by SDS-PAGE and immunoblotting with streptavidin HRP.
Identification of Cysteine Residues of Keap1 Labeled by Sulfoxythiocarbamate 8f.
To identify specific modification site(s) in Keap1 labeled by 8f, immobilized Keap1 was biotinylated, eluted with FLAG peptide, and subjected to exhaustive trypsin digestion. The tryptic digest was exposed to avidin-coated beads, and the captured peptides were eluted with acetonitrile and analyzed by MALDI and separately by linear trap quadrupole (LTQ) mass spectrometry. MALDI MS showed the appearance of two prominent peaks not present in control samples that were in precise agreement with molecular weights for modified Cys 273 (m/z 1569, C*HALTPR) and Cys 288 (m/z 1790, C*EILQADAR) peptides (Fig. 4C). Tandem MS analysis confirmed these assignments and revealed further that Cys 613 (SGVGVAVTMEPC*R) was also labeled by 8f (Fig. S7).
Transfected FLAG-Keap1 Cys151Ala, Cys273Ala, and Cys288Ala proteins were studied as single mutants and in combinations to dissect the importance of these mutations in sulfoxythiocarbamate labeling. In transgenic mouse and cell transfection experiments (17, 37–39), each of these cysteines in Keap1 has been shown to be important for repressing Nrf2 activity or conferring resistance to inducers. Whereas single and paired cysteine Keap1 mutants showed minimal or modestly diminished labeling by 8f, the triple mutant showed a sharper reduction in modification by 8f (Fig. 4D). This is consistent with the mass spectrometry findings and underscores that multiple cysteine sites are significant in the targeting of Keap1 by inducers.
Exploring the Target Proteins Labeled by Sulfoxythiocarbamate 8f in Cells.
To explore the potential scope of other protein targets of 8f, cells were treated with 8f for 30 min, and cell extracts were reacted with biotin azide and then blotted with streptavidin. As shown in Fig. 4F, a large number of bands were detected in cells treated with 8f. Many of these bands could be effectively competed by 100 μM 8a and sulforaphane, and to a lesser degree with TP225 at 1 μM. These surprising results underscore that there are many potential pathways that can be influenced by these electrophilic compounds that could lead to complex pharmacology, not readily recapitulated with single gene knockouts.
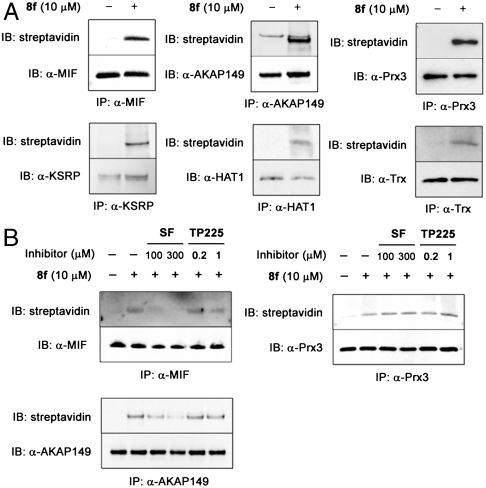
We then employed a mass spectrometry-based proteomic approach to identify candidate proteins modified by 8f. After treating HEK293 cells with 8f, cell extracts were reacted with biotin azide under click conditions, immobilized on streptavidin beads, and subjected to trypsin followed by mass spectrometric analysis. Over 100 proteins were identified in this fashion (Table S1). Using immunoprecipitation-Western analysis (Fig. 5), we showed that at least six of these appear to be bona fide cellular targets of 8f, including macrophage migration inhibitory factor (MIF), peroxiredoxin 3 (Prx3), histone acetyltransferase 1 (HAT1), A-kinase anchoring protein 149 (AKAP149), KH-type splicing regulatory protein (KSRP), and thioredoxin (Trx) (Fig. 5A). We further showed that, like Keap1, labeling of MIF and AKAP149 by 8f can be blocked by pretreatment with sulforaphane or TP225, whereas Prx3 labeling is not competed by these inducers (Fig. 5B). This is consistent with the concept that there are overlapping but distinct pharmacodynamic profiles for 8f vs. other chemopreventive inducers.
Fig. 5.
Validation of selected target proteins labeled by sulfoxythiocarbamate analog 8f in cells. (A) Target proteins labeled by 8f. HEK293 cells were treated with 8f or vehicle for 2.5 h. (B) Competition by sulforaphane or TP225. HEK293 cells were pretreated with sulforaphane, TP225, or vehicle for 0.5 h before incubation with 8f for 0.5 h. Each protein was immunoprecipitated from cell lysates, subjected to click reaction with biotin azide on beads. Eluted samples were immunoblotted with streptavidin as well as protein specific antibodies.
Discussion
Here we have developed a series of sulforaphane analogs that show promising chemoprotective properties and proved to be useful in establishing Keap1 as a direct target of these agents in cells. A number of mass spectroscopic studies have examined recombinant Keap1 labeling in vitro by dexamethasone mesylate (15), iodoacetamide-biotin (40–43), and several other agents (44). Iodoacetamide-biotin has also been studied in cells (41, 42). There are major structural differences between sulforaphane and these other agents, particularly with respect to the electrophilic carbon, raising concerns about their mechanistic relevance in predicting the behavior of the plant natural product. Moreover, the complexity of isolating and purifying recombinant Keap1 together with the lack of a compelling bioassay to ensure its functional integrity limit the potential significance of in vitro modification studies. Nevertheless, each of the sites identified here in the cellular assays with 8f (Cys 273, Cys 288, and Cys 613) have been also identified in the in vitro dexamethasone mesylate analyses (15). This convergence along with competitive reduction in labeling by the established agents, sulforaphane and TP225, as well as transgenic mouse studies highlighting the importance of Cys 273 and Cys 288 (38), strongly support the role of these residues in mediating repressor activity. As pointed out previously (15, 17), these cysteine side chains may have significantly increased chemical reactivity because of the presence of neighboring basic residues that can lower the pKa values of their interactive cysteine thiols (45). Left unsettled is the precise consequence of these cysteine modifications on the interaction of Keap1 with Nrf2. The current model that cysteine modification of Keap1 influences protein-protein interactions in the Keap1-Nrf2-Cullin-3 complex is consistent with the literature.
The structure-potency data in this paper show that the small variation of structural modifications on analogs (e.g., 8a and 8b) appears to give less significant difference in their potencies. These data as well as the fact that many structurally distinct compounds are known to be inducers of NQO1 (14) may imply that the electrophile sensor protein Keap1 has evolved to respond to a wide range of compounds with less defined structural specificity in order to facilitate the protective response of cells to exogenous oxidants and electrophile toxicity. Nevertheless, structural modifications of sulfoxythiocarbamate analogs prepared here displayed a range of potencies for NQO1 induction (CD 2.1–94 μM), which suggest that there are important structural requirements for phase 2 enzyme induction.
The distinctive relative inducing potentials of sulforaphane vs. the sulfoxythiocarbamates in the three cell lines, murine hepatoma, retinal pigment epithelial, and murine keratinocytes, illustrate the necessity for caution in drawing conclusions about chemoprotective properties from single cell assays. Such differential effects may be due to the specific nature of the ARE pathway in individual cell types, but are perhaps more likely to be related to the range of protein targets modified by these compounds. As revealed here, sulfoxythiocarbamate analogs are considerably less reactive electrophiles with thiols than is sulforaphane, which might give a more restricted set of adducts in cells. Nevertheless, the widespread labeling by 8f in HEK293 cells provides an indication of the many proteins that can be carbamylated by this agent, and it is difficult to imagine that some of these will not have significant functional implications. Using a proteomics approach, we have discovered at least six protein targets of 8f other than Keap1, MIF, AKAP149, Prx3, Trx, HAT1, and KSRP. Inhibition of MIF, a well-established chemokine (46), has previously been associated with a derivative of sulforaphane, phenethyl isothiocyanate (47), and may be important in blocking inflammation. Modification of AKAP149 by 8f could influence protein kinase cell signaling, whereas modification of Prx3 and Trx could affect the antioxidant response. The identification and characterization of such protein targets of 8f and other agents facilitates our understanding of the chemoprotection mosaic associated with specific compounds. Finally, these studies further highlight the utility of the sulfoxythiocarbamate functionality for proteomic and pharmacologic investigation.
Methods
NQO1 Assays.
Cells were grown for 24 h in 96-well plates (10,000 per well for Hepa1c1c7 and ARPE-19 cells; 30,000 per well for PE cells). Culture media were then replaced with fresh media containing serial dilutions of compounds dissolved in acetonitrile (final 0.5%). After incubation for 48 h, the spent culture media were discarded, and adhered cells were washed three times with PBS and lysed with 0.08% digitonin (75 μL/well). A fraction of the cell lysates (20 μL) was used for determining protein concentration by the bicinchoninic acid (BCA) assay. The remaining cell lysates were used for measuring NQO1 activity by the Prochaska method (26).
NQO1 Induction of SKH-1 Hairless Mouse Skin.
Eight-week-old female SKH-1 hairless mice (n = 3) were treated three times at 24-h intervals on their backs with 0, 0.5, 2, and 8 μmol of 8a dissolved in 80% aqueous (aq.) acetone (vol/vol, 40 μL) over ca. 2.5 cm2 area. Mice were euthanized 24 h after the final dose. Each treated segment of their dorsal skin was removed, pulverized in liquid N2, and the resulting powder was homogenized in 10 volumes of 0.25 M sucrose in 10 mM Tris buffer (pH 7.4). After three freeze-thaw cycles, centrifugation at 14,000 rpm and 4 °C for 30 min yielded clear supernatant fractions, which were analyzed for protein concentration (BCA) assay and NQO1 activity. Experiments were in compliance with the National Institutes of Health Guidelines and were approved by The Johns Hopkins University Animal Care and Use committee.
Other Methods.
Detailed experimental procedures for synthesis of compounds, cell culture, transfection and protein labeling, and mass analysis can be found in SI Text.
Supplementary Material
Acknowledgments.
We thank M. Yamamoto (Tohoku University) and T. W. Kensler (Johns Hopkins University) for the generous gift of the FLAG-Keap1 plasmid and for the mouse embryonic fibroblasts, M. B. Sporn, T. Honda, and G. Gribble (Dartmouth School of Medicine) for providing triterpenoid TP225, and J. D. Hayes (University of Dundee) for the antibody against NQO1. This work was supported by National Institutes of Health Grants CA094076, GM62437, U54 RR020839, and CA093780, American Cancer Society Grants IRG5800543 and RSG-07-157-01-CNE, the Lewis B. and Dorothy Cullman Foundation, the Kaufman Foundation, the American Institute for Cancer Research, and Cancer Research UK (C20953/A10270)
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004104107/-/DCSupplemental.
References
- 1.Talalay P, Fahey JW. Phytochemicals from cruciferous plants protect against cancer by modulating carcinogen metabolism. J Nutr. 2001;131:3027S–3033S. doi: 10.1093/jn/131.11.3027S. [DOI] [PubMed] [Google Scholar]
- 2.Surh YJ. Cancer chemoprevention with dietary phytochemicals. Nat Rev Cancer. 2003;3:768–780. doi: 10.1038/nrc1189. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Talalay P, Cho CG, Posner GH. A major inducer of anticarcinogenic protective enzymes from broccoli–isolation and elucidation of structure. Proc Natl Acad Sci USA. 1992;89:2399–2403. doi: 10.1073/pnas.89.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Juge N, Mithen RF, Traka M. Molecular basis for chemoprevention by sulforaphane: A comprehensive review. Cell Mol Life Sci. 2007;64:1105–1127. doi: 10.1007/s00018-007-6484-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, et al. Anticarcinogenic activities of sulforaphane and structurally related synthetic norbornyl isothiocyanates. Proc Natl Acad Sci USA. 1994;91:3147–3150. doi: 10.1073/pnas.91.8.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kensler TW, et al. Effects of glucosinolate-rich broccoli sprouts on urinary levels of aflatoxin-DNA adducts and phenanthrene tetraols in a randomized clinical trial in He Zuo township, Qidong, People’s Republic of China. Cancer Epidem Biomar. 2005;14:2605–2613. doi: 10.1158/1055-9965.EPI-05-0368. [DOI] [PubMed] [Google Scholar]
- 7.Cornblatt BS, et al. Preclinical and clinical evaluation of sulforaphane for chemoprevention in the breast. Carcinogenesis. 2007;28:1485–1490. doi: 10.1093/carcin/bgm049. [DOI] [PubMed] [Google Scholar]
- 8.Conaway CC, et al. Phenethyl isothiocyanate and sulforaphane and their N-acetylcysteine conjugates inhibit malignant progression of lung adenomas induced by tobacco carcinogens in A/J mice. Cancer Res. 2005;65:8548–8557. doi: 10.1158/0008-5472.CAN-05-0237. [DOI] [PubMed] [Google Scholar]
- 9.Kong L, et al. Delay of photoreceptor degeneration in tubby mouse by sulforaphane. J Neurochem. 2007;101:1041–1052. doi: 10.1111/j.1471-4159.2007.04481.x. [DOI] [PubMed] [Google Scholar]
- 10.Talalay P, et al. Sulforaphane mobilizes cellular defenses that protect skin against damage by UV radiation. Proc Natl Acad Sci USA. 2007;104:17500–17505. doi: 10.1073/pnas.0708710104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinkova-Kostova AT, et al. Protection against UV-light-induced skin carcinogenesis in SKH-1 high-risk mice by sulforaphane-containing broccoli sprout extracts. Cancer Lett. 2006;240:243–252. doi: 10.1016/j.canlet.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 12.Dinkova-Kostova AT, et al. Induction of the phase 2 response in mouse and human skin by sulforaphane-containing broccoli sprout extracts. Cancer Epidem Biomar. 2007;16:847–851. doi: 10.1158/1055-9965.EPI-06-0934. [DOI] [PubMed] [Google Scholar]
- 13.Kong L, et al. Molecular mechanisms underlying cochlear degeneration in the tubby mouse and the therapeutic effect of sulforaphane. Neurochem Int. 2009;54:172–179. doi: 10.1016/j.neuint.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dinkova-Kostova AT, Fahey JW, Talalay P. Chemical structures of inducers of nicotinamide quinone oxidoreductase 1 (NQO1) Method Enzymol. 2004;382:423–448. doi: 10.1016/S0076-6879(04)82023-8. [DOI] [PubMed] [Google Scholar]
- 15.Dinkova-Kostova AT, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinkova-Kostova AT, et al. Potency of Michael reaction accepters as inducers of enzymes that protect against carcinogenesis depends on their reactivity with sulfhydryl groups. Proc Natl Acad Sci USA. 2001;98:3404–3409. doi: 10.1073/pnas.051632198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wakabayashi N, et al. Protection against electrophile and oxidant stress by induction of the phase 2 response: Fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci USA. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heiss E, et al. Nuclear factor kappa B is a molecular target for sulforaphane-mediated anti-inflammatory mechanisms. J Biol Chem. 2001;276:32008–32015. doi: 10.1074/jbc.M104794200. [DOI] [PubMed] [Google Scholar]
- 19.Hong F, Freeman ML, Liebler DC. Identification of sensor cysteines in human Keap1 modified by the cancer chemopreventive agent sulforaphane. Chem Res Toxicol. 2005;18:1917–1926. doi: 10.1021/tx0502138. [DOI] [PubMed] [Google Scholar]
- 20.Zipper LM, Mulcahy RT. The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J Biol Chem. 2002;277:36544–36552. doi: 10.1074/jbc.M206530200. [DOI] [PubMed] [Google Scholar]
- 21.Li XC, Zhang D, Hannink M, Beamer LJ. Crystal structure of the Kelch domain of human Keap1. J Biol Chem. 2004;279:54750–54758. doi: 10.1074/jbc.M410073200. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Kolm RH, Mannervik B, Talalay P. Reversible conjugation of isothiocyanates with glutathione catalyzed by human glutathione transferases. Biochem Biophys Res Commun. 1995;206:748–755. doi: 10.1006/bbrc.1995.1106. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Talalay P. Anticarcinogenic activities of organic isothiocyanates—chemistry and mechanisms. Cancer Res. 1994;54:S1976–S1981. [PubMed] [Google Scholar]
- 24.Hwang Y, et al. A selective chemical probe for coenzyme A—Requiring enzymes. Angew Chem Int Edit. 2007;46:7621–7624. doi: 10.1002/anie.200702485. [DOI] [PubMed] [Google Scholar]
- 25.Prochaska HJ, Santamaria AB. Direct measurement of NAD(P)H—quinone reductase from cells cultured in microtiter wells—a screening assay for anticarcinogenic enzyme inducers. Anal Biochem. 1988;169:328–336. doi: 10.1016/0003-2697(88)90292-8. [DOI] [PubMed] [Google Scholar]
- 26.Fahey JW, Dinkova-Kostova AT, Stephenson KK, Talalay P. The “Prochaska” microtiter plate bioassay for inducers of NQO1. Method Enzymol. 2004;382:243–258. doi: 10.1016/S0076-6879(04)82014-7. [DOI] [PubMed] [Google Scholar]
- 27.Posner GH, et al. Design and synthesis of bifunctional isothiocyanate analogs of sulforaphane—correlation between structure and potency as inducers of anticarcinogenic detoxication enzymes. J Med Chem. 1994;37:170–176. doi: 10.1021/jm00027a021. [DOI] [PubMed] [Google Scholar]
- 28.Dinkova-Kostova AT, et al. Extremely potent triterpenoid inducers of the phase 2 response: Correlations of protection against oxidant and inflammatory stress. Proc Natl Acad Sci USA. 2005;102:4584–4589. doi: 10.1073/pnas.0500815102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prochaska HJ, Santamaria AB, Talalay P. Rapid detection of inducers of enzymes that protect against carcinogens. Proc Natl Acad Sci USA. 1992;89:2394–2398. doi: 10.1073/pnas.89.6.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerhauser C, et al. Cancer chemopreventive potential of sulforamate, a novel analogue of sulforaphane that induces phase 2 drug-metabolizing enzymes. Cancer Res. 1997;57:272–278. [PubMed] [Google Scholar]
- 31.Gao X, Talalay P. Induction of phase 2 genes by sulforaphane protects retinal pigment epithelial cells against photooxidative damage. Proc Natl Acad Sci USA. 2004;101:10446–10451. doi: 10.1073/pnas.0403886101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuspa SH, et al. Cultivation and characterization of cells derived from mouse skin papillomas induced by an initiation promotion protocol. Carcinogenesis. 1986;7:949–958. doi: 10.1093/carcin/7.6.949. [DOI] [PubMed] [Google Scholar]
- 33.Wang XJ, Hayes JD, Wolf CR. Generation of a stable antioxidant response element-driven reporter gene cell line and its use to show redox-dependent activation of Nrf2 by cancer chemotherapeutic agents. Cancer Res. 2006;66:10983–10994. doi: 10.1158/0008-5472.CAN-06-2298. [DOI] [PubMed] [Google Scholar]
- 34.Liu H, Dinkova-Kostova AT, Talalay P. Coordinate regulation of enzyme markers for inflammation and for protection against oxidants and electrophiles. Proc Natl Acad Sci USA. 2008;105:15926–15931. doi: 10.1073/pnas.0808346105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q, et al. Bioconjugation by copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J Am Chem Soc. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 36.Sun XL, Stabler CL, Cazalis CS, Chaikof EL. Carbohydrate and protein immobilization onto solid surfaces by sequential Diels-Alder and azide-alkyne cycloadditions. Bioconjugate Chem. 2006;17:52–57. doi: 10.1021/bc0502311. [DOI] [PubMed] [Google Scholar]
- 37.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto T, et al. Physiological significance of reactive cysteine residues of keap1 in determining Nrf2 activity. Mol Cell Biol. 2008;28:2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobayashi M, et al. The antioxidant defense system Keap1-Nrf2 comprises a multiple sensing mechanism for responding to a wide range of chemical compounds. Mol Cell Biol. 2009;29:493–502. doi: 10.1128/MCB.01080-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eggler AL, et al. Modifying specific cysteines of the electrophile-sensing human Keap1 disrupt binding to the protein is insufficient to Nrf2 domain Neh2. Proc Natl Acad Sci USA. 2005;102:10070–10075. doi: 10.1073/pnas.0502402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong F, Sekhar KR, Freeman ML, Liebler DC. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. J Biol Chem. 2005;280:31768–31775. doi: 10.1074/jbc.M503346200. [DOI] [PubMed] [Google Scholar]
- 42.Rachakonda G, et al. Covalent modification at Cys151 dissociates the electrophile sensor Keap1 from the ubiquitin ligase CUL3. Chem Res Toxicol. 2008;21:705–710. doi: 10.1021/tx700302s. [DOI] [PubMed] [Google Scholar]
- 43.Eggler AL, Luo Y, van Breemen RB, Mesecar AD. Identification of the highly reactive cysteine 151 in the chemopreventive agent-sensor Keap1 protein is method-dependent. Chem Res Toxicol. 2007;20:1878–1884. doi: 10.1021/tx700217c. [DOI] [PubMed] [Google Scholar]
- 44.Luo Y, et al. Sites of alkylation of human Keap1 by natural chemoprevention agents. J Am Soc Mass Spectr. 2007;18:2226–2232. doi: 10.1016/j.jasms.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snyder GH, Cennerazzo MJ, Karalis AJ, Field D. Electrostatic influence of local cysteine environments on disulfide exchange kinetics. Biochemistry. 1981;20:6509–6519. doi: 10.1021/bi00526a001. [DOI] [PubMed] [Google Scholar]
- 46.Cooke G, Armstrong ME, Donnelly SC. Macrophage migration inhibitory factor (MIF), enzymatic activity and the inflammatory response. Biofactors. 2009;35:165–168. doi: 10.1002/biof.27. [DOI] [PubMed] [Google Scholar]
- 47.Brown KK, et al. Direct Modification of the Proinflammatory Cytokine Macrophage Migration Inhibitory Factor by Dietary Isothiocyanates. J Biol Chem. 2009;284:32425–32433. doi: 10.1074/jbc.M109.047092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.