Abstract
Despite the key role of the linker histone H1 in chromatin structure and dynamics, its location and interactions with nucleosomal DNA have not been elucidated. In this work we have used a combination of electron cryomicroscopy, hydroxyl radical footprinting, and nanoscale modeling to analyze the structure of precisely positioned mono-, di-, and trinucleosomes containing physiologically assembled full-length histone H1 or truncated mutants of this protein. Single-base resolution •OH footprinting shows that the globular domain of histone H1 (GH1) interacts with the DNA minor groove located at the center of the nucleosome and contacts a 10-bp region of DNA localized symmetrically with respect to the nucleosomal dyad. In addition, GH1 interacts with and organizes about one helical turn of DNA in each linker region of the nucleosome. We also find that a seven amino acid residue region (121–127) in the COOH terminus of histone H1 was required for the formation of the stem structure of the linker DNA. A molecular model on the basis of these data and coarse-grain DNA mechanics provides novel insights on how the different domains of H1 interact with the nucleosome and predicts a specific H1-mediated stem structure within linker DNA.
Keywords: nucleosome structure, chromatin higher order structure
The nucleosome is the fundamental repeating unit of chromatin in the nucleus of eukaryotic cells. The composition and the basic organization of the nucleosome is well established, and the structure of the nucleosomal core particle (NCP) has been described with nearly atomic precision by X-ray diffraction (1). However, similar information for the structure of a complete nucleosome, i.e., the NCP with associated linker DNA segments and a linker histone, is still lacking. Electron microscopy and electron cryomicroscopy (ECM) imaging have provided a relatively low-resolution picture of the complete nucleosome, both native (2) and reconstituted (3). However, important features of the structure remain obscure.
Linker histones are typically ∼200 aa in length with a rather short nonstructured N terminus, followed by a ∼70–80 aa structured (“globular”) domain, and a ∼100 aa long apparently unstructured C terminal domain, highly enriched in lysines. The globular domain of the linker histone appears to be internally located in the 30-nm chromatin fiber (4, 5), but its exact position within the nucleosome remains a subject of debate (for review, see ref. 6). A second question not yet resolved concerns the interactions and location of the linker histone C terminus. These issues have their origin in difficulties related to the preparation of well-defined nucleosomal samples. Indeed, direct binding of linker histone to nucleosomes in vitro is inefficient and complicated by the formation of large aggregates because of the nonspecific association of linker histones with DNA (7, 8). The situation can be considerably improved by using chaperones for linker histone deposition in vitro, a mechanism that is likely used in vivo (9). It was recently shown that NAP-1 could be used to efficiently and properly incorporate the somatic linker histone H1 as well as the embryonic linker histone B4 into a dinucleosome reconstituted on a DNA template containing a tandem repeat of the Xenopus borealis 5S RNA gene (8). The DNase I footprinting analysis of the 5S dinucleosome indicated that both B4 and H1 protected linker DNA. However, because histone octamers do not precisely position on this DNA sequence (10), details regarding the interaction of the linker histone with the nucleosomes core were not apparent from this experiment (8).
In this study we have used 601 DNA repeats to reconstitute precisely positioned mono-, di-, and trinucleosomes and NAP-1 to properly incorporate either wild-type histone H1 or NH2 or COOH terminus truncated mutants. The structure of the H1-containing nucleosomal templates was analyzed by ECM, •OH footprinting, and coarse-grain molecular modeling. Our results provide a strikingly clear picture of how histone H1 binds to the nucleosome and indicate a specific H1-mediated organization of the linker DNA.
Results
NAP-1 Mediated Assembly of H1 and Truncation Mutants into Reconstituted Nucleosomes.
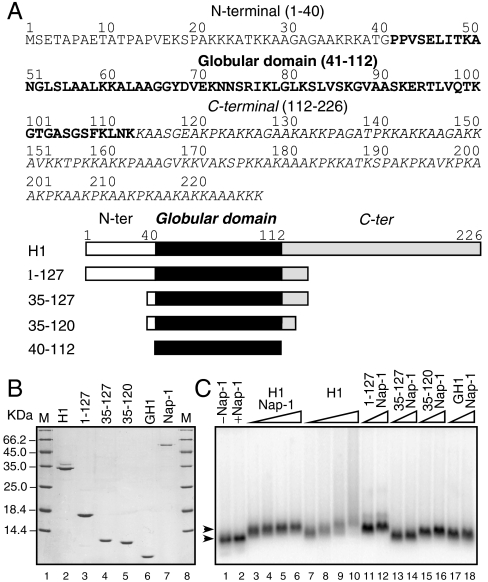
To investigate the complex interactions of H1 with nucleosomes, we reconstituted mono-, di-, and trinucleosomes by using templates on the basis of the 601 nucleosome positioning sequence to ensure that histone octamers were precisely positioned with respect to the DNA sequence. We also prepared full-length linker histone H1.5 and truncated mutants of this protein (Fig. 1A) and purified the recombinant proteins to homogeneity (Fig. 1B). We have used the H1.5 histone isoform because it is ubiquitously expressed in different tissues (for simplicity, we will refer to this protein as H1). We incubated H1 with the nucleosomes in either the presence or the absence of NAP-1, and the binding of histone H1 was evaluated by EMSA (Fig. 1C). Upon incubation with increasing amounts of H1, in the absence of NAP-1 the dinucleosome band exhibited shifts consistent with the binding of histone H1 and a smear throughout the lane of the gel indicative of the formation of aggregates at higher H1:dinucleosome ratios (Fig. 1C, lanes 7–10). This result is in complete agreement with the reported data and reflects the superstoichiometric association of histone H1 with the dinucleosomes (8). However, when NAP-1 was present in the reaction, the binding of histone H1 resulted in sharp, well-defined bands indicating the homogenous formation of dinucleosomes initially bound by one H1 and, upon increasing the NAP-1-H1 concentration, bound by two H1s (Fig. 1C, lanes 3–6). Importantly, further increases in the amount of NAP-1-H1 in the reaction did not change either the shape or the mobility of the band, consistent with the reported capacity of NAP-1 to act as an histone H1 chaperone, facilitating proper H1 binding to nucleosomes in a 1∶1 stoichiometry (8, 9). Importantly, NAP-1 was also able to mediate the binding of the H1 truncation mutants, including the globular domain of histone H1, GH1 (AA 40–112), to nucleosomes (Fig. 1C, lanes 11–18).
Fig. 1.
NAP-1 facilitates binding of linker histone H1 and truncation mutants to 601 dinucleosomes. (A) Primary structure of histone H1 (Upper) and schematics of the histone H1 deletion mutants (Lower). (B) 15% SDS-PAGE of purified recombinant full-length H1, truncation mutants, and NAP-1. (C) Agarose gel electrophoresis of 601 dinucleosomes incubated with increasing amounts of either full-length histone H1 alone (lanes 7–10), NAP-1-histone H1 complex (lanes 3–6), or a complex of NAP-1 and the indicated H1 truncation mutants (lanes 11–18). Lanes 1 and 2, control dinucleosome without H1 and dinucleosomes incubated with NAP-1 only.
ECM Imaging of Trinucleosomes Containing Either Full-Length Histone H1 or Truncation Mutants.
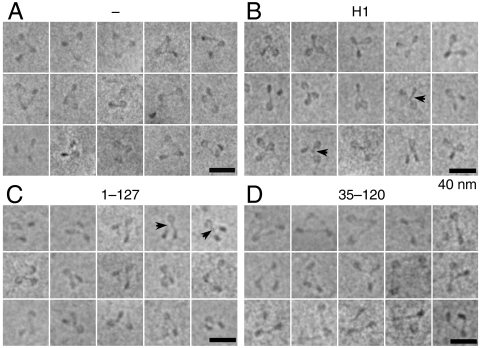
To evaluate the overall structure of nucleosomes containing H1, we examined the conformation of trinucleosomes by using ECM. Trinucleosomal particles were used in order to best approximate the situation in native chromatin, where nucleosomes are surrounded by neighbors. The central nucleosome in the trinucleosomal particle thus experiences an environment more similar to that in native chromatin than a mononucleosome.
Fig. 2 shows a gallery of trinucleosomes without H1 (Fig. 2A) and trinucleosomes bound by full-length H1 or selected H1 truncation mutants in the presence of NAP-1 (Fig. 2 B–D). The nucleosomes without H1 adopt an open conformation with diverging DNA segments, most easily visualized on the central nucleosome, where DNA is entering and exiting the octamer at different sites (Fig. 2A). In cases with convenient projections, the short DNA segments on external nucleosomes also can be seen. In contrast, upon H1 association the structure of the nucleosome closes and the formation of a stem structure is clearly visible (Fig. 2B, Arrowheads). The structural properties of the stem are visually identical to that observed in native chromatin particles (see ref. 2). We conclude that the NAP-1 assisted incorporation of histone H1 results in the reconstitution of native-like chromatin structures.
Fig. 2.
H1 binding to nucleosomes organizes linker DNA into a stem structure. Representative ECM images of reconstituted 601 trinucleosomes. Shown are trinucleosomes in the absence of H1 (A) and trinucleosomes assembled with full-length histone H1 (B), the 1–127 H1 truncation mutant (C), or the 35–120 truncation mutant (D). NAP-1 was present in the experiments shown in A. The arrowheads indicate selected examples of the stem. (Scale bar, 40 nm.)
Intriguingly, the association of the H1 truncated mutants 1–177 or 1–127 (which lack either the last 50 or 100 aa from the H1 C terminus; see Fig. 1A) leads to a structure very similar to that obtained upon association of full-length H1 with the trinucleosome (Fig. 2; compare B with C and Fig. S1), although the statistical analysis showed that the amount of stem structure, formed in the central nucleosome of trinucleosomes reconstituted with the H1 truncation mutant 1–127, was ∼15% less than that of trinucleosomes reconstituted with either full-length H1 or with the H1 truncation mutant 1–177 (Fig. S1B). In contrast, the 3D organization of the trinucleosomes assembled with the 35–120 mutant consisting of the globular domain and 5 and 8 aa from the NH2 and COOH termini of H1, respectively, was similar to that of trinucleosomes without H1 (Fig. 2; compare A with D). We conclude that the H1 globular domain alone is not able to organize the linker DNA into a stem-like structure.
Hydroxyl Radical Footprinting of Histone H1 Bound to Nucleosomes.
ECM reveals the overall structure and 3D conformation of the nucleosomal particles. To correlate the generation of this structure with the interaction of the different domains of histone H1 with the nucleosomal DNA, we have used both DNase I and hydroxyl radical footprinting techniques. Initially, we applied these techniques to study the organization of dinucleosomes. The presence of full-length histone H1 but not of the globular domain affected the accessibility of the linker DNA to DNase I (Fig. S2). This result is in agreement with reported data (8, 11) and suggests an interaction of either the NH2 and COOH termini of histone H1 or both with linker DNA. DNase I footprinting, however, did not resolve the precise localization of H1 on the nucleosome.
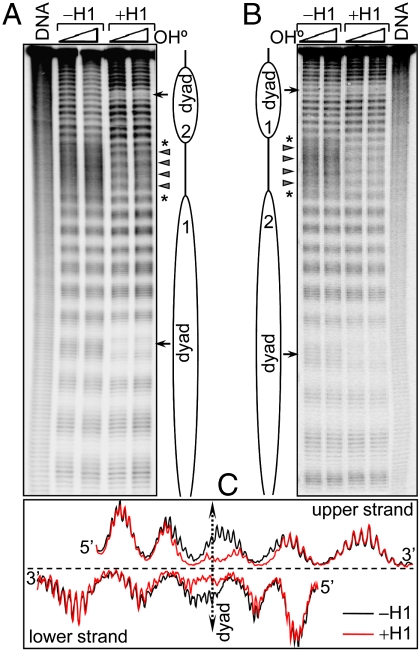
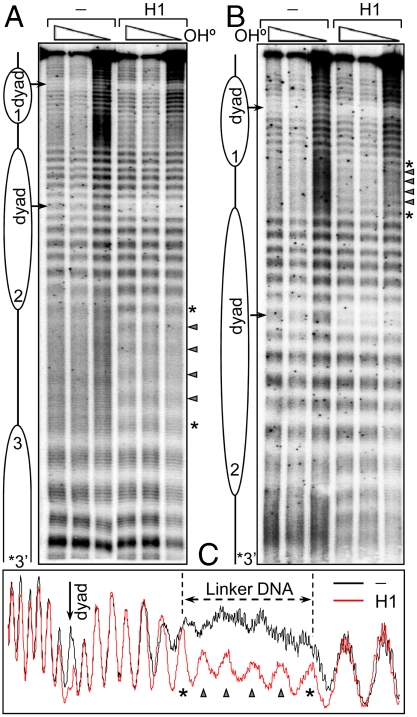
To more accurately identify regions of DNA interacting with H1 within the nucleosome, we employed footprinting with hydroxyl radicals, which provides single-base resolution information on protein–DNA contacts (12) (see SI Materials and Methods). Each of the nucleosomes within the dinucleosome without H1 showed a well-defined 10-bp repeat, because of the wrapping of the nucleosome core DNA around the histone octamer, whereas the linker DNA exhibited a uniform •OH cleavage pattern, similar to that of naked DNA (Fig. 3, lane DNA). However, in contrast to the DNase I experiments, the presence of H1 induced two major alterations in the •OH cleavage pattern of the dinucleosomal DNA (Figs. 3 and 4): (i) a strong decrease in the accessibility of DNA at the dyad axis of each individual nucleosome, where a 10-bp stretch of DNA symmetrically located about the dyad was protected by histone H1 (see Fig. 3C for details) and (ii) a clear 10-bp repeat in the cleavage pattern of the linker DNA.
Fig. 3.
Hydroxyl radical footprinting of control and H1-containing dinucleosomes. (A) Sequencing gel analysis of •OH cleavage of dinucleosomes with 32P label incorporated at the 3′ end of the upper DNA strand. Lane 1, •OH cleavage pattern of the naked DNA template; lanes 2–5, •OH cleavage pattern of control and H1-containing dinucleosomes. The triangle and the asterisk highlight the digestion products of the central part and the ends of the linker DNA region, respectively. (B) Same as A but for dinucleosomes reconstituted with 32P 3′-end-labeled lower DNA strand. (C) Scans of the •OH digestion pattern in the vicinity of the nucleosome dyad of control (Black) and H1-containing (Red) dinucleosomes.
Fig. 4.
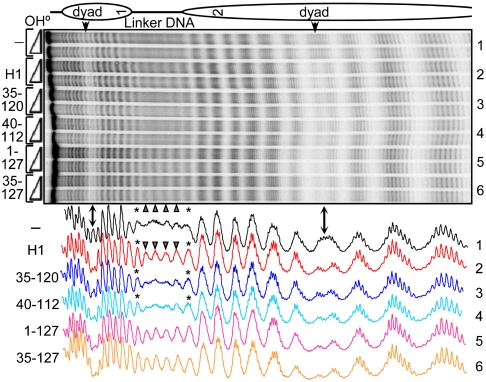
Hydroxyl radical footprinting of control and H1 truncation mutants bound to dinucleosomes. The gel shows dinucleosomes in the absence of H1 (-) and in the presence of H1 or truncation mutants, as indicated on the left. Scans of the •OH cleavage patterns depicted in the gel are shown (Bottom). Cleavage products within the central part of the linker DNA are indicated by triangles, whereas asterisks indicate the 10 bp at either end of the linker DNA adjacent to the core region.
To more closely approximate the physiological H1 binding environment and to correlate the binding of H1 with the 3D organization observed by ECM, we also carried out •OH footprinting with trinucleosomes without and with H1 (Fig. 5). The same types of alterations in the •OH cleavage pattern were observed upon histone H1 incorporation in these particles, namely, a clear pattern of protection at the dyad of each individual nucleosome and the appearance of a 10-bp repeat within the linker DNA (Fig. 5). Note that in this case the central nucleosome has two bona fide linkers and each linker exhibited the 10-bp repeat pattern. These types of structural changes because of the NAP-1 assisted incorporation of histone H1 were also observed with mononucleosomes, indicating that the protections were caused by H1 binding per se, and not H1-induced internucleosome interactions (Fig. S3).
Fig. 5.
Hydroxyl radical footprinting of H1 bound to trinucleosomes. (A) •OH cleavage pattern of trinucleosomes without H1 (lanes 1–3) and with H1 (lanes 4–6). (B) shows the same samples as in A except run longer for better resolution of the linker region. (C) Scans of the •OH cleavage pattern of control, without H1 (Black) and H1-containing (Red) trinucleosomes.
The H1 Globular Domain Alone Protects 10 bp of DNA at the Nucleosome Dyad and DNA Beyond the Edge of the Nucleosome Core Region.
To determine the domain(s) of histone H1 required for the observed protection of the nucleosome against •OH cleavage, we employed truncation mutants in the footprinting assay. We first concentrated on the globular domain of histone H1, GH1 (aa 40–112; see Fig. 1). As seen in Fig. 4, the association of GH1 with dinucleosomes resulted in a clear protection of the dyad similar to that observed with full-length H1, with ∼10 bp of DNA located symmetrically about the dyad protected against •OH cleavage. Note that the binding of the slightly larger (compared to GH1) 35–120 mutant of H1 (containing an additional 5 and 8 aa from the H1 NH2 and the COOH terminus, respectively) resulted in a footprint identical to that of GH1 (Fig. 4). Importantly, the association of this mutant with the mononucleosome (Fig. S3) led to the same pattern of protection of the dyad. Therefore, the globular domain of histone H1 interacts specifically with the central 10 bp of DNA in the nucleosome.
In addition to the protection of the dyad, the presence of either GH1 (aa 40–112) or the 35–120 H1 mutant in the dinucleosome also resulted in a symmetrical ∼10 bp extension of DNA protection at both ends of the nucleosome core (Fig. 4, Asterisks). This additional protection of linker DNA was also observed in mononucleosomes bound by both GH1 and the 35–120 H1 mutant (Fig. S3). Taken together, the data described above demonstrate highly specific binding of the globular domain of H1 with 10 bp of DNA located precisely at the nucleosome dyad and an additional interaction with a total of ∼20 bp of linker DNA distributed evenly at either end of the nucleosome core region.
Amino Acid Residues 121–127 of the H1 COOH Terminus Are Necessary for the Generation of the 10-bp Repeat in the •OH Cleavage Pattern of Linker DNA.
The footprints of nucleosomes associated with the histone H1 globular domain did not show a 10-bp repeat in the linker DNA, as was observed with full-length histone H1-bound particles. This suggested that either the NH2 or the COOH termini of H1 or both are required for the generation of this repeat. We initially approached this question by studying the •OH cleavage pattern of dinucleosomes bound by H1 C terminus truncation mutant 1–127 (see Fig. 1 for details). Mono- and dinucleosomes bound by the deletion protein exhibited a 10-bp repeat in the linker DNA (Fig. 4 and Fig. S3). This indicated that either the NH2 terminus or the portion of the C terminus remaining in the mutant or both are required for the generation of the repeat. To differentiate between these possibilities, we next carried out •OH footprinting of mono- and dinucleosomes assembled with the 35–127 truncation mutant of histone H1 in which the majority of the NH2 terminus (35 aa) and 100 aa from the COOH terminus of histone H1 were removed. Both mononucleosomal and dinucleosomal particles bound by the 35–127 mutant showed a clear 10-bp repeat in the cleavage pattern of the linker DNA (Fig. 4 and Fig. S3). These results show that the NH2 terminus is not required for the 10-bp repeat of the linker. Furthermore, because the linker repeat was not detected in 35–120 H1 mutant-associated particles, we conclude that a stretch of only seven aa (aa 121–127) within the COOH terminus of H1 plays the predominant role in the generation of the 10-bp repeat and thus in the structuring of the linker DNA.
Discussion
Our data resolve a long-standing issue in defining the structure of the nucleosome, the fundamental repeating subunit of eukaryotic chromatin, and shed light on how H1 binds to and organizes nucleosomal DNA. Despite numerous studies, the location and the interactions of the different domains of the linker histone with nucleosomal DNA have remained an unresolved and controversial issue. Many studies have focused on locating the binding site for the globular domain, which is responsible for structure-specific recognition of the nucleosome (13, 14). Past studies on the basis of the digestion of native chromatin with micrococcal nuclease and DNase I suggested a symmetrical model of the interaction of the linker histone with the nucleosome (11, 13, 15). According to this model the linker histone interacts with both the dyad and the entering and exiting DNA from the core particle. More recently, cross-linking studies of the globular domain (GH5) of the linker histone H5 to nucleosomal DNA pointed to a “bridging model,” where GH5 interacts with DNA near the dyad and with only one (either the exiting or entering) of the linker DNA arms (16). Other studies using cross-linking and site-directed cleavage methods to map DNA contacts of H1 within reconstituted positioned 5S nucleosomes led to a proposal for an asymmetric location of the globular domain inside the gyres of DNA at a distance of ∼65 bp from the dyad (17, 18). A recent experiment employing in vivo photobleaching microscopy supported the existence of two distinct DNA binding sites within the globular domain of the linker histone H1° and suggested that GH1° interacts with the DNA major grove about 10 bp from the dyad and with only one of the linker DNA arms adjacent to the nucleosome core (19).
Several factors likely contribute to the incongruence among the reported data. The previous in vitro studies used salt dialysis or direct binding to deposit histone H1 on the nucleosomes, which may have led to only a fraction of the nucleosomes exhibiting a proper 1∶1 stoichiometric association with H1. In addition, the reconstitution on 5S DNA results in the formation of nucleosomes exhibiting multiple translational positions, which, in turn, would interfere with the mapping of histone H1:nucleosomal DNA contacts (10). The in vivo photobleaching studies suggested two regions for nucleosome interaction on the globular domain of H1° but modeling was constrained by assumptions regarding interactions with the linker DNA.
In this work we have overcome the above problems by using (i) a physiologically relevant linker histone chaperone (NAP-1) for deposition of histone H1, (ii) the 601 DNA sequence for nucleosome reconstitution, and (iii) a combination of ECM and •OH footprinting techniques. These approaches have allowed the reconstitution of precisely positioned nucleosomal templates containing physiologically assembled histone H1 or truncated mutants, mapping histone H1∶DNA interactions within mono-, di-, and trinucleosomal templates at single-base resolution, and dissection of the role of distinct H1 domains in the 3D organization of the structures. The ECM data demonstrated that our reconstituted H1-containing trinucleosomes were visually indistinguishable from native trinucleosomes (2). Importantly, the presence of either full-length H1 or the 1–127 COOH terminus truncation mutant led to the generation of the characteristic stem structure of the linker DNA observed in native fibers (2). Hydroxyl radical footprinting showed that binding of full-length H1 caused the appearance of a clear 10-bp repeat in the •OH cleavage pattern along the entire length of the linker DNA. We attribute this repeat pattern as reflecting the H1-induced stem structure of the linker. Interestingly, in addition to the globular domain (35–120), only a seven amino acid residue stretch (aa 121–127) of the C terminus of H1 enriched in basic residues appeared to be necessary and sufficient for the induction of the 10-bp repeat and thus for the structuring of the linker DNA.
A second critical feature of the •OH cleavage patterns was the striking H1-dependent protection of DNA at the nucleosome dyad. In mono-, di-, and trinucleosomes, 10 bp of DNA located symmetrically about the dyad were clearly and consistently protected against •OH cleavage. This highly specific protection was also observed with all samples assembled with GH1. The corresponding patterns of protection on opposite DNA strands indicate that GH1 interacts with the minor groove of DNA in the center of the nucleosome, with the 10-bp binding site centered on the nucleosome dyad. Importantly, in addition to the protection of the dyad, GH1 binding alone also resulted in the protection of ∼1 additional helical turn of DNA at each edge of the nucleosome core region, suggesting a direct and simultaneous interaction with both the DNA helices entering and exiting the nucleosome (see below).
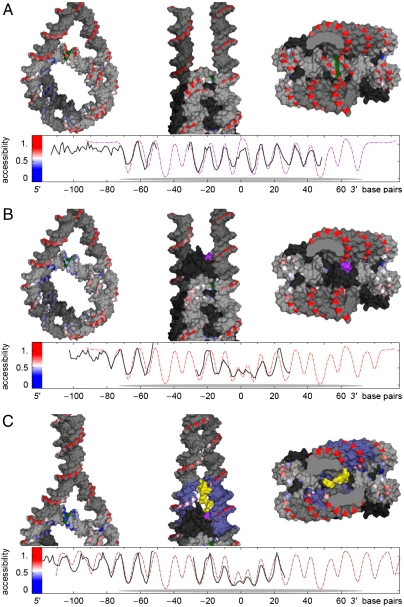
The •OH radicals used in the footprinting are known to primarily attack the C5′ carbon atoms of the backbone sugars (20), allowing us to pinpoint protected sites in 3D molecular models of the nucleosome with Angstrom resolution. As a complementary test of proposed structures, we have determined protection patterns from the accessible surfaces of the C5′ unified atoms (SI Materials and Methods and Fig. S4). The organization of the nucleosome without H1 bound is presented in Fig. 6A (Upper and Movie S1). Sites protected from •OH cleavage (in blue) are located exclusively on the inside of the DNA superhelix and correspond to the DNA–histone octamer interface, whereas the outward-facing DNA (including the region around the dyad and the linkers) is freely accessible to •OH (in red). The predicted accessibility profile (Fig. 6A, Lower) accurately reproduces the experimentally determined accessibilities (linear correlation coefficient R = 0.79), thus validating our approach.
Fig. 6.
Molecular models for the nucleosome particle. (A) Model of the nucleosome without H1. The model of the nucleosomal DNA alone and two views of the nucleosome with histones are shown in the top panel. The experimental •OH-accessibility profile is depicted on the three-dimensional nucleosome structure by color coding the DNA deoxyribose C5′ atoms from blue (maximal protection) to white (partial protection) to red (maximal accessibility). DNA C5′ atoms without footprinting data and all other DNA are shown in gray, the dyad in green. Protein is shown in black (omitted in the left column). Views shown, from left to right, are (i) rotated sideways and up by 30° from the NCP superhelical axis, (ii) at right angles to the superhelical and dyad axes, and (iii) along the dyad axis. The bottom panel shows plots of the experimental •OH-accessibility profile (Solid Line) and the corresponding model-derived accessibility profile (Dashed Line). Nucleosome core region is indicated by the gray oval. (B) Modeling of the nucleosome associated with the globular domain (GH1) of histone H1. The upper panel illustrates the location of GH1 in the nucleosome in the three-contact model, as described in the text. Note that GH1 protects the dyad and directly interacts with 10 bp of each linker DNA. The C terminus of GH1 is highlighted in magenta. (C) Model of the nucleosome associated with 35–127 H1 mutant. The location of the 35–127 H1 mutant and the 3D organization of the linker DNA stem obtained from constrained DNA elastic relaxation were determined as described in the text. Note the strong protection of the dyad and the presence of the 10-bp repeat within the linker DNA (Bottom). DNA within a 30 Å radius of the GH1 C terminus is colored blue. A hypothetical conformation of aa 112–127 is shown in yellow. Because the location of 5 aa (35–39) sequence of the NH2 terminus is not known, this sequence was not shown in the model.
To discriminate between models for the structure of nucleosomes containing the globular domain of the linker histone (16, 17, 19, 21), we built corresponding structures by manually placing a GH1 solution structure (22) into the nucleosome (see SI Materials and Methods and Fig. S5). The calculated footprinting pattern of a three-contact GH1-nucleosome particle on the basis of a model for the placement of H5 (GH5) by Fan and Roberts (21) (Fig. 6B, Upper and Movie S2) matches very well the experimental pattern: Both the dyad and ∼1 helical turn of the linker DNA to either side of the core region are protected against •OH cleavage (Fig. 6B, Lower). In contrast, the suggested two-contact models (16, 17, 19) were incompatible with the strong protection observed at the dyad (see Movies S3 and S4 and Fig. S5).
For the linker DNA stem, which is formed with full-length H1 or the truncated mutants 1–127 or 35–127, models on the basis of high-resolution structural studies are not available. In this case the detailed register of the protected sites along the stem provides valuable structural information. To model the stem structure, we have aligned the linkers in space in such a way that their mutual protection reproduces the measured accessibility profile. We have assumed that the alignment was facilitated by neutralization of the linker DNA phosphates through interactions of the COOH terminus of H1 and NH2 terminus of histone H3 [which is known to associate with the linker DNA (23)] and that the most likely stem structure has minimal DNA elastic energy (see SI Materials and Methods). The resulting calculated stem structure, which satisfies these requirements, is shown in Fig. 6C, Upper. By construction, the structure-derived accessibility profile matches well the experimental profile (Fig. 6C, Lower). In the minimal-energy configuration, the linkers come together along ∼20–30 bases outside the core particle, slightly curving into a two-start superhelical stem with a large pitch of around 100–120 bp (Fig. 6C, Upper; see also Movie S5). This structure has, as the core particle itself, a twofold symmetry.
The footprinting and ECM data also allowed us to predict how the stem structure might be generated and maintained. The short sequence of H1 (aa 121–127) required for the formation of the stem contains three positively charged amino acids residues, namely, K122, K124, and K125 (see Fig. 1A). These three lysines, together with the neighboring lysine K120, would interact with both DNA linkers in the vicinity of the binding site of the COOH end of GH1 (Fig. 6C). The binding of these additional four lysine residues together with the binding of GH1 would be sufficient to clamp the exiting and entering DNA and to form the stem structure (Fig. 6C, Upper and Movie S5). We hypothesize that the remaining part of the COOH terminus of H1 (aa 128–227; see Fig. 1A) serves to further neutralize the DNA phosphates and to facilitate transient contacts between the linkers resulting in a partial protection against •OH cleavage beyond the spatial extension of the COOH terminus of H1. In agreement with this hypothesis, our statistical analysis (Fig. S1B) shows that the linker DNA stem formed with the truncated mutant 35–127 was less stable compared to that formed with either full-length H1 or the H1 truncated mutant 1–177.
Materials and Methods
Preparation of Proteins and Nucleosomal Substrate.
Mouse NAP-1 and histones were bacterially expressed and purified by anion and cation exchange chromatography, respectively. Mononucleosomal 601-DNA was PCR amplified, whereas dinucleosomal and trinucleosomal constructs were subcloned from the 33x 200–601 DNA array (kindly provided by Daniela Rhodes, Cambridge, UK). DNA substrates were 32P end-labeled (see SI Text). Nucleosomal substrates were prepared by salt dialysis method and H1 was deposited by using NAP-1 as described in ref. 8.
Hydroxyl Radical Footprinting and ECM.
Control and H1 (or deletion mutant) assembled nucleosomal samples were buffer exchanged into quencher-free buffer (5 mM Tris, pH 7.5, 5 mM NaCl, and 0.25 mM EDTA) by repeated filtration through 100 kDa cutoff centricons. The samples were treated with hydroxyl radicals (see SI Text) and analyzed on sequencing gel. ECM of the samples was performed on 200 ng/μL samples as described earlier (2).
Coarse-Grained Modeling.
Hydroxyl radical accessibility of the different nucleosome models was computed and compared with the experimental data (16, 19, 21). The •OH accessibility was visualized by the Chimera software (24). Energy minimization was performed to improve the modeling of linker DNA organization. For detailed procedure, see SI Text.
Supplementary Material
Acknowledgments.
This work was supported by grants from Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, Agence Nationale de Recherches “EPIVAR” 08-BLAN-0320-02 (to S.D.), ANR-09-BLAN-NT09-485720 “CHROREMBER” (to D.A., J.B., and S.D.), the Association pour la Recherche sur le Cancer [Grant 4821 (to D.A.)], the Région Rhône-Alpes [Convention CIBLE 2008 (to D.A. and S.D.)], and the European Community’s Seventh Framework Program FP7/2007-2013, Grant 222008S. J.B. acknowledges the support of the Czech Grants LC535, MSM0021620806, and AV0Z50110509. J.J.H. acknowledges National Institutes of Health Grant GM52426.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000309107/-/DCSupplemental.
References
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Bednar J, et al. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci USA. 1998;95(24):14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamiche A, Schultz P, Ramakrishnan V, Oudet P, Prunell A. Linker histone-dependent DNA structure in linear mononucleosomes. J Mol Biol. 1996;257(1):30–42. doi: 10.1006/jmbi.1996.0144. [DOI] [PubMed] [Google Scholar]
- 4.Dimitrov SI, Russanova VR, Pashev IG. The globular domain of histone H5 is internally located in the 30 nm chromatin fiber: An immunochemical study. EMBO J. 1987;6(8):2387–2392. doi: 10.1002/j.1460-2075.1987.tb02516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russanova VR, Dimitrov SI, Makarov VL, Pashev IG. Accessibility of the globular domain of histones H1 and H5 to antibodies upon folding of chromatin. Eur J Biochem. 1987;167(2):321–326. doi: 10.1111/j.1432-1033.1987.tb13339.x. [DOI] [PubMed] [Google Scholar]
- 6.Zlatanova J, Seebart C, Tomschik M. The linker-protein network: Control of nucleosomal DNA accessibility. Trends Biochem Sci. 2008;33(6):247–253. doi: 10.1016/j.tibs.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Clark DJ, Thomas JO. Salt-dependent co-operative interaction of histone H1 with linear DNA. J Mol Biol. 1986;187(4):569–580. doi: 10.1016/0022-2836(86)90335-9. [DOI] [PubMed] [Google Scholar]
- 8.Shintomi K, et al. Nucleosome assembly protein-1 is a linker histone chaperone in Xenopus eggs. Proc Natl Acad Sci USA. 2005;102(23):8210–8215. doi: 10.1073/pnas.0500822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saeki H, et al. Linker histone variants control chromatin dynamics during early embryogenesis. Proc Natl Acad Sci USA. 2005;102(16):5697–5702. doi: 10.1073/pnas.0409824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howe L, Ausio J. Nucleosome translational position, not histone acetylation, determines TFIIIA binding to nucleosomal Xenopus laevis 5S rRNA genes. Mol Cell Biol. 1998;18(3):1156–1162. doi: 10.1128/mcb.18.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staynov DZ, Crane-Robinson C. Footprinting of linker histones H5 and H1 on the nucleosome. EMBO J. 1988;7(12):3685–3691. doi: 10.1002/j.1460-2075.1988.tb03250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tullius TD. DNA footprinting with hydroxyl radical. Nature. 1988;332(6165):663–664. doi: 10.1038/332663a0. [DOI] [PubMed] [Google Scholar]
- 13.Allan J, Hartman PG, Crane-Robinson C, Aviles FX. The structure of histone H1 and its location in chromatin. Nature. 1980;288(5792):675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- 14.Crane-Robinson C. How do linker histones mediate differential gene expression? Bioessays. 1999;21(5):367–371. doi: 10.1002/(SICI)1521-1878(199905)21:5<367::AID-BIES2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 15.Simpson RT. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- 16.Zhou YB, Gerchman SE, Ramakrishnan V, Travers A, Muyldermans S. Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature. 1998;395(6700):402–405. doi: 10.1038/26521. [DOI] [PubMed] [Google Scholar]
- 17.Pruss D, et al. An asymmetric model for the nucleosome: A binding site for linker histones inside the DNA gyres. Science. 1996;274(5287):614–617. doi: 10.1126/science.274.5287.614. [DOI] [PubMed] [Google Scholar]
- 18.Hayes JJ, Kaplan R, Ura K, Pruss D, Wolffe A. A putative DNA binding surface in the globular domain of a linker histone is not essential for specific binding to the nucleosome. J Biol Chem. 1996;271(42):25817–25822. doi: 10.1074/jbc.271.42.25817. [DOI] [PubMed] [Google Scholar]
- 19.Brown DT, Izard T, Misteli T. Mapping the interaction surface of linker histone H1(0) with the nucleosome of native chromatin in vivo. Nat Struct Mol Biol. 2006;13(3):250–255. doi: 10.1038/nsmb1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balasubramanian B, Pogozelski WK, Tullius TD. DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone. Proc Natl Acad Sci USA. 1998;95(17):9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan L, Roberts VA. Complex of linker histone H5 with the nucleosome and its implications for chromatin packing. Proc Natl Acad Sci USA. 2006;103(22):8384–8389. doi: 10.1073/pnas.0508951103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerf C, et al. Homo- and heteronuclear two-dimensional NMR studies of the globular domain of histone H1: Full assignment, tertiary structure, and comparison with the globular domain of histone H5. Biochemistry. 1994;33(37):11079–11086. doi: 10.1021/bi00203a004. [DOI] [PubMed] [Google Scholar]
- 23.Stefanovsky V, Dimitrov SI, Russanova VR, Angelov D, Pashev IG. Laser-induced crosslinking of histones to DNA in chromatin and core particles: implications in studying histone-DNA interactions. Nucleic Acids Res. 1989;17(23):10069–10081. doi: 10.1093/nar/17.23.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.