Abstract
When unfolded proteins accumulate in the endoplasmic reticulum (ER), the unfolded protein response is activated. This ER stress response restores ER homeostasis by coordinating processes that decrease translation, degrade misfolded proteins, and increase the levels of ER-resident chaperones. Ribonuclease inositol-requiring protein–1 (IRE-1), an endoribonuclease that mediates unconventional splicing, and its target, the XBP-1 transcription factor, are key mediators of the unfolded protein response. In this study, we show that in Caenorhabditis elegans insulin/IGF-1 pathway mutants, IRE-1 and XBP-1 promote lifespan extension and enhance resistance to ER stress. We show that these effects are not achieved simply by increasing the level of spliced xbp-1 mRNA and expression of XBP-1’s normal target genes. Instead, in insulin/IGF-1 pathway mutants, XBP-1 collaborates with DAF-16, a FOXO-transcription factor that is activated in these mutants, to enhance ER stress resistance and to activate new genes that promote longevity.
Keywords: aging, daf-2, insulin signaling, unfolded protein response
The endoplasmic reticulum (ER) is the site where newly synthesized secreted and cell-surface proteins fold and mature. ER stress response regulators monitor the flux of proteins through the ER. Increased flux and accumulation of misfolded proteins within the ER creates ER stress, which in turn, triggers signaling pathways collectively termed the unfolded protein response (UPR). The UPR helps restore ER homeostasis by coordinating processes that decrease translation, degrade misfolded proteins, and increase the levels of ER-resident chaperones (1).
In Caenorhabditis elegans, three proteins are known to sense ER stress and activate the UPR: the ribonuclease inositol-requiring protein–1 (IRE-1), the PERK kinase homologue PEK-1, and activating transcription factor–6 (ATF-6) (2–5). Of the three, IRE-1 is the most conserved. When activated by ER stress, IRE-1’s endonuclease activity is switched on, and it then removes an intron from xbp-1 (X-box binding protein–1) mRNA through unconventional splicing (2, 4). Spliced xbp-1 encodes a transcription factor that activates expression of downstream genes, such as genes encoding chaperones and ER-associated degradation proteins (2, 4, 5), that expand the ER’s folding capacity and increase degradation of misfolded proteins.
A functional and well balanced ER is critical for maintaining the integrity of the cell and the organism and for protecting the organism from heart disease, neurodegenerative disorders, and diabetes (6–8). Hence, genes that regulate the ER stress response machinery are potential candidate longevity genes. In this study, we have investigated the role of components of the ER stress response in the regulation of aging.
C. elegans mutants carrying reduction-of-function mutations in daf-2, which encodes the animal's insulin/IGF-1 receptor, are long-lived. Multiple stress-protective transcription factors, including the FOXO family member DAF-16, are activated in daf-2 mutants, where they alter the expression of downstream metabolic and stress-protective genes (9–14). The combined activities of these genes produce large changes in lifespan (15, 16). Although many cell-protective genes are induced in daf-2 mutants, we find that most known ER stress response genes are not. Nevertheless, in this study, we demonstrate that ire-1 and xbp-1 make a large contribution to the long lifespans of daf-2 receptor mutants and also increase their ER stress resistance. We find that XBP-1 contributes to the long lifespan of daf-2 mutants, at least in part, by collaborating with DAF-16 to up-regulate the expression of a gene called dox-1. In addition, we find that the increased ER stress resistance of daf-2 mutants, which is dependent on xbp-1, also requires DAF-16. Together, our findings suggest that XBP-1 and DAF-16 interact, directly or indirectly, to promote ER stress resistance and to activate new longevity genes in daf-2 mutants.
Results and Discussion
ire-1 and xbp-1 Promote the Longevity of Insulin/IGF-1 Pathway Mutants.
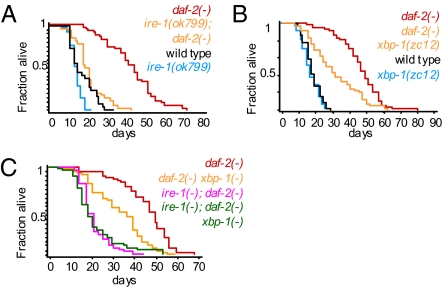
To ask whether ER stress-response proteins contribute to the increased longevity of animals with reduced insulin/IGF-1 signaling, we individually inactivated each of the three ER stress-response genes that comprise the UPR: ire-1, pek-1, and atf-6 (2–5). We found that inactivation of ire-1 by RNAi or by a deletion mutation shortened the lifespan of daf-2 mutants (Fig. 1A and Fig. S1A), but that inactivation of atf-6 or pek-1 did not (Fig. S1 B and C). In both wild-type and daf-2(−) backgrounds, animals subjected to ire-1 RNAi appeared normal and healthy, whereas ire-1(ok799)–null mutants had morphological and behavioral defects. Thus, whereas complete loss of ire-1 compromised the health of the animal, partial loss of ire-1, through RNAi, could shorten lifespan without making the animals appear unhealthy. Together, these findings implicated IRE-1, but not other ER stress–sensing proteins, in the longevity of insulin/IGF-1 pathway mutants.
Fig. 1.
ire-1 and xbp-1 contribute to the longevity of daf-2(−) animals. (A) Reduction of ire-1 activity by the null mutation ok799 shortened the lifespan of daf-2(e1370) mutants more than it shortened the lifespan of wild type. (B) The xbp-1(zc12) mutation shortened the lifespan of daf-2(e1370) mutants more than it shortened wild-type lifespan. (C) The xbp-1(zc12) mutation did not further shorten the lifespan of ire-1(−); daf-2(−) double mutants, suggesting that these genes function in a common pathway. [Compare survival curves of ire-1(−); daf-2(−) and ire-1(−); daf-2(−) xbp-1(zc12) mutants.] Note that xbp-1(zc12) mutation shortened the lifespan of daf-2(e1370) mutants, but not as much as did an ire-1(ok799) deletion. [Compare survival curves of ire-1(−); daf-2(−) and daf-2(−) xbp-1(zc12) mutants.] As xbp-1(zc12) is likely to eliminate xbp-1 function, this finding implies that ire-1 contributes to the longevity of daf-2 mutants in part through an xbp-1–independent pathway. For additional lifespan data, see Table S1. Also, see Fig. S1 for data showing that other ER stress–response genes, namely abu-11, pek-1, and atf-6, are not involved in this pathway.
One could imagine that IRE-1 promotes the longevity of all worms to the same extent that it promotes the longevity of insulin/IGF-1 pathway mutants; however, we found that this was not the case. Indeed, ire-1 knockdown shortened the lifespan of wild-type animals as well as eat-2 mutants (which live long as a result of caloric restriction) and isp-1 mutants (which live long as a result of reduced mitochondrial respiration). However, the extent of lifespan shortening by ire-1 inactivation was significantly more pronounced for the daf-2 mutants than for the other strains tested: inactivation of ire-1 in any of three daf-2 mutant strains (e1370, e1368, and mu150) shortened lifespan by an average of 40%, whereas inactivation of ire-1 shortened wild-type lifespan by an average of 22% (Fig. 1A and Table S1), and ire-1 knockdown shortened the extended lifespan caused by the eat-2 mutation or the isp-1 mutation by an average of less than 20% (Fig. S1 E and F and Table S1). We conclude that IRE-1 plays a particularly important role in promoting the longevity of insulin/IGF-1 pathway mutants.
We hypothesized that IRE-1 might contribute to the long lifespan of insulin/IGF-1 signaling mutants by acting through its known target, the transcription factor xbp-1, as it does under conditions of ER stress. Consistent with this hypothesis, we found that a putative xbp-1–null mutation, which causes truncation of the XBP-1 protein before its DNA binding domain, shortened the lifespan of daf-2 mutants more than it shortened the lifespan of wild type (Fig. 1B) without affecting the apparent health of the animals. Significantly, this xbp-1 mutation did not further shorten the lifespan of ire-1; daf-2 double mutants (Fig. 1C), suggesting that xbp-1 did not act independently of ire-1 to lengthen the lifespan of daf-2 mutants.
The ire-1/xbp-1 Pathway Is Set at a Lower Level in daf-2 Mutants.
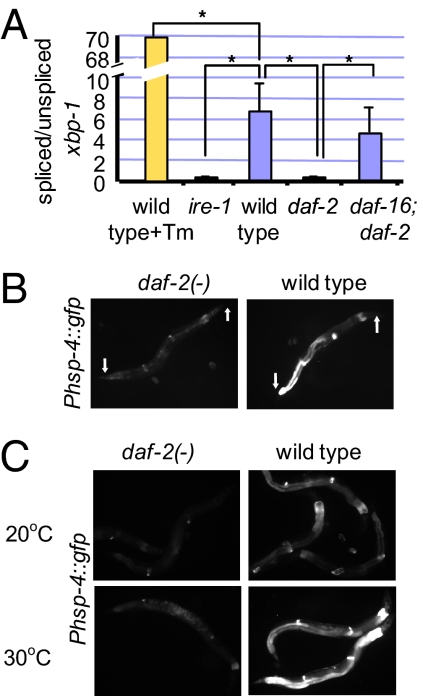
The simplest scenario to account for the major contribution of the ire-1/xbp-1 branch of the UPR to the longevity of daf-2 mutants was that the ire-1/xbp-1 pathway is activated constitutively in daf-2 mutants. To test this idea, we examined the level of spliced xbp-1 mRNA, which correlates with the level of activation of this UPR pathway in wild-type animals. Surprisingly, by using RT-PCR, we found that, in daf-2(−) mutants, the level of spliced xbp-1 mRNA was lower, not higher, than it was in wild-type animals (Fig. 2A and Fig. S2). Likewise, the basal expression level of a GFP fusion to the promoter of the XBP-1 target gene hsp-4, which encodes an ER chaperone, was lower in daf-2 mutants than it was in wild type (Fig. 2B). Moreover, when exposed to the ER stress–inducing agent tunicamycin (Fig. S3A) or to global protein misfolding caused by heat shock (Fig. 2C), adult daf-2 mutants induced less Phsp-4::gfp expression than did wild-type animals. The low level of Phsp-4::gfp in daf-2 mutants could be further reduced upon inactivation of xbp-1 by RNAi. Thus, the ire-1/xbp-1 pathway appears to be set at a very low level in daf-2 mutants, yet is still active enough to influence gene expression.
Fig. 2.
The ire-1/xbp-1 pathway is set at a lower level in daf-2(−) mutants. (A) Bar graph representing the mean ratio of spliced/unspliced xbp-1 transcripts, multiplied by a factor of 100, in three independent biological experiments. Control lane (yellow bar) presents the xbp-1 transcripts ratio in wild-type animals treated with tunicamycin. The rest of the bars present steady-state xbp-1 transcripts ratio in day-1 wild-type, ire-1, daf-2, and daf-16; daf-2 animals. These animals have not been treated with tunicamycin (blue bars). Error bars represent SEM. Asterisks mark Student t test values of P < 0.01. See Fig. S2 for data showing a representative RT-PCR experiment. (B) Representative fluorescence micrographs (100-fold magnification) of day-3 adults harboring an integrated Phsp-4::gfp transgene in a wild-type or daf-2(e1370) background. daf-2 mutants expressed lower levels of the Phsp-4::gfp transgene than did wild type. wild type, n = 9, relative mean fluorescence intensity (m) = 1 ± 0.14; daf-2(e1370), n = 10, m = 0.34 ± 0.07, P = 0.0012. (C) daf-2(e1370) mutants expressed less Phsp-4::gfp transgene than did wild type even upon heat shock. Phsp-4::gfp expression at 20 °C or upon 3 h of heat shock at 30 °C. Arrows point at anterior and posterior ends of the animals.
The ire-1/xbp-1 Pathway Confers Enhanced ER Stress Resistance to daf-2 Mutants.
Is the lower setting of the ire-1/xbp-1 pathway in daf-2 mutants a result of direct inhibition of this pathway, or is it a reflection of a reduced demand on the ER in these mutants? Down-regulation of the ire-1/xbp-1 pathway (for example, by loss-of-function mutations in these genes) is associated with increased sensitivity to ER stress. If this ER stress response pathway were simply suppressed in daf-2 mutants, we would expect daf-2 mutants to be more sensitive to ER stress than are wild-type animals. However, if the lower setting of the ire-1/xbp-1 pathway were a reflection of a superior ER homeostatic mechanism in daf-2 animals, we would expect the daf-2 animals to be more resistant to ER stress. We exposed young animals to toxic concentrations of two ER stress–inducing agents, tunicamycin and DTT, and found that daf-2 mutants were more resistant to both treatments than were wild-type animals (Fig. 3 and Fig. S3B). We interpret this to mean that the lower setting of the ire-1/xbp-1 pathway in daf-2 mutants reflects an improved ER homeostasis state compared with wild type. Moreover, this improved state of ER homeostasis is not achieved by up-regulating xbp-1 mRNA splicing.
Fig. 3.
daf-2(−) mutants are resistant to ER stress. Eggs from wild-type or daf-2(e1370) animals containing xbp-1(zc12), ire-1(ok799), pek-1(ok275), or atf-6(ok551) mutations were grown in the presence of 0, 2, or 5 μg/mL tunicamycin (corresponding to the blue, red, and off-white bars in the graph). Percentage of eggs that developed into mature adults was scored. Student t test values were calculated between daf-2(+) and daf-2(−) backgrounds. Each experiment was repeated independently with similar effects. Each strain was scored on three independent plates. Error bars represent SEM for repeat plates within the experiment. Asterisks mark Student t test values of P < 0.01.
Could it be that alternative mechanisms that do not rely on the ire-1/xbp-1 pathway are responsible for the improved ER homeostasis in daf-2 mutants? We addressed the possibility that the alternative ER stress response pathways involving pek-1 or atf-6 may promote ER homeostasis in daf-2 mutants. However, this was not the case, as null mutations in pek-1 or atf-6 genes did not affect daf-2 mutants’ resistance to tunicamycin (Fig. 3). We also considered the possibility that translation is reduced in daf-2 mutants, slowing the flow of proteins into the ER and thereby reducing the need for XBP-1 and IRE-1. However, translation rates in the daf-2(e1370) mutant have been measured under these growth conditions, and they appear normal (17). Finally, we considered the general possibility that the physiology of daf-2 mutants reduced the level of unfolded proteins, and hence placed a lower demand on the ER. However, if daf-2 mutants simply had fewer unfolded proteins, then they would be more resistant to tunicamycin even in the absence of ire-1 and xbp-1, but they were not. Instead, mutations in ire-1 or xbp-1 genes completely abrogated the enhanced ER stress resistance of daf-2 mutants, such that daf-2(−) mutants had no advantage over daf-2(+) animals (Fig. 3). This finding was significant, as it indicated that the ire-1/xbp-1 branch of the UPR played an essential, integral role in conferring enhanced ER stress resistance to daf-2 mutants.
How could the ire-1/xbp-1 pathway confer enhanced longevity to daf-2 mutants, and yet be set at a lower level than in wild type? We hypothesized that, instead of simply raising the level of spliced xbp-1 and its known target genes, lowering insulin/IGF-1 signaling may be affecting the very nature of the IRE-1/XBP-1 pathway, for example, by changing the transcriptional outputs of the XBP-1 transcription factor. How could this be achieved? One possible model was that some of the multiple stress-protective transcription factors that are switched on in daf-2 mutants (9–14) act in a combinatorial fashion with XBP-1 to activate the expression of new longevity and stress-resistance genes. In such a case, these new XBP-1 target genes could specifically enhance the daf-2 mutant's longevity and stress resistance in a manner that is completely dependent on ire-1 and xbp-1 without necessarily increasing the level of spliced XBP-1.
xbp-1 and daf-16 Collaborate to Express dox-1, a New Longevity Gene, in daf-2 Mutants.
To identify genes that function downstream of XBP-1 in this pathway, we used microarray analysis. We compared the transcription profiles of two daf-2 mutants (e1370 and e1368) versus those of the corresponding daf-2(−) xbp-1(−) double mutants on the first day of adulthood. We identified genes whose expression changed in daf-2 mutants upon inactivation of xbp-1 in a highly consistent fashion, regardless of the degree to which their expression was changed Table S2). We then carried out a functional analysis of these genes using RNA interference to identify those that significantly affected lifespan.
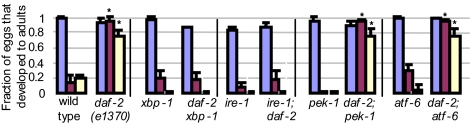
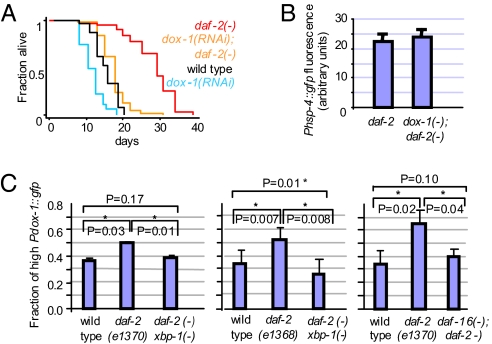
Using this approach, we identified a gene, Y52B11A.9 (which we named dox-1 for “downstream of xbp-1”) that encodes an evolutionarily conserved Zn-finger protein. Based on its orthologues and conserved protein domains, dox-1 is predicted to function in RNA splicing or processing. Inactivation of dox-1 had a substantial effect on the lifespan of daf-2 mutants without affecting their apparent health. In four of six trials, dox-1 RNAi shortened the lifespan of daf-2 animals substantially more than it shortened wild-type lifespan (Fig. 4A and Table S1). Thus, dox-1 contributes significantly to the longevity of daf-2 mutants.
Fig. 4.
dox-1 promotes longevity, but not ER stress resistance, in daf-2 mutants. (A) Reduction of dox-1 activity by RNAi shortened the lifespan of daf-2(mu150) mutants more than it shortened the lifespan of daf-2(+) animals. Animals were in a fer-15(b26); fem-1(hc17) background to prevent reproduction. Similar results were seen in daf-2(e1370) mutants (Table S1). (B) Reduction of dox-1 did not increase the set-point of the ire-1/xbp-1 branch of the UPR in daf-2 mutants. Bar graph showing the mean intensity of day-2 adults expressing a Phsp-4::gfp transgene. daf-2(e1370), n = 13, relative mean fluorescence intensity (m) = 21.9 ± 2.2; dox-1(RNAi); daf-2(e1370), n = 17, m = 23.3 ± 2.3, P = 0.677. See Fig. S4 for representative micrographs and wild-type data. (C) dox-1 expression was up-regulated in daf-2 mutants. Bar graph showing the fraction of animals expressing high levels of Pdox-1::gfp. Day-1 adults of each strain were classified as expressing high or low GFP levels (Fig. S4). The fraction of animals expressing high levels of Pdox-1::gfp was 1.35 fold higher in transgenic daf-2(e1370) mutants than in wild-type animals (P = 0.037), but was not significantly increased in daf-2(e1370) xbp-1(tm2457) double mutants. The fraction of animals expressing high levels of Pdox-1::gfp was 1.52 fold higher in transgenic daf-2(e1368) mutants than in wild-type animals (P = 0.007), but was not increased in daf-2(e1368) xbp-1(tm2457) double mutants. The fraction of animals expressing high levels of Pdox-1::gfp was higher by 1.8 fold in transgenic daf-2(e1370) mutants than in wild-type animals (P = 0.026), but was not significantly increased in daf-16(mu86); daf-2(e1370) double mutants. Error bars represent SEM of at least two independent trials. Asterisks mark Student t test values of P < 0.05.
Next, we investigated the regulation of dox-1 in more detail. dox-1 was identified in our microarray analysis as a gene whose expression in daf-2 mutants was significantly dependent on xbp-1. By comparing the levels of the dox-1 transcripts in microarrays profiling daf-2 and wild-type animals (Table S3), we found that the levels of dox-1 transcripts were higher in daf-2 mutants than in wild type. [The levels of dox-1 transcript were increased by a factor of 1.37 in daf-2 mutants (t test, P = 0.028).] We validated the microarray results by analyzing a GFP reporter fused to the dox-1 promoter. We found that a significantly higher fraction of transgenic daf-2 mutants expressed high levels of Pdox-1::GFP than did their wild-type counterparts and that loss of xbp-1 significantly reduced the proportion of animals expressing high levels of Pdox-1::GFP (Fig. 4C). In addition, we found that loss of daf-16 also significantly reduced the proportion of daf-2 animals expressing high levels of Pdox-1::GFP (Fig. 4C). Thus, consistent with our model, in daf-2 mutants, the expression of a powerful new longevity gene, dox-1, is up-regulated in a manner that requires both the transcription factor XBP-1 and the stress-protective transcription factor DAF-16, which is activated when insulin/IGF-1 signaling is low.
We asked whether inactivation of dox-1, which was sufficient to affect longevity, also affected ER homeostasis in daf-2 mutants. To test this, we used the reduced level of the ER chaperone reporter Phsp-4::gfp as a reflection of the improved ER homeostasis in daf-2 mutants. Unexpectedly, we found that inactivation of dox-1 did not elevate the levels of this reporter (Fig. 4B). Thus, even though dox-1 expression is dependent on the ER stress–related transcription factor xbp-1, dox-1 appears to contribute specifically to daf-2’s longevity but not to the improved ER homeostasis of daf-2 mutants.
daf-16 Promotes the Enhanced ER Stress Response in daf-2 Mutants.
How is ER homeostasis improved in insulin/IGF-1 mutants? In daf-2 mutants, multiple stress-protective transcription factors, such as DAF-16/FOXO, HSF-1, ELT-3, and SKN-1, switch on downstream genes that promote longevity (9–14). We wondered if any of these daf-2–related transcription factors acted in a combinatorial fashion with xbp-1, as in the case of the dox-1 gene, to turn on new ER stress–response genes that are essential to the enhanced ER stress response in daf-2 mutants. As outlined earlier, if this were the case, the enhanced ER stress resistance of daf-2 mutants would be completely xbp-1–dependent (as the expression of these genes would require xbp-1), and yet specific to daf-2 mutants (as the expression of these genes would also require a transcription factor that is activated only when insulin/IGF-1 signaling is reduced). Moreover, if one or more of these transcription factors contributed to the expression of new ER stress response genes in daf-2 mutants, its inactivation should compromise ER homeostasis in daf-2 mutants. This reduced ER homeostasis, in turn, would activate the classical ire-1/xbp-1 ER stress response pathway, raising the set-point of XBP-1 and its normal downstream targets. We found that these predictions were met for DAF-16. Using the reduced level of Phsp-4::gfp as a reflection of the improved ER homeostasis in daf-2 mutants, we asked whether inactivation of any of these transcription factors would reduce ER homeostasis (i.e., elevate the expression of Phsp-4::gfp) in daf-2 mutants. Inactivation of the transcription factors hsf-1, skn-1, and elt-3 by RNAi had a weak and inconsistent effect on the level of Phsp-4::gfp expression. In contrast, the level of this reporter was consistently increased in daf-2 mutants upon inactivation of daf-16 (Fig. 5A). Consistent with this finding, by using PAGE analysis of previously published daf-2 and daf-16; daf-2 microarray data (18), we found that a group of 17 genes (including hsp-4) that previously had been found to be induced in an xbp-1–dependent fashion upon tunicamycin treatment (3, 5) was significantly up-regulated in daf-16; daf-2 double mutants compared with daf-2 single mutants (P = 2.328E-06; Table S4). Finally, by using RT-PCR, we found that the level of spliced xbp-1 mRNA was consistently higher in daf-2 mutants upon inactivation of daf-16 (Fig. 2A and Fig. S2).
Fig. 5.
daf-16 contributes to the ER stress resistance in daf-2 mutants. (A) Representative fluorescence micrographs of day-1 adults harboring an integrated Phsp-4::gfp transgene in a daf-2(e1370) or daf-16(mu86); daf-2(e1370) background. daf-16 deletion (and inactivation of daf-16 by RNAi) increased the level of Phsp-4::gfp in daf-2(−) mutants. daf-2(e1370), n = 20, relative mean fluorescence intensity (m) = 1 ± 0.06; daf-16(mu86); daf-2(e1370), n = 28, m = 2.70 ± 0.2, P < 0.0001. Error bars represent SEM. Arrows point at anterior and posterior ends of the animals. (B) Tunicamycin resistance developmental assay. Eggs from wild-type, daf-2(e1370), and daf-16(mu86); daf-2(e1370) animals were grown in the presence of 0 or 5 μg/mL tunicamycin (corresponding to the blue and off-white bars in the graph). Percentage of eggs that developed into mature adults was scored. Student t test values were calculated relative to daf-16(−); daf-2(−). Asterisks mark Student t test values of P < 0.05. This experiment was repeated independently with similar effects. (C) Model for how XBP-1 extends lifespan, increases ER stress resistance, and also lowers XBP-1 levels when insulin/IGF-1 signaling is reduced. In daf-2 mutants, multiple transcription factors, such as DAF-16/FOXO, HSF-1, ELT-3, and SKN-1 are activated. These can act in a combinatorial fashion with XBP-1 to turn on new longevity genes and new ER stress response genes (both in purple), which, unlike the “normal” ER stress–protective genes (in red), cannot be induced by XBP-1 alone. This combinatorial control results in a wider array of longevity and ER stress–protective genes, both of which contribute to the longevity of daf-2 mutants. In addition, as part of a negative feedback loop, the wider array of ER stress protective genes expressed in daf-2 mutants by xbp-1 can better reduce the level of unfolded proteins in the ER and improve ER homeostasis. Improved ER homeostasis would, in turn, dictate a lower basal activity level of the ire-1/xbp-1 pathway. In this model, the enhanced ER stress resistance of daf-2 mutants is completely xbp-1–dependent, and yet, because the ER stress response is more efficient, the set point of XBP-1 and its normal downstream targets is lower than in wild type.
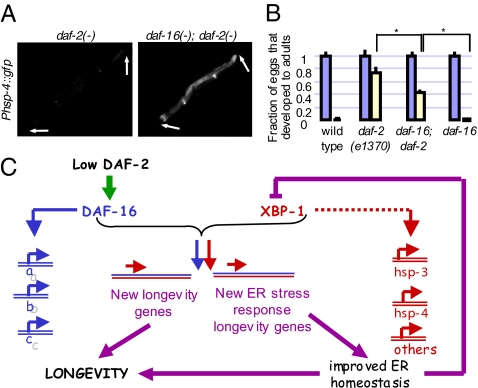
As loss of daf-16 activates this UPR pathway in daf-2 mutants, these findings may bring to mind the possibility that daf-16 is simply a direct repressor of the ire-1/xbp-1 pathway. However, this possibility is inconsistent with the enhanced (and xbp-1–dependent) ER stress resistance of daf-2 mutants. Instead, this finding is consistent with DAF-16’s being a factor that collaborates with XBP-1 to induce new stress-protective gene expression in daf-2 mutants. One prediction of this model would be that daf-16 contributes to the ER stress resistance of daf-2 mutants. We found that this was the case, as inactivation of daf-16 in daf-2 mutants significantly compromised the ability of daf-2 mutants to develop into adults when treated with tunicamycin (Fig. 5B). This finding, too, argues against the possibility that daf-16 simply represses the ire-1/xbp-1 pathway: if this were the case, loss of daf-16 should increase, not decrease, ER stress resistance.
Together these findings support the model that DAF-16 collaborates with XBP-1 to up-regulate the expression of genes that promote ER homeostasis in daf-2 mutants (Fig. 5C). In this model, loss of daf-16 results in loss of ER stress protection in daf-2(−) mutants, compromising ER function and homeostasis and eliciting ER stress, which in turn increases the splicing of xbp-1 as well as the expression of XBP-1’s “normal” ER stress response genes. We note that the ER stress resistance of daf-2 mutants was not completely dependent on daf-16 (Fig. 5B). This implies that DAF-16 is one of several factors, rather than the only factor, that collaborates with XBP-1 to positively regulate the expression of genes that promote ER homeostasis in daf-2 mutants.
Discussion
Reduced insulin/IGF-1 signaling, which happens naturally in response to stressful environmental conditions, is thought to extend lifespan by catalyzing a physiological shift toward pathways that promote cell maintenance and protection. Insulin/IGF-1 pathway mutants are resistant to a variety of environmental stressors, including heat, paraquat, hydrogen peroxide, DNA damaging agents, hypoxia, pathogens, and xenobiotic agents. Many genes encoding proteins predicted to increase environmental stress resistance are expressed at higher levels when insulin/IGF-1 signaling is reduced. These genes encode chaperones, innate-immunity proteins, metabolic genes, proteins that detoxify reactive oxygen species, and even proteins that protect the chromatin of the germline (16, 19). In general, the loss of any one of these stress-related genes shortens the lifespan of daf-2 mutants more than it shortens the lifespan of wild-type animals (15, 16). Thus it was not surprising to find that daf-2 mutants also exhibited increased ER stress resistance and that loss of the ER stress response genes ire-1 or xbp-1 shortened the lifespan of daf-2 mutants substantially.
This finding suggested a very simple model: namely that, as with other stress-protection pathways, the IRE-1/XBP-1 pathway was constitutively active in daf-2 mutants, and its enhanced ability to reduce ER stress prevented age-related imbalances in protein homeostasis that accelerate aging. However, this simple model was incorrect. By using RT-PCR to measure the level of spliced (i.e., active) xbp-1 mRNA and an in vivo GFP reporter to assay the expression of a representative ER stress–response gene activated by XBP-1, we found that this pathway was not constitutively activated in daf-2 mutants. Instead, its activity was actually lowered.
To explain these paradoxical findings, we considered the model that daf-2 mutants reduce the load on their ER by activating ER stress–protective pathways that are independent of XBP-1 and IRE-1. A reduced load on the ER would improve ER homeostasis and increase ER stress resistance while also lowering the set point of the ire-1/xbp-1 pathway. However, this model predicts that the improved ER homeostasis of daf-2 animals should be, at least in part, independent of the ire-1/xbp-1 pathway. However, all the increased ER stress resistance in these animals was XBP-1–dependent. Thus, this model was flawed too.
To resolve this conundrum, we devised a model that reconciles all these findings. In this model, XBP-1 collaborates with one or more of the transcription factors that are activated when insulin/IGF-1 signaling is reduced to stimulate the expression of new cell-protective genes. As the expression of these genes requires xbp-1, this model accounts for why the increased longevity and ER stress resistance require xbp-1. Likewise, as the expression of these genes also requires a transcription factor that is activated only when insulin/IGF-1 signaling is reduced, this model accounts for the fact that xbp-1 promotes a greater degree of ER stress resistance and longevity in daf-2 mutants than it does in wild type. Finally, the expression of new ER stress–response genes in daf-2 mutants would increase ER homeostasis, and thus could account for the lower set-point of XBP-1 and its normal downstream targets.
By looking for such genes by means of microarrays, we tested and validated this model for the role of XBP-1 in longevity. We identified dox-1 as a lifespan-extending gene that is up-regulated in daf-2 mutants by both XBP-1 and a transcription factor activated in daf-2 mutants, DAF-16/FOXO. Thus, directly or indirectly, XBP-1 and DAF-16 collaborate to activate dox-1 expression. Within the first 300 bp upstream of the dox-1 coding sequence, we identified two potential DAF-16/FOXO binding sites (20, 21) and a potential XBP-1 ETS-like binding site (22). Thus, it is possible that both of these proteins bind to the dox-1 promoter to activate dox-1 expression. Alternatively, one or both transcription factors could act on intermediate components of a more complex regulatory pathway.
Loss of dox-1 reduces the lifespan of daf-2 mutants substantially, much more than does the loss of most of the DAF-16 target genes that have been examined. The sequence of dox-1 suggests that it encodes a regulatory protein; specifically, a Zn-finger protein whose orthologues in other species are implicated in RNA splicing or processing. The fact that dox-1 could potentially influence multiple downstream targets is consistent with its having a relatively large effect on lifespan.
The finding that ire-1 and xbp-1 promote both ER stress resistance and longevity in daf-2 mutants suggested that they extend lifespan by enhancing the ER stress response, thereby improving protein homeostasis. Thus it was surprising to find that loss of dox-1 activity influenced lifespan but not ER homeostasis. This finding indicates that, when insulin/IGF-1 signaling is low, IRE-1 and XBP-1 can engage in processes that extend lifespan without affecting ER homeostasis.
Loss of daf-16 does reduce the ER stress resistance of daf-2 mutants, suggesting that XBP-1 and DAF-16 together may control the expression of other genes that do affect ER stress resistance. We propose that genes regulated in this combinatorial fashion may allow daf-2 mutants to achieve better ER homeostasis under normal conditions, and to restore ER homeostasis more effectively when confronted with ER stress. In principle, these two transcription factors could regulate one or a few potent ER stress–response genes, or they could affect the expression of many ER stress–response genes that have small effects singly but together produce large increases in ER homeostasis in daf-2 mutants. Recently, Kapahi and coworkers showed that IRE-1 and XBP-1 contribute to the longevity of animals with altered levels of the oxygen sensor hypoxia-inducible factor–1 (23). The mechanism here may be different, as this longevity pathway does not require DAF-16. It will be interesting to learn whether, and to what extent, these pathways may converge on similar targets.
It will also be interesting to learn whether this pathway has been conserved in higher organisms. This seems possible, as many insulin/IGF-1 pathway mutations extend lifespan in mice, and the DAF-16 orthologue FOXO3A has been linked to exceptional longevity in humans (24).
In summary, our data suggest that, in C. elegans, the transcription factors responsible for the global, cell-protective physiological shift that takes place in daf-2 mutants collaborate with classical ER stress response proteins to promote longevity and ER stress resistance. We hope these findings will stimulate additional studies in higher organisms, as these may form the basis of new therapeutic strategies to combat diseases that are associated with protein misfolding in the ER.
Materials and Methods
Strains and Lifespan Analysis.
Strain maintenance and lifespan analysis were performed as described previously (25) and are detailed further in SI Materials and Methods.
RNAi.
The identity of RNAi clones was verified by sequencing. RNAi clones were obtained from the Ahringer genomic RNAi library. For all RNAi experiments, animals were grown from hatching on bacteria expressing dsRNA, unless indicated otherwise.
SI Materials and Methods includes details about the systematic RNAi lifespan screen, which was carried out using RNAi clones corresponding to the approximately 600 candidate genes identified in the microarray study.
RT-PCR.
Total RNA isolation and reverse transcription using random hexamer primers of synchronized day-1 adult C. elegans RNA were carried out as described (26). Experiments were performed on serial dilutions of the cDNA from three independent biological replicates per strain, within the linear range of the PCR. See SI Materials and Methods for xbp-1 primer design.
ER Stress Resistance.
Eggs of different genetic backgrounds were placed onto plates containing 0.1% DMSO and 0, 2, or 5 μg/mL tunicamycin (Sigma) or 0, 2, or 5 mM DTT. The number of eggs was noted and compared with the number of animals that reached the L4/adult stage within 72 to 96 h. For each strain, typically 50 to 200 eggs were placed on each of three plates.
Microscopy and Quantification.
Young adults expressing Phsp-4::GFP were immobilized on agarose pads containing 2 mM levamisole. Whole worm images were captured using a Retiga EXi Fast1394 CCD digital camera (QImaging) attached to an Axioplan 2 compound microscope (Zeiss), using the OpenLab program (version 4.0.2). For each individual trial, exposure time was calibrated to avoid saturated pixels for the set of animals. OpenLab 4.0.2 software was used to measure and quantify fluorescence intensity within the entire animal. Total fluorescence was calculated by the OpenLab program as measured by intensity of each pixel in the selected area of the worm.
Microarray Analysis.
Standard RNA purification, labeling using the Agilent QuickAmp Kit, and hybridization on Agilent 4 × 44K C. elegans arrays using the Hi-RPM hybridization kit were performed by the University Health Network Microarray Centre (Toronto, ON, Canada). See SI Materials and Methods for analysis and sample description.
Supplementary Material
Acknowledgments
We thank members of the Kenyon laboratory. We thank the Caenorhabditis Genetics Center, the International C. elegans Gene Knockout Consortium, David Ron, and Ian Hope for strains. Most of the experiments were performed by S.H.-K. P.Z. contributed to some of the lifespan and stress resistance experiments. S.H.-K. and M.H. identified ire-1 in a longevity screen. S.H.-K., M.M., S.-J.L., and M.C. contributed to the microarray experiments. S.H.K. was supported in part by a Human Frontier Science Program Fellowship, P.Z. by the Larry Hillblom Foundation. S.J.L. by the Ellison Medical Foundation Fellow of the Life Sciences Research Foundation and the American Heart Association, and M.H. by the Ellison/American Federation for Aging Research Foundation. This work was supported by National Institutes of Health Grant 5R37AG011816 (to C.K.). C.K. is an American Cancer Society Research Professor and the director of the University of California San Francisco Hillblom Center for the Biology of Aging.
Footnotes
The authors declare no conflicts of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE20148).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002575107/-/DCSupplemental.
References
- 1.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 2.Shen X, et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- 3.Shen X, Ellis RE, Sakaki K, Kaufman RJ. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet. 2005;1:e37. doi: 10.1371/journal.pgen.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 5.Urano F, et al. A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J Cell Biol. 2002;158:639–646. doi: 10.1083/jcb.200203086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin JH, Walter P, Yen TS. Endoplasmic reticulum stress in disease pathogenesis. Annu Rev Pathol. 2008;3:399–425. doi: 10.1146/annurev.pathmechdis.3.121806.151434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshida H. ER stress and diseases. FEBS J. 2007;274:630–658. doi: 10.1111/j.1742-4658.2007.05639.x. [DOI] [PubMed] [Google Scholar]
- 8.Lindholm D, Wootz H, Korhonen L. ER stress and neurodegenerative diseases. Cell Death Differ. 2006;13:385–392. doi: 10.1038/sj.cdd.4401778. [DOI] [PubMed] [Google Scholar]
- 9.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 10.Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- 12.Ogg S, et al. The fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 13.Tullet JM, et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee RY, Hench J, Ruvkun G. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr Biol. 2001;11:1950–1957. doi: 10.1016/s0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- 15.Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- 16.Murphy CT, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 17.Hansen M, et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 18.McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem. 2004;279:44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- 19.McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 20.Pierrou S, Hellqvist M, Samuelsson L, Enerbäck S, Carlsson P. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. EMBO J. 1994;13:5002–5012. doi: 10.1002/j.1460-2075.1994.tb06827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furuyama T, Nakazawa T, Nakano I, Mori N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J. 2000;349:629–634. doi: 10.1042/0264-6021:3490629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Acosta-Alvear D, et al. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 23.Chen D, Thomas EL, Kapahi P. HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in Caenorhabditis elegans. PLoS Genet. 2009;5:e1000486. doi: 10.1371/journal.pgen.1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 25.Apfeld J, Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature. 1999;402:804–809. doi: 10.1038/45544. [DOI] [PubMed] [Google Scholar]
- 26.Taubert S, Van Gilst MR, Hansen M, Yamamoto KR. A Mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 2006;20:1137–1149. doi: 10.1101/gad.1395406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.