Abstract
The tumor suppressor p53 is a canonical inducer of cellular senescence (irreversible loss of proliferative potential and senescent morphology). p53 can also cause reversible arrest without senescent morphology, which has usually been interpreted as failure of p53 to induce senescence. Here we demonstrate that p53-induced quiescence actually results from suppression of senescence by p53. In previous studies, suppression of senescence by p53 was masked by p53-induced cell cycle arrest. Here, we separated these two activities by inducing senescence through overexpression of p21 and then testing the effect of p53 on senescence. We found that in p21-arrested cells, p53 converted senescence into quiescence. Suppression of senescence by p53 required its transactivation function. Like rapamycin, which is known to suppress senescence, p53 inhibited the mTOR pathway. We suggest that, while inducing cell cycle arrest, p53 may simultaneously suppress the senescence program, thus causing quiescence and that suppression of senescence and induction of cell cycle arrest are distinct functions of p53. Thus, in spite of its ability to induce cell cycle arrest, p53 can act as a suppressor of cellular senescence.
Keywords: cell cycle, p21, rapamycin, mTOR, aging
Induction of p53 can cause apoptosis, reversible cell cycle arrest, and cellular senescence (1–5). In contrast to reversible cell cycle arrest (quiescence), cellular senescence is defined by irreversible loss of proliferative potential, acquisition of characteristic morphology (large, flattened cells), and expression of specific biomarkers (e.g., senescence-associated β-galactosidase, SA-β-Gal) (6). Since p53 appeared to induce senescence in some situations, observations of p53-induced quiescence have usually been interpreted as failure of p53 to activate the senescence program, which remains poorly understood. We recently reported that in two cell lines in which ectopic expression of p21 caused senescence, activation of endogenous p21 by endogenous p53 caused quiescence (7). The simplest conventional explanation for this result is that p53 failed to activate p21 to the degree required for induction of senescence, although it was sufficient for induction of quiescence. However, an alternative possibility is that p53 acts as a suppressor of the senescence program. This model leads to the testable prediction that induction of p53 would suppress p21-mediated senescence and convert it into quiescence. Here we demonstrate that this model is indeed correct, which indicates that, despite its ability to induce cell cycle arrest, p53 is a suppressor, not an inducer, of cellular senescence. Retrospectively, this result is not completely unexpected. First, it is known that p53 inhibits the mTOR (mammalian target of rapamycin) pathway (8–12). Second, it is known that inhibition of mTOR by rapamycin converts senescence into quiescence (13–15). In turn, this predicts that p53, like rapamycin, may suppress senescence. Here we confirmed this prediction.
Results
The p53 Activator Nutlin-3a Suppresses p21-Induced Senescence.
As previously shown, induction of p21 in HT1080-derived HT-p21-9 cells carrying an isopropyl-thio-galactosidase (IPTG)-inducible p21expression construct caused senescence (7, 16). In the same cells, induction of p53 by nutlin-3a caused reversible cell cycle arrest (quiescence) and cells resumed proliferation after removal of nutlin-3a (7). We used nutlin-3a, an inhibitor of p53-Mdm2 binding, in these experiments because it induces p53 at physiological levels without DNA damage and is highly specific (17). Thus, physiological levels of p53 induced quiescence, whereas ectopic expression of p21 induced senescence (7). There are two alternative models that could explain these results. First, the conventional model suggests that the physiological levels of p53 induced by nutlin-3a are not sufficient to induce p21 to the extent required to activate the senescent program in this cell line. Then addition of nutlin-3a to IPTG may only intensify senescence. A second, alternative model is that p53 actually suppresses senescence. In this case, activation of p53 by nutlin-3a in concert with IPTG-mediated induction of p21 would be expected to convert senescence into quiescence.
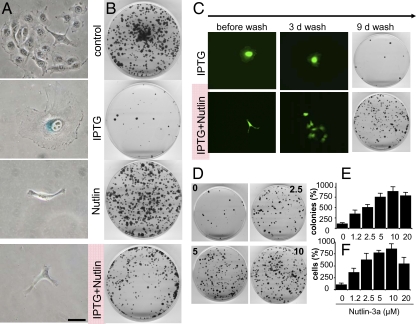
As shown in Fig. 1 and reported previously (7), IPTG- and nutlin-3a-treated cells are positive controls for senescence and quiescence, respectively. IPTG treatment induced characteristic senescent morphology (large, flat, SA-β-Gal-positive cells), whereas nutlin-3a treated cells remained small, lean and SA-β-Gal-negative (Fig. 1A). In addition, colony formation assays showed that IPTG treatment resulted in irreversible loss of proliferative potential (only a few cells formed colonies upon removal of IPTG), whereas nutlin-3a treatment caused reversible arrest (substantial colony formation upon nutlin-3a removal) (Fig. 1B).
Fig. 1.
Nutlin-3a converted senescence into quiescence. (A) HT-p21-9 cells were treated with IPTG, 10 μM nutlin-3a, and IPTG plus nutlin-3a for 3 days. Cells were stained for β-Gal and photographed (original magnification, ×400). (Scale bar, 50 μm.) (B) HT-p21-9 cells were treated with IPTG, 10 μM nutlin-3a, and IPTG plus nutlin-3a for 3 days. After 3 days, cells were washed to remove IPTG and nutlin-3a. Then cells were cultured in fresh medium until colonies became visible. Dishes were stained with crystal violet and photographed on day 4 (control) or on day 9 (IPTG- and nutlin-3a-treated). (C) HT-p21-9 cells were treated with IPTG in the presence or absence of 10 μM nutlin-3a for 3 days. Before washing, live HT-p21-9 cells (expressing GFP for better visualization of live cells) were photographed (original magnification ×100) under blue light. Three (3) d wash, 3 days after drug removal; 9 d wash, 9 days after drug removal, cells were stained with crystal violet and photographed. (D) Nutlin-3a dose–response. HT-p21-9 cells were plated with IPTG and 0, 2.5, 5, or 10 μM nutlin-3a. After 3 days, the plates were washed and cells were incubated for an additional 9 days in fresh medium, stained with crystal violet, and photographed. (E) Colonies per dish. HT-p21-9 cells were plated with IPTG and 0, 1.2, 2.5, 5, 10, or 20 μM nutlin-3a, as indicated. After 3 days, the plates were washed and cells were incubated for an additional 9 days in fresh medium. Colonies were counted and results are shown as percentage of control (IPTG alone). (F) Cells per dish, as in E. Cells were trypsinized and counted. Results are shown as percentage of control (IPTG alone).
The key question is what is dominant? Would addition of nutlin-3a to IPTG convert senescence into quiescence? The result of this key experiment showed that treatment with nutlin-3a prevented the senescent morphology caused by IPTG: cells remained small, lean and negative for SA-β-Gal-staining (Fig. 1A). Furthermore, such cells retained the proliferative potential and clonogenicity (Fig. 1B). So the effect of nutlin-3a on IPTG-induced senescence was dominant. Importantly, nutlin-3a neither abrogated nor diminished the levels of p21 (see immunoblots throughout the paper). Nutlin-3a did not abrogate the cytostatic effect of IPTG. In agreement with previous results, IPTG caused instant cell cycle arrest, manifested as solitary cells with senescent morphology at low cell density (Fig. S1). In the presence of nutlin-3a alone, cells typically underwent one division and did not proliferate further, as illustrated by colonies of two adjusted cells with nonsenescent morphology (Fig. S1). In the presence of both nutlin-3a and IPTG, cells were arrested immediately without a single division, but did not acquire senescent morphology (Fig. S1). Thus, without abrogating cell cycle arrest caused by IPTG, nutlin-3a converted senescence into a reversible condition (quiescence). When IPTG and nutlin-3a were washed out of the cultures, the cells resumed proliferation, forming microcolonies (Fig. 1C) and then macrocolonies (Fig. 1C). These results indicate that nutlin-3a prevented cells from undergoing IPTG-induced senescence. Suppression of senescence by nutlin-3a was observed at a range of active concentrations of nutlin-3a in a dose-dependent manner (Fig. 1 D and E). The most quantitative way to measure preservation of proliferative potential (PP) is the total cell number per dish (15). Nutlin-3a preserved PP in a dose-dependent manner (Fig. 1F). We have measured the number of cells per colony versus the number of colonies per dish (Fig. S2). Thus, nutlin-3a increased the number of cells with normal PP. The preservation of proliferative potential by nutlin-3a in IPTG-arrested cells was confirmed in both IPTG-regulated p16- and p21-expressing cells (Fig. S3).
Suppression of Senescence Requires the Transactivation Function of p53.
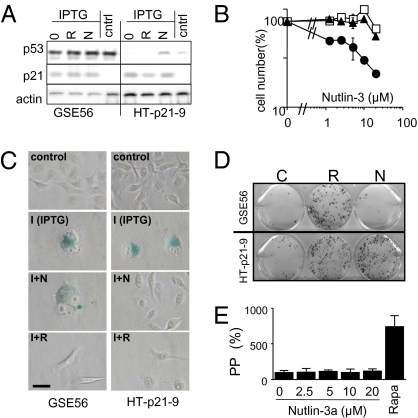
Nutlin-3a is a highly specific activator of p53 and no off-target effects of the compound have been reported (5). In fact, nutlin-3b, an optimer of nutlin-3a that does not block Mdm-2/p53 interaction, was not able to convert senescence into quiescence (Fig. S3 B and C). To directly test whether nutlin-3a inhibits senescence by a p53-dependent mechanism, we used HT-p21-GSE56 cells, a derivative of the HT-p21cell line in which p53 function is blocked by a transdominant inhibitor, GSE56 (18). As expected, p53 was expressed at very high levels in these cells because inhibition of its transactivation function results in stabilization of the protein (analogous to mutant p53). Although nutlin-3a induced p53 in HT-p21 cells, it did not affect p53 levels in HT-p21-GSE56 cells (Fig. 2A). IPTG strongly induced p21 in HT-p21-GSE56 cells and nutlin-3a did not affect this induction (Fig. 2A). As expected, nutlin-3a failed to inhibit proliferation of HT-p21-GSE56 cells (Fig. 2B), thereby confirming that the model was adequate for testing whether suppression of senescence by nutlin-3a depends on p53. In addition, it was important to employ a positive control for p53-independent suppression of senescence. We have previously demonstrated that activation of mTOR (mammalian target of rapamycin) was required for cellular senescence (13, 19). Deactivation of mTOR by rapamycin prevented senescence, causing quiescence instead (14). Rapamycin did not induce p53 (Fig. 2A) in agreement with its p53-independent inhibition of mTOR. Rapamycin suppressed IPTG-induced senescence in HT-p21-GSE56 cells (Fig. 2C). In contrast, nutlin-3a suppressed senescence in IPTG-treated HT-p21-9 cells only and not in similarly treated HT-p21-GSE56 cells (Fig. 2C). Consistent with these findings, nutlin-3a (unlike rapamycin) did not preserve colony formation and PP in IPTG-treated HT-p21-GSE56 cells lacking functional p53 (Fig. 2 D and E). These data demonstrate that the transcriptional activity of p53 is required for suppression of senescence by nutlin-3A. In contrast, rapamycin inhibited senescence without relying on p53, as illustrated by its ability to prevent senescent morphology (Fig. 2C) and to preserve proliferative potential (Fig. 3 D and E) in IPTG-treated HT-p21-GSE56 cells.
Fig. 2.
p53-dependent effects of nutlin-3a. (A) HT-p21-GSE56 and HT-p21-9 cells were treated with IPTG alone (0) or IPTG plus rapamycin (R) and nutlin-3a (N). Control cells were left untreated (no IPTG). After 1 day, cells were lysed and immunoblot was performed. (B) HT-p21-GSE56 (open circles) and HT-p21-9 cells (closed circles) were treated with nutlin-3a for 5 days and then counted. As a negative control, parental cells were treated with nutlin-3b (open squares). (C and D) HT-p21-GSE56 and HT-p21-9 cells were treated with IPTG alone or with IPTG + rapamycin (I+R) or IPTG + nutlin-3a (I+N), as indicated. Control cells were left untreated (no IPTG). (C) Morphology. After 3 days, cells were stained for β-Gal. (Scale bars, 50 μm.) (D) Colony formation. After 3 days, cells were washed and incubated in fresh medium w/o drugs for an additional 9 days. Plates were stained with crystal violet and photographed. (E) Proliferative potential (PP). After 3 days, HT-p21-GSE56 cells were washed and incubated in fresh medium w/o drugs. Cells were counted and results are shown as percentage of IPTG alone.
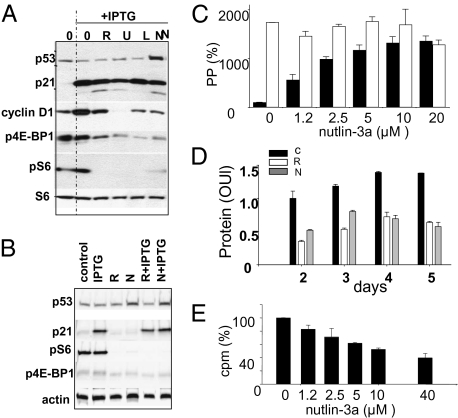
Fig. 3.
Effects of nutlin-3a on the mTOR pathway and protein synthesis. (A) Immunoblot. HT-p21 cells were treated with IPTG alone or with IPTG plus 500 nM rapamycin (R), 25 μM LY-294002 (L), 10 μM U0126 (U), or 10 μM nutlin-3a (N) for 24 h. Immunoblot was performed as described in Methods. (B) Immunoblot. HT-p21 cells were treated with rapamycin (R) and nutlin-3a (N) in the presence or absence of IPTG for 18 h. Immunoblot was performed as described in Methods. (C) Effects of nutlin-3a on PP (proliferative potential) of IPTG-treated HT-p21-9 cells in the absence (black bars) or presence (open bars) of rapamycin (500 nM). After 3 days, cells were washed and incubated in fresh medium w/o drugs for an additional 7 days. Cells were counted and are shown as percentage of IPTG alone. (D) Effects of nutlin-3a and rapamycin on cellular hypertrophy caused by IPTG. Cells were treated with either IPTG alone (black bars) or IPTG plus rapamycin (white bars) or plus nutlin-3a (gray bars). On days 2, 3, 4, and 5, cells were lysed and protein content per well was measured. The numbers presented correspond to protein content per cell, because the cells did not proliferate and their numbers were unchanged during the course of the experiment. (E) Effects of nutlin-3a on protein synthesis ([35S]methionine/cysteine incorporation). Cells were labeled with [35S]methionine/cysteine as described in Methods.
Inhibition of the mTOR Pathway by Nutlin-3a.
We previously reported that inhibitors of mTOR (rapamycin), PI-3K (LY294002), and MEK (U0126) all deactivate the mTOR pathway in HT-p21-9 cells, as measured by lack of phosphorylation of the S6 ribosomal protein, and suppress cellular senescence (19). Like all of these agents, nutlin-3a inhibited S6 phosphorylation and partially inhibited phosphorylatation of 4E-BP1, another downstream target of the mTOR pathway (Fig. 3A). Nutlin-3a also normalized elevated levels of cyclin D1, associated with cellular senescence. Like rapamycin, nutlin-3a inhibited the mTOR pathway both in the presence and absence of IPTG and did not prevent induction of p21 by IPTG (Fig. 3B). Importantly, IPTG-induced p21 did not affect S6 and 4E-BP1 phosphorylation (Fig. 3 A and B).
Rapamycin and nutlin-3a were equally potent in suppression of senescence (preservation of proliferative potential) in IPTG-treated HT-p21-9 cells (Fig. 3C). Moreover, in the presence of rapamycin at doses that completely inhibit mTOR (19), nutlin-3a could not further suppress senescence, as measured by preservation of proliferative potential (PPP) of IPTG-arrested cells (Fig. 3C). This supports the notion that nutlin-3a and rapamycin affect either the same or overlapping pathways. The mTOR pathway stimulates protein synthesis. Importantly, protein synthesis remained high in IPTG-arrested cells and is inhibited by rapamycin (Fig. S4), thus explaining cellular hypertrophy associated with senescence (15). Both nutlin-3a and rapamycin decreased the protein content per cell in IPTG-treated HT-p21-9 cells (Fig. 3D). To evaluate whether this decrease involved inhibition of protein synthesis, we measured [35S]methionine/cysteine incorporation into nascent proteins in the presence of nutlin-3a (Fig. 3E). Nutlin-3a inhibited [35S]methionine/cysteine incorporation in IPTG-treated HT-p21-9 cells in a dose-dependent manner (Fig. 3E).
Suppression of Senescence by Ectopic Expression of p53.
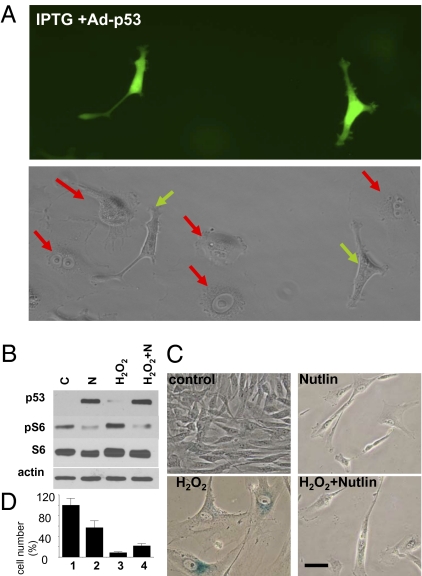
To confirm our results without reliance on nutlin-3a to activate p53, we tested whether expression of exogenous p53 would also lead to suppression of p21-induced senescence. We used an adenovirus that directs constitutive expression of p53 along with GFP (Ad-p53-GFP) (20) such that infected cells can be easily identified by fluorescence microscopy. (Note: In these experiments, we used HT-p21-a cells that unlike HT-p21-9, do not express internal GFP and therefore are not green). At low titers, Ad-p53-GFP infected ~20% of HT-p21-a cells; therefore, we were able to compare p53-overexpressing and noninfected cells on the same slide. As expected, in noninfected cells, IPTG treatment caused senescent morphology (Fig. 4A, red arrows in Lower panel). In contrast, Ad-p53-GFP-infected cells did not acquire senescent morphology (Fig. 4A). To test a different means of inducing senescence, we used infection with a p21-expressing adenovirus (Ad-p21) rather than IPTG to induce p21. Ad-p21 infected cells rapidly acquired senescent morphology, whereas Ad-p53-GFP infected cells did not (Fig. S5A). Furthermore, Ad-p53-GFP suppressed senescence caused by Ad-p21 (Fig. S5B) and by IPTG-induced p16 (Fig. S6).
Fig. 4.
Effects of ectopic and endogenous p53 on senescence in HT-p21-9 and WI-38-tert. (A) p53-expressing adenovirus (Ad-p53) suppresses senescent morphology caused by IPTG in HT-p21-a cells. HT-p21-a cells were treated with IPTG and infected with Ad-p53. After 3 days, cells were photographed (original magnification ×200): (Upper) Under blue light to visualize cells expressing p53 (green cells). (Lower) Under visible light to visualize all cells. Red arrows indicate cells lacking p53 expression. All of these cells show large, flat cell morphology. Green arrows indicate cells expressing p53. (B–D) Effects of nutlin-3a on cellular senescence in WI-38-tert fibroblasts, WI-38-tert cells were treated with 200 μM H202 for 30 min in serum-free medium. Then, the medium was replaced for complete medium (10% serum) with or without 10 μM nutlin-3a. (B) After 1 day, cells were lysed and immunoblot was performed as described in Methods. N, nutlin-3a. (C) After 3 days, the cells were washed (nutlin-3a was removed) and grown for 3 additional days in fresh complete medium. Cells were then stained for β-Gal activity and microphotographed. (Scale bar, 50 μm.) (D) After 3 days, the cells were washed (nutlin-3a was removed) and grown for 6 additional days in fresh complete medium. Cells were then trypsinized and counted. In control, cells reached confluence by day 5 and did not proliferate further. Results are shown as percentage of control.
Suppression of Stress-Induced Senescence in Fibroblasts.
To extend our observation of p53-mediated suppression of senescence to cells unrelated to HT1080, we used telomerase-immortalized human WI-38 fibroblasts (WI-38-tert cells). As shown in Fig. S7, infection of these cells with Ad-p53 also resulted in quiescent morphology (slim, β-Gal-negative cells); however, infection with Ad-p21 induced senescent morphology. Most importantly, coinfection of the cells with Ad-p53 and Ad-p21 demonstrated that p53 suppressed p21-induced senescence (Fig. S7). Because Ad-p53 infection resulted in excessive levels of p53, the observed effect was limited by concomitant induction of apoptosis. Therefore, we used nutlin-3a to induce p53 at physiological levels in this system. We have previously shown that treatment of WI-38-tert cells with nutlin-3a caused quiescence (7). Importantly, nutlin-3a (at concentrations that induce p53) inhibited S6K and S6 phosphorylation (Fig. S8). In contrast, doxorubicin does not inhibit mTOR (13). This may explain why nutlin-3a induced quiescence in WI-38-tert cells, whereas doxorubicin caused senescence in WI-38-tert cells (7). We next investigated whether nutlin-3a could suppress senescence caused by hydrogen peroxide (H2O2), a canonical inducer of cellular senescence in fibroblasts. In WI-38-tert cells, H2O2 inhibited cell proliferation without induction of p53 and without affecting S6 phosphorylation (Fig. 4B). This results in senescent morphology (Fig. 4C). Nutlin-3a induced p53, inhibited S6 phosphorylation (Fig. 4B), and suppressed senescence induced by H2O2 (Fig. 4C). Furthermore, nutlin-3 partially preserved proliferative potential in H2O2-treated cells (Fig. 4D). Thus, we have used different cell lines, as well as various means of inducing cellular senescence and of activating p53, to demonstrate that p53 suppresses senescence.
Discussion
We have demonstrated that p53, thought of as an emblematic inducer of cellular senescence, actually suppresses cellular senescence. In previous studies, suppression of senescence by p53 was apparently masked by p53-induced cell cycle arrest, which (if prolonged) can lead to senescence. As previous studies relied on p53 itself to cause cell cycle arrest (and subsequent senescence or quiescence), it was not possible to distinguish whether p53 actively suppressed senescence or merely failed to induce it in some experimental situations. Here, we were able to differentiate between these two scenarios by testing the effect of p53 on senescence induced by p21 or p16 rather than p53 itself. We found that in either p21- or p16-arrested cells, p53 converted senescence (irreversible arrest with senescent morphology) into quiescence (reversible arrest with preservation of proliferation capacity and no senescent morphology). We showed previously that, when the cell cycle is blocked, activation of mTOR is required for induction of senescence. Rapamycin and inhibitors of PI-3K and MEK all inhibited the mTOR pathway and cellular hypertrophy and converted p21-induced senescence into quiescence (13, 19). It was previously shown that p53 inhibits the mTOR pathway by multiple mechanisms (8–12). As shown here, nutlin-3a-induced p53 inhibited the mTOR pathway and protein synthesis, preventing cellular hypertrophy and senescent morphology. In addition, we found that suppression of senescence by p53 required its ability to function as a transactivator. A number of known p53 transcriptional targets inhibit the mTOR pathway, including AMPK, TSC2, PTEN, IGF-BP3, and sestrins (8–12). Moreover, p53 causes cleavage or deactivation of eIF4GI, eIF4B, eIF4E, and eIF4F, thereby inhibiting protein synthesis downstream of mTOR (21–23). Finally, p53 inhibits the NF-κB pathway (24, 25). Furthermore, sestrin, a target of p53, by inhibiting TOR prevents age-related pathologies in Drosophila (26).The multiplicity of overlapping potential mechanisms by which p53 suppresses senescence is not surprising, but rather may be analogous to the multiple mechanisms by which p53 causes cell cycle arrest and apoptosis.
Remarkably, although p53 has long been thought of as an inducer of senescence, it has never been shown that p53 actually activates the senescence program (namely, that induction of p53 can convert quiescence into senescence). To avoid misunderstanding, we emphasize that activation of p53 in proliferating cells does lead to senescence in many cell lines. For example, nutlin-3a causes senescence in mouse cells of fibroblast origin (27). Also, induction of p53 associated with DNA damage may favor senescence. For example, doxorubicin, a DNA damaging drug, causes cellular senescence in cell lines used herein (7, 13, 28, 29). However, interpretation of this as if p53 “drives” senescence appears to have been illusive. According to our current understanding, p53 simply creates conditions, by causing cell cycle arrest, that enable the senescence program to start. The success of this program depends on whether p53 in a given cell type can exercise its senescence suppressor function. Suppression of senescence by p53 can be explained, at least in part, by its ability to inhibit the mTOR pathway (Fig. S9). This may, in turn, determine the choice between senescence and quiescence. We are currently testing the hypothesis that senescence occurs in those cells in which p53 is incapable of suppressing mTOR. In agreement with this hypothesis, it was shown that mTOR activators—oncogenic Ras and MEK—facilitate senescence in the presence of p53 (27, 30).
Our finding that p53 suppresses senescence can explain prior observations showing that loss of p53 resulted in amplification of the senescence-associated secretory and proinflammatory phenotype (24, 31) and organismal proinflammatory state (usually associated with aging) in mice (24). In addition, it has been noted in model organisms that oncoproteins accelerate aging, whereas tumor suppressors suppress aging (32). p53 was previously considered to be a notable exception to this paradigm. Our findings suggest that p53 may indeed fit the rule and behave like other tumor suppressors, which suppress aging (e.g., PTEN, AMPK, LKB1, and TSC1/2), rather than like oncoproteins, which accelerate aging (e.g., PI-3K and Ras). This illuminates the significance of observations of declining p53 function with age (33, 34) and explains why mild activation of p53 may increase life span in mice (35).
Methods
Cell Lines and Reagents.
HT-p21-9 and HT-p21-a cells are derivatives of HT1080 human fibrosarcoma cells, where p21 expression can be turned on or off using IPTG (7, 16, 28, 29, 36). HT-p21-9 cells express GFP, whereas HT-p21-a cells do not. HT-p16 cells are derivatives of HT1080 cells in which p16 expression can be turned on or off using IPTG (16, 36). WI-38-tert, WI-38 fibroblasts immortalized by telomerase, were described previously (7). HT-p21-GSE56 cells: p53 inhibiting peptide GSE56 (18) was introduced into HT1080 p21-9 cells via a retroviral vector LXSE (37). Cells were grown in high glucose DMEM with 10% FC2 serum. WI-38-tert cells were grown in low glucose DMEM with 10% FCS. Rapamycin was obtained from LC Laboratories. IPTG (final concentration of 50 μg/mL) and FC2 were obtained from Sigma-Aldrich. Nutlin-3-a and -b were obtained from Sigma-Aldrich and Roche (38). p53, p21, and p53-GFP expressing adenoviruses (Ad-p53, Ad-p21, and Ad-p53-GFP) were described previously (20, 39) and obtained from Wafik El-Deiry (University of Pennsylvania).
Colony Formation Assay.
Plates were fixed and stained with 1.0% methylene blue or with crystal violet (13).
Immunoblot Analysis.
Proteins were separated on 4–15% gradient Tris-HCl gels (Bio-Rad). The following antibodies were used: mouse anti-actin from Santa Cruz Biotechnology, rabbit anti-phospho-S6 (Ser240/244) and (Ser235/236), mouse anti-S6, mouse anti-phospho-p70 S6 kinase (Thr389), mouse anti-p21 and anti-p53, rabbit anti-phospho-4E-BP1 (Thr37/46) from Cell Signaling, mouse anti-4E-BP1 from Invitrogen, and mouse anti-p53 (Ab-6) from Calbiochem.
β-Galactosidase Staining.
β-Gal staining was performed using senescence-galactosidase staining kit (Cell Signaling Technology).
Metabolic Labeling.
HT-p21-9 cells were seeded at 25,000 cells/well in 12-well plates. On the next day, cells were treated with drugs. After 24 h, cells were labeled with 30 μCi [35S]methionine/cysteine (Amersham) per milliliter of Met/Cys-free DMEM (Invitrogen) for 1 h at 37 °C. Cells were washed with PBS and lysed in 1% SDS, with 0.5% BSA. To determine 35S incorporation, total protein was precipitated with 0.5 mL 10% TCA and collected on nitrocellulose filters. Filters were air dried and counted using liquid scintillation counter.
Supplementary Material
Acknowledgments
We thank Wafik S. El-Deiry (University of Pennsylvania) for providing Ad-p21 and Ad-p53. We also thank Olga Leontieva for help with Fig. 4B. This work was supported in part by National Institutes of Health Grant CA 75179 and CA 060730 (to A.V.G.).
Footnotes
*This Direct Submission article had a prearranged editor.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002298107/-/DCSupplemental.
References
- 1.Vogelstein B, Lane DP, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Levine AJ, Feng Z, Mak TW, You H, Jin S. Coordination and communication between the p53 and IGF-1-AKT-TOR signal transduction pathways. Genes Dev. 2006;20:267–275. doi: 10.1101/gad.1363206. [DOI] [PubMed] [Google Scholar]
- 3.Levine AJ, Oren M. The first 30 years of p53: Growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vousden KH, Prives C. Blinded by the light: The Growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 5.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: Drugging the p53 pathway. Nat Rev Cancer. 2009;9:862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 6.Itahana K, Campisi J, Dimri GP. Methods to detect biomarkers of cellular senescence: The senescence-associated beta-galactosidase assay. Methods Mol Biol. 2007;371:21–31. doi: 10.1007/978-1-59745-361-5_3. [DOI] [PubMed] [Google Scholar]
- 7.Korotchkina LG, Demidenko ZN, Gudkov AV, Blagosklonny MV. Cellular quiescence caused by the Mdm2 inhibitor nutlin-3A. Cell Cycle. 2009;8:3777–3781. doi: 10.4161/cc.8.22.10121. [DOI] [PubMed] [Google Scholar]
- 8.Stambolic V, et al. Regulation of PTEN transcription by p53. Mol Cell. 2001;8:317–325. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 9.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci USA. 2005;102:8204–8209. doi: 10.1073/pnas.0502857102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drakos E, et al. Stabilization and activation of p53 downregulates mTOR signaling through AMPK in mantle cell lymphoma. Leukemia. 2009;23:784–790. doi: 10.1038/leu.2008.348. [DOI] [PubMed] [Google Scholar]
- 11.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braunstein S, Badura ML, Xi Q, Formenti SC, Schneider RJ. Regulation of protein synthesis by ionizing radiation. Mol Cell Biol. 2009;29:5645–5656. doi: 10.1128/MCB.00711-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demidenko ZN, Blagosklonny MV. Growth stimulation leads to cellular senescence when the cell cycle is blocked. Cell Cycle. 2008;7:3355–3361. doi: 10.4161/cc.7.21.6919. [DOI] [PubMed] [Google Scholar]
- 14.Demidenko ZN, et al. Rapamycin decelerates cellular senescence. Cell Cycle. 2009;8:1888–1895. doi: 10.4161/cc.8.12.8606. [DOI] [PubMed] [Google Scholar]
- 15.Demidenko ZN, Blagosklonny MV. Quantifying pharmacologic suppression of cellular senescence: Prevention of cellular hypertrophy versus preservation of proliferative potential. Aging (Albany NY) 2009;1:1008–1016. doi: 10.18632/aging.100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang BD, et al. p21Waf1/Cip1/Sdi1-induced growth arrest is associated with depletion of mitosis-control proteins and leads to abnormal mitosis and endoreduplication in recovering cells. Oncogene. 2000;19:2165–2170. doi: 10.1038/sj.onc.1203573. [DOI] [PubMed] [Google Scholar]
- 17.Huang B, Deo D, Xia M, Vassilev LT. Pharmacologic p53 activation blocks cell cycle progression but fails to induce senescence in epithelial cancer cells. Mol Cancer Res. 2009;7:1497–1509. doi: 10.1158/1541-7786.MCR-09-0144. [DOI] [PubMed] [Google Scholar]
- 18.Ossovskaya VS, et al. Use of genetic suppressor elements to dissect distinct biological effects of separate p53 domains. Proc Natl Acad Sci USA. 1996;93:10309–10314. doi: 10.1073/pnas.93.19.10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demidenko ZN, Shtutman M, Blagosklonny MV. Pharmacologic inhibition of MEK and PI-3K converges on the mTOR/S6 pathway to decelerate cellular senescence. Cell Cycle. 2009;8:1896–1900. doi: 10.4161/cc.8.12.8809. [DOI] [PubMed] [Google Scholar]
- 20.Wang W, Takimoto R, Rastinejad F, El-Deiry WS. Stabilization of p53 by CP-31398 inhibits ubiquitination without altering phosphorylation at serine 15 or 20 or MDM2 binding. Mol Cell Biol. 2003;23:2171–2181. doi: 10.1128/MCB.23.6.2171-2181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Constantinou C, Clemens MJ. Regulation of the phosphorylation and integrity of protein synthesis initiation factor eIF4GI and the translational repressor 4E-BP1 by p53. Oncogene. 2005;24:4839–4850. doi: 10.1038/sj.onc.1208648. [DOI] [PubMed] [Google Scholar]
- 22.Constantinou C, et al. p53-induced inhibition of protein synthesis is independent of apoptosis. Eur J Biochem. 2003;270:3122–3132. doi: 10.1046/j.1432-1033.2003.03687.x. [DOI] [PubMed] [Google Scholar]
- 23.Constantinou C, Elia A, Clemens MJ. Activation of p53 stimulates proteasome-dependent truncation of eIF4E-binding protein 1 (4E-BP1) Biol Cell. 2008;100:279–289. doi: 10.1042/BC20070121. [DOI] [PubMed] [Google Scholar]
- 24.Komarova EA, et al. p53 is a suppressor of inflammatory response in mice. FASEB J. 2005;19:1030–1032. doi: 10.1096/fj.04-3213fje. [DOI] [PubMed] [Google Scholar]
- 25.Dey A, Wong ET, Bist P, Tergaonkar V, Lane DP. Nutlin-3 inhibits the NFkappaB pathway in a p53-dependent manner: Implications in lung cancer therapy. Cell Cycle. 2007;6:2178–2185. doi: 10.4161/cc.6.17.4643. [DOI] [PubMed] [Google Scholar]
- 26.Lee JH, et al. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science. 2010;327:1223–1228. doi: 10.1126/science.1182228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Efeyan A, et al. Induction of p53-dependent senescence by the MDM2 antagonist nutlin-3a in mouse cells of fibroblast origin. Cancer Res. 2007;67:7350–7357. doi: 10.1158/0008-5472.CAN-07-0200. [DOI] [PubMed] [Google Scholar]
- 28.Chang BD, et al. Role of p53 and p21waf1/cip1 in senescence-like terminal proliferation arrest induced in human tumor cells by chemotherapeutic drugs. Oncogene. 1999;18:4808–4818. doi: 10.1038/sj.onc.1203078. [DOI] [PubMed] [Google Scholar]
- 29.Chang BD, et al. A senescence-like phenotype distinguishes tumor cells that undergo terminal proliferation arrest after exposure to anticancer agents. Cancer Res. 1999;59:3761–3767. [PubMed] [Google Scholar]
- 30.Ferbeyre G, et al. Oncogenic ras and p53 cooperate to induce cellular senescence. Mol Cell Biol. 2002;22:3497–3508. doi: 10.1128/MCB.22.10.3497-3508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coppé JP, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinkston JM, Garigan D, Hansen M, Kenyon C. Mutations that increase the life span of C. elegans inhibit tumor growth. Science. 2006;313:971–975. doi: 10.1126/science.1121908. [DOI] [PubMed] [Google Scholar]
- 33.Feng Z, et al. Declining p53 function in the aging process: A possible mechanism for the increased tumor incidence in older populations. Proc Natl Acad Sci USA. 2007;104:16633–16638. doi: 10.1073/pnas.0708043104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng Z, Hu W, Rajagopal G, Levine AJ. The tumor suppressor p53: Cancer and aging. Cell Cycle. 2008;7:842–847. doi: 10.4161/cc.7.7.5657. [DOI] [PubMed] [Google Scholar]
- 35.Matheu A, et al. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- 36.Broude EV, et al. p21(Waf1/Cip1/Sdi1) mediates retinoblastoma protein degradation. Oncogene. 2007;26:6954–6958. doi: 10.1038/sj.onc.1210516. [DOI] [PubMed] [Google Scholar]
- 37.Kandel ES, et al. Applications of green fluorescent protein as a marker of retroviral vectors. Somat Cell Mol Genet. 1997;23:325–340. doi: 10.1007/BF02674280. [DOI] [PubMed] [Google Scholar]
- 38.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 39.Blagosklonny MV, Prabhu NS, El-Deiry WS. Defects in p21WAF1/CIP1, Rb, and c-myc signaling in phorbol ester-resistant cancer cells. Cancer Res. 1997;57:320–325. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.