Abstract
The mammalian gastrointestinal (GI) tract is colonized by a complex consortium of bacterial species. Bacteria engage in chemical signaling to coordinate population-wide behavior. However, it is unclear if chemical sensing plays a role in establishing mammalian host–bacterial commensal relationships. Enterohemorrhagic Escherichia coli (EHEC) is a deadly human pathogen but is a member of the GI flora in cattle, its main reservoir. EHEC harbors SdiA, a regulator that senses acyl-homoserine lactones (AHLs) produced by other bacteria. Here, we show that SdiA is necessary for EHEC colonization of cattle and that AHLs are prominent within the bovine rumen but absent in other areas of the GI tract. We also assessed the rumen metagenome of heifers, and we show that it is dominated by Clostridia and/or Bacilli but also harbors Bacteroidetes. Of note, some members of the Bacteroidetes phyla have been previously reported to produce AHLs. SdiA-AHL chemical signaling aids EHEC in gauging these GI environments, and promotes adaptation to a commensal lifestyle. We show that chemical sensing in the mammalian GI tract determines the niche specificity for colonization by a commensal bacterium of its natural animal reservoir. Chemical sensing may be a general mechanism used by commensal bacteria to sense and adapt to their mammalian hosts. Additionally, because EHEC is largely prevalent in cattle herds, interference with SdiA-mediated cattle colonization is an exciting alternative to diminish contamination of meat products and cross-contamination of produce crops because of cattle shedding of this human pathogen.
Keywords: bovine, enterohemorrhagic Escherichia coli, metagenomics, rumen
Bacteria thrive in complex multispecies communities within the gastrointestinal (GI) tracts of mammals (1). Mammals and bacteria have amicable and detrimental interactions, and enterohemorrhagic Escherichia coli (EHEC) can behave as a commensal or a pathogen depending on its host. EHEC is a commensal in the GI tract of cattle, its main reservoir, but is a human pathogen (2). EHEC causes bloody diarrhea, and it colonizes the large intestine of humans to form attaching and effacing (AE) lesions that are thought to be largely responsible for promoting disease (2). The genes for AE lesion formation are encoded within the locus of enterocyte effacement (LEE) (2). The LEE and AE lesion formations are also necessary for EHEC colonization of the recto-anal junction (RAJ) of cattle, facilitating its shedding to the environment (3–5). Whereas AE lesion formation leads to disease in humans, it is innocuous in adult cattle. The site of AE lesion formation in the GI tract may be responsible for the different outcomes. AE lesions occur in the large intestine in humans, leading to diarrheal disease, whereas AE lesion formation in the RAJ does not compromise the electrolyte balance in the bovine GI tract (2, 6).
To adapt to different hosts and environments, EHEC exploits chemical signaling (7). Quorum sensing (QS) is a signaling mechanism that allows bacteria to respond to chemicals by altering gene expression. EHEC uses several QS systems for intercellular signaling (7), including the autoinducer-3 (AI-3)/epinephrine/norepinephrine (8) system and the LuxR homolog SdiA that senses acyl-homoserine lactones (AHLs) (9–12). AHLs have a conserved homoserine lactone connected to a variable acyl chain. Different acyl chains ensure that different AHLs will be recognized by different LuxR-type proteins. LuxR-type proteins are transcription factors that regulate transcription of their target genes when binding to AHL. AHL binding to most of these proteins stabilizes them; otherwise, in the absence of signal, they are targeted to degradation (13–16). Congruent with AHLs being used as folding switches by these proteins, the NMR structure of SdiA shows that SdiA requires binding of this signal for folding and function (16). EHEC encodes SdiA but does not contain a luxI (AHL synthase) gene, and it does not produce AHLs (11, 12, 17). The LuxR signaling system has been primarily associated with intraspecies signaling, but there are examples of LuxR-type proteins (such as SdiA) that are primarily involved in interspecies signaling (9–12). Although the AI-3/epinephrine/norepinephrine QS system is important to activate EHEC virulence in animal models of pathogenesis (18), the role of the SdiA system in EHEC–host associations has not been established.
Here, we report that SdiA-AHL signaling regulates expression of EHEC genes known to facilitate the commensal EHEC colonization of cattle. We show that AHLs are present within the bovine rumen but absent in other areas of the GI tract and that rumen AHLs through SdiA modulate expression of these EHEC genes.
Results and Discussion
SdiA-AHL Represses Transcription of the LEE Genes.
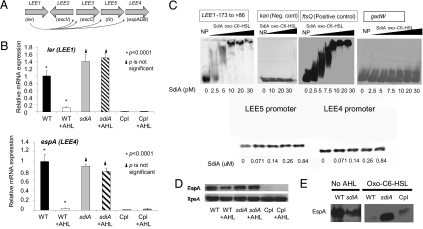
Transcriptome studies were conducted to investigate SdiA-AHL signaling in EHEC. SdiA-AHL signaling altered the expression of 49 genes. Within these 49 genes, the LEE and glutamate decarboxylase (gad) acid-resistance system are present. Both the LEE and gad system have been shown to be essential for EHEC colonization of cattle (4, 19, 20). There was no difference in transcription of the LEE genes between wild type (WT) and ΔsdiA in the absence of AHLs (EHEC produces no endogenous AHLs). However, when oxo-C6-homoserine lactone was added, transcription of the LEE genes was decreased in the WT strain but not in ΔsdiA (Fig. 1B and Fig. S1). These results suggested that AHLs repress transcription of the LEE genes and that this repression is mediated through SdiA. These results were also consistent with the role of AHLs in stabilizing the SdiA protein, given that, like TraR, SdiA will misfold and be targeted for degradation in the absence of AHLs (16). Transcription of the LEE genes was repressed in the complemented strain both in the presence and absence of AHLs (Fig. 1B), suggesting that enough SdiA is produced to overcome the lack of the AHL through even mild plasmid overexpression (pACYC177 is a low copy-number vector). Of note, these results are consistent with a report that expression of SdiA from a high copy-number plasmid in EHEC caused reduced expression of the LEE genes (21). However, no sdiA mutant was constructed and tested in this previous study (21). SdiA repressed transcription of the LEE genes by directly repressing transcription of LEE1 (Fig. 1C). LEE1 encodes the LEE-encoded regulator (Ler), which is required for the expression of all LEE genes (22).
Fig. 1.
SdiA-mediated AHL regulation of the EHEC LEE genes. (A) Schematic depiction of the LEE pathogenicity island. (B) Quantitative RT-PCR (qRT-PCR) of the LEE genes (ler/LEE1 and espA/LEE4) in WT EHEC (86-24), isogenic sdiA mutant, and complemented (Cpl) strain (ΔsdiA with pDH6; sdiA cloned in pACYC177) grown in DMEM to an OD600 of 1.0 in the absence and presence of AHL (10 μM oxo-C6-homoserine lactone). (C) EMSAs of LEE1, LEE4, LEE5, gadW, ftsQ (positive control), and kanamycin-resistant gene, kan promoter, (negative control) with SdiA-AHL. (D) Western blots of whole-cell lysates of WT, sdiA mutant, and sdiA mutant complemented with pDH6 probed with antisera against EspA and RpoA (loading control). (E) Western blots of the secreted proteins of WT, sdiA mutant, and sdiA mutant complemented with pDH6 strains in the absence and presence of AHL (10 μM oxo-C6-HSL) probed with an antiserum against EspA. In conditions where no AHLs were added, the same amount of the ethyl-acetate solvent was added to ensure that the solvent had no effect in gene expression.
The LEE encodes for a type III secretion system (TTSS), a needle-like structure used to inject bacterial effectors into host cells, as well as the translocon of this system comprised of the EspA, EspB, and EspD proteins, which are themselves secreted through this TTSS (Fig. 1A) (2). There was no difference in the expression of EspA between the WT and ΔsdiA in the absence of AHLs (Fig. 1D). However, expression of EspA was reduced in the WT in the presence of AHL and decreased in the complemented strain in both the presence and absence of signal (Fig. 1D). These results were consistent with the SdiA-AHL repression of LEE transcription (Fig. 1B). Type III secretion of EspA on WT and ΔsdiA in the absence of AHLs was similar; however, type III secretion in the presence of AHL was strikingly reduced in WT and complemented strains but not in the sdiA mutant (Fig. 1E). SdiA-AHL controls the expression of all LEE genes, including the genes encoding the TTSS. Hence, one would expect a more striking phenotype on type III secretion of EspA, where one observes defects in secretion coupled to lesser expression of espA.
SdiA-AHL Activates Transcription of the gad Genes.
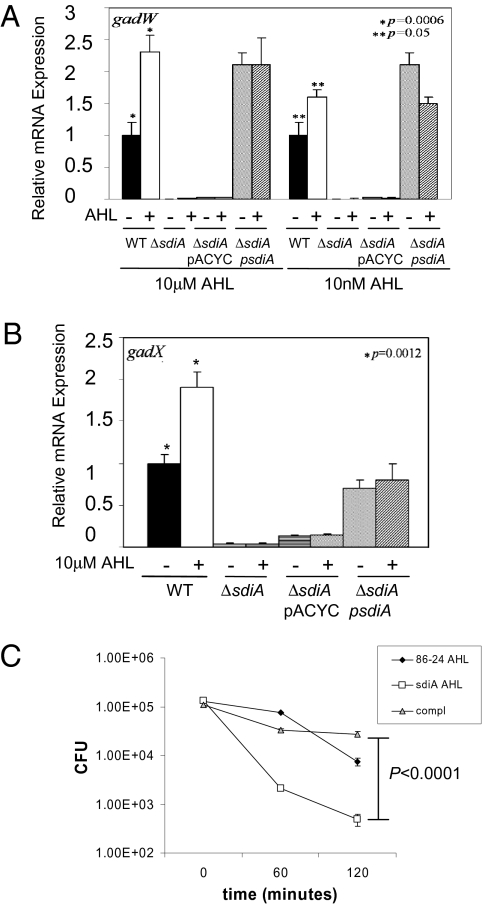
Congruent with the transcriptome studies, AHLs activated expression of the gad acid-resistance genes (Fig. 2 A and B). EHEC has other acid-resistance systems, but only the gad system is necessary for EHEC survival within the acidic stomachs of the cow (19). Of note, the arginine acid-resistance system (adi) was not regulated by SdiA or the addition of AHLs (Fig. S2). Expression of the gad genes was significantly enhanced by AHLs (Fig. 2 A and B). However, in contrast with the regulation of the LEE genes, the expression of the gad genes was dramatically decreased in ΔsdiA, even in the absence of AHLs (Fig. 2 A and B). The observation that the gad system can be activated by SdiA even in the absence of AHLs is consistent with a previous report from Dyszel et al. (9). A potential explanation is that within the bacterial cell, SdiA may have a half-life in the absence of AHLs that is sufficient to exert an activation role in the transcription of the gad genes. SdiA alone did not directly activate transcription of the gad system (Fig. 1C), suggesting that SdiA-AHL either requires a second factor to directly activate transcription of gad or activates expression of an unidentified transcription factor that directly activates transcription of the gad system. Consequently, the sdiA mutant was less resistant to acidic environments than WT EHEC (Fig. 2C), and this diminished acid resistance was caused by the down-regulation of the gad system (Fig. 2 A and B).
Fig. 2.
SdiA regulation of the gad acid-resistance system. (A) qRT-PCR of the gadW gene encoding the master regulator of the gad system in WT EHEC (86-24), sdiA mutant, sdiA mutant with pACYC177, and sdiA mutant complemented with pDH6 (sdiA in pACYC177) strains grown in DMEM to an OD600 of 1.0 in the absence and presence of AHL (10 μM oxo-C8-HSL and 10 nM oxo-C8-HSL). (B) qRT-PCR of the gadX gene in WT EHEC (86-24), sdiA mutant, sdiA mutant with pACYC177, and sdiA mutant complemented with pDH6 (sdiA in pACYC177) strains grown in DMEM to an OD600 of 1.0 in the absence and presence of AHL (10 μM oxo-C8-HSL). (C) Acid resistance (survival in acidic pH) through the gad system in the absence and presence of 10 μM oxo-C8-HSL. In conditions where no AHLs were added, the same amount of the ethyl-acetate solvent was added to ensure that the solvent had no effect in gene expression.
AHLs Are Present in the Bovine Rumen.
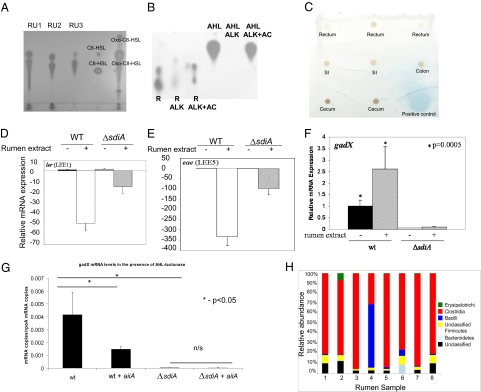
Because SdiA is active in the presence of AHLs (16) and previous reports of AHLs state that they are present within the cattle rumen (23, 24), the presence of AHLs in rumen fluid was assessed. AHLs were extracted from the rumen fluid of cannulated cattle and tested with an Agrobacterium tumefaciens (25) reporter strain for their presence. This strain has the traI gene (activated by TraR-AHL) fused to lacZ that, in the presence of AHLs and its substrate, converts to blue on a reverse-phase TLC overlay assay. This detection strain was chosen, because it detects a wide range of AHLs (25). AHLs were detected in all rumen extracts (Fig. 3A), further showing that AHLs are indeed present within the rumen of cattle. Alkalinization (alkaline pH hydrolyzes the homoserine lactone ring of AHLs and inactivates these signals) caused loss of activity of the rumen-extracted AHLs, confirming that these signals were AHLs (Fig. 3B). The activity of these signals was restored in the rumen sample on acidification of this reaction (Fig. 3B), which allows for reformation of the lactone ring. Although AHLs were detected in all rumen extracts (Fig. 3A), these signals were not detected in other portions of the ruminant GI tract, suggesting that these signals are restricted to the rumen (Fig. 3C and Fig. S3). These results are consistent with the chemistry of AHLs. The homoserine ring of these signals is hydrolyzed at alkaline pH (26), which is the pH in the intestine; thus, it inactivates these signals. These signals, however, are stable in acidic pH, such as the pH in the rumen (the average rumen pH is 5.98 in animals on a grain diet) (Tables S1–S3).
Fig. 3.
Rumen AHLs affect expression of EHEC genes. (A) TLC from AHLs extracted from 50 mL (evaporated to 5 μL that were added to the TLC) of rumen fluid (RU) collected from three different cows (RU1, RU2, and RU3). C6-HSL (402 pmol), C8-HSL (15.8 pmol), oxo-C6-HSL (400 pmol), and oxo-C8-HSL (15 pmol) were used as controls. (B) TLC from AHLs extracted from 50 mL (evaporated to 5 μL that were added to the TLC) of rumen fluid (R) that has been subjected to alkaline treatment (ALK; hydrolyses the homoserine lactone of AHLs) and acidification (AC; ALK+AC restores the homoserine lactone); oxo-C8-HSL (AHL) undergoing the same treatments was used as a control. (C) Preparative TLC from AHLs extracted from 50 mL (evaporated to 5 μL that were added to the TLC) of other portions of the GI of ruminants. The positive control turns blue. (D) qRT-PCR of ler (LEE1) from WT and sdiA mutant in the presence of rumen AHLs (5 μL extract). In conditions where no rumen extracts were added, the same amount of the dichloromethane solvent was added to ensure that the solvent had no effect in gene expression. (E) qRT-PCR of eae (LEE5) gene from WT and sdiA mutant in the presence of rumen AHLs (5 μL extract). (F) qRT-PCR of gadX from WT and sdiA mutant in the presence of rumen AHLs (5 μL extract). (G) qRT-PCR of gadX from WT and sdiA mutant containing either empty vector or the aiiA gene from B. cereus (encodes a lactonase that inactivates AHLs) cloned into pBADMYcHisA grown in filtered, nonconcentrated rumen fluid in the presence of arabinose. The presence of AHLs in this rumen fluid was previously confirmed (Fig. S3). (H) Graph depicting the bacterial composition of the rumen of eight heifers on a grain diet.
Rumen AHLs Modulate Transcription of EHEC Genes.
Given that AHLs repressed expression of the LEE, activated expression of the gad genes (Figs. 1 and 2) and that the rumen harbors AHLs (Fig. 3A), we then tested if AHLs extracted from the rumen could mimic the effect of purified AHLs on transcription. The AHLs extracted from the rumen repressed transcription of the LEE (Fig. 3 D and E) and activated expression of gadX (Fig. 3F). However, not all rumen AHL effects on LEE transcription occurred through SdiA (Fig. 3 D and E). There are two potential explanations for this observation: (i) the presence of another receptor for AHLs in EHEC or (ii) another(s) molecule(s) is responsible for this further repression. Indeed, de Sablet et al. (27) showed that, in gnotobiotic rats colonized with human microbiota, these bacteria produce chemical signals that repress expression of the genes encoding Shiga toxin in EHEC in a SdiA-independent manner. These studies highlight that complex microbial communities may produce different combinations of signals. Conversely, gadX activation by rumen-extracted AHLs was completely abrogated in the sdiA mutant (Fig. 3F).
Because one cannot directly assess the levels of AHLs within the rumen fluid, one potential caveat of the experiments using rumen extracts could be that these signals in the extracts are concentrated to levels that are not physiologically relevant. To address this issue, the aiiA gene from Bacillus cereus, which encode for a lactonase that specifically hydrolyses the lactone ring from AHLs (28), was cloned into WT EHEC and the sdiA mutant. Expression of gadX in WT grown in filtered, nonconcentrated, rumen fluid was decreased in the presence of the aiiA-encoded lactonase (which inactivates any AHLs present in the rumen fluid) (Fig. 3G), whereas expression of gadX in the sdiA mutant did not change with or without this lactonase (Fig. 3G). These data support that AHLs at physiological levels in the rumen fluid activate transcription of gadX and that this activation is SdiA-dependent.
Rumen Metagenomic Studies.
The AHLs in the rumen are probably synthesized by the microbial flora that reside in this GI compartment. However, there is limited information on the composition of the rumen microbial flora, and previous metagenomic studies were conducted only in forage- (grass-legume hay) fed steers (29). To further assess the composition of the rumen flora, we performed metagenomic studies on DNA extracted from the ruminant contents of eight ruminally cannulated heifers on a grain diet. Forage is the usual diet of cattle on farms, whereas grain is the preferred diet on feedlots before they are sent to the abattoir.
The general sequence data obtained from this study are summarized in Table S4. More than 40,000 sequences with acceptable coverage and quality were included in further analysis with a mean of >100 per rumen sample and a range from 4,534 to 5,887 reads. Using a 97% similarity threshold value, we identified a wide range of operational taxonomic units (OTUs) for each sample. The rumen sample group displayed significant variability among the number of OTUs that could be identified (640 in sample 8 and 2,576 in sample 1), suggesting that there is variability between the rumen contents of each of the animals. This finding is similar to the level of variability observed in the human gastrointestinal tract (1). The calculations on the species richness using ACE and Chao (30, 31) and diversity estimators, Shannon and Weaver (32), indicated that there is significant variation between each of eight rumen samples (Tables S4–S6).
We also examined the most abundant phyla in the rumen microbiota. In seven of eight samples, the Clostridia class was dominant in the rumen microbiota, similar to the findings in the rumen metagenomic studies of steers on a forage diet (29). However, in sample 4, the Bacilli were dominant with Clostridia, the second most prevalent class. Other phyla encountered in these rumen samples include Erysipelotrichi and Bacteroidetes. Of note, members of the Bacteroidetes phyla have been previously reported to produce AHLs (33) (Fig. 3H). The analysis on this phylogenetic level shows that the rumen constituents are stable among these eight samples.
The variability in the number and quantity of OTU that are observed in the rumen samples is similar to the level of variability observed in the human GI tract (1). Not surprisingly, the bacterial community that is present is different. The microbiota of the human colonic GI tract has been characterized by the examination of the fecal material and is dominated by Bacteroidetes and Firmicutes, whereas the bovine rumen samples are dominated by Clostria or Bacilli, both within the Firmicutes (Fig. 3H). This difference in the microbiota is most likely linked to the difference in environmental conditions (pH, anaerobiosis, etc.) and tissue type. However, both bovine rumen and human colonic samples show sample-to-sample variation. Additionally, although this study represented the deep sequencing of bovine rumen samples, we still have not explored the rare microbiome. Further deep sequencing of these environments will provide insight into the population structure in these animals and may provide a mechanism of preventing or interrupting the carriage of these important human pathogens.
The current analysis does not allow the determination of the species of the members of the microbiota. Although we do not know which members of the rumen microbiota are producing AHLs, there has been one species of Bacteroidetes associated with fish that was found to produce AHLs (33). Future studies of these complex communities and their interactions will further illuminate the role of chemical signaling within the rumen.
sdiA Mutant Is Defective for Colonization of Cattle.
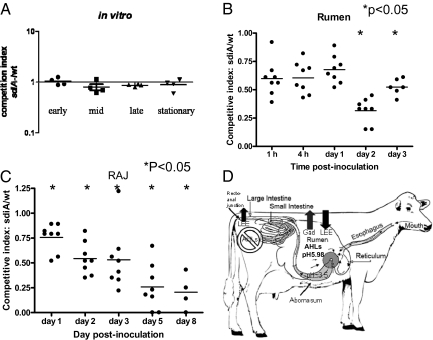
Inasmuch as rumen AHLs in a SdiA-dependent fashion modulate expression of EHEC genes necessary for cattle colonization, we assessed the contribution of SdiA to EHEC survival within the rumen and subsequent colonization of the RAJ. For this purpose, a competition trial was done in ruminally cannulated heifers on a grain diet (we used the same eight animals in which the rumen pH was assessed and the metagenomic studies were performed). Competition studies were performed to avoid issues concerning individual variation between different heifers in scoring the ability of these strains to establish themselves in the GI tract. As an initial control, an in vitro competition experiment between the WT and the sdiA mutant was performed to ensure that there were no growth defects in the sdiA mutant (Fig. 4A). The ratio of WT to mutant bacteria was determined in the inoculum, rumen, and RAJ mucosal swab (RAMS) samples. The competitive index for each day was determined by dividing the ratio of mutant to WT bacteria in the rumen and RAJ. Fig. 4 B and C shows that WT EHEC outcompeted the sdiA mutant in both the rumen and RAJ of cattle, confirming that SdiA is important for cattle colonization (further details on these infection studies can be found in SI Results and Discussion).
Fig. 4.
Competition of WT and sdiA mutant in the bovine rumen and RAJ. Eight 1.5-year-old fully ruminant and ruminally cannulated Charolais heifers were inoculated with equal cfus of WT and sdiA mutant. (A) As a control, equal cfus of WT and sdiA were grown in coculture in vitro, and their competitive indexes were determined throughout growth (early, mid, late, and stationary phases of growth). A competitive index of 1 means no difference, a competitive index <1 means that WT was in higher numbers than the sdiA mutant, and a competitive index >1 means that the mutant was in higher numbers than WT. The ratio of WT to mutant bacteria was determined in the (B) rumen and (C) RAJ. (D) Schematic model of SdiA-AHL–dependent EHEC gene expression in the bovine GI tract.
This study shows that AHLs present in the rumen down-regulate expression of the LEE genes whose expression in this GI compartment would constitute a superfluous expenditure of energy (Fig. 3 D and E). However, expression of the LEE is necessary for colonization of the RAJ (20), where AHLs are absent (Fig. 3C), allowing for LEE expression and efficient AE lesion formation. Conversely, rumen AHLs activate expression of the gad acid-resistant system (Fig. 3 F and G) necessary for survival within the bovine rumen and the subsequent acidic stomachs (Fig. 4D).
Conclusions
In this study, we show that the rumen of cattle harbor AHLs and that these chemical signals can be sensed in part through SdiA to modulate gene expression in EHEC, leading to successful colonization of these animals. However, EHEC also uses other chemical signaling systems to modulate expression of its virulence and colonization genes (8, 34). In contrast to the SdiA-AHL system, the AI-3/epinephrine/norepinephrine system activates virulence traits in EHEC, and in animal infection models thought to mimic pathogenic interactions, such as the rabbit infant model (18), mutants that cannot sense these chemicals are attenuated for disease. QS contribution to the pathogenic and/or commensal lifestyle of EHEC may constitute an example of yin-and-yang relationship in chemical signaling in bacteria.
EHEC is carried by an estimated 70–80% of the cattle herds in the United States, and thus, interference with AHL signaling in cattle colonization may engender a strategic alternative to diminish EHEC shedding and consequently, human disease. Chemical sensing may be an important mechanism used by commensal bacteria to sense and adapt to specific niches in a complex host environment.
Materials and Methods
Detailed materials and methods describing microbiological examination, molecular biology techniques, AHL extraction from rumen samples, microarray and metagenomic studies, and animal infection studies can be found in SI Materials and Methods. Detailed strains and plasmid constructions can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank J. Kaper for anti-EspA antisera. We also thank S. McKnight, L. Hooper, M. V. Norgard, and E. Olsen for reading the manuscript. The contents of this work are solely the responsibility of the authors and do not represent the official views of the National Institutes of Health National Institute of Allergy and Infectious Diseases. This work was supported by National Institutes of Health Grant AI053067, the Burroughs Wellcome Fund, and the National Cattlemen’s Beef Association. D.T.H. was supported through National Institutes of Health Training Grant 5-T32-AI007520-07. We would like to thank Lonie Austin for cattle care and handling. This work was supported, in part, by the Idaho Agriculture Experiment Station and PHS grant P20-RR16454 (to C.J.H.) from the NIH NCRR.
Footnotes
The authors declare no conflict of interest.
Data deposition: This sequence reported in this paper has been deposited in the National Center of Biotechnology Information Gene Expression Omnibus database (accession no. GAE13562).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1002551107/-/DCSupplemental.
References
- 1.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 3.Sheng H, Lim JY, Watkins MK, Minnich SA, Hovde CJ. Characterization of an Escherichia coli O157:H7 O-antigen deletion mutant and effect of the deletion on bacterial persistence in the mouse intestine and colonization at the bovine terminal rectal mucosa. Appl Environ Microbiol. 2008;74:5015–5022. doi: 10.1128/AEM.00743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheng H, Lim JY, Knecht HJ, Li J, Hovde CJ. Role of Escherichia coli O157:H7 virulence factors in colonization at the bovine terminal rectal mucosa. Infect Immun. 2006;74:4685–4693. doi: 10.1128/IAI.00406-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naylor SW, et al. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect Immun. 2003;71:1505–1512. doi: 10.1128/IAI.71.3.1505-1512.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naylor SW, et al. Impact of the direct application of therapeutic agents to the terminal recta of experimentally colonized calves on Escherichia coli O157:H7 shedding. Appl Environ Microbiol. 2007;73:1493–1500. doi: 10.1128/AEM.01736-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes DT, Sperandio V. Inter-kingdom signalling: Communication between bacteria and their hosts. Nat Rev Microbiol. 2008;6:111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria-host communication: The language of hormones. Proc Natl Acad Sci USA. 2003;100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyszel JL, et al. Salmonella enterica serovar Typhimurium can detect acyl homoserine lactone production by Yersinia enterocolitica in mice. J Bacteriol. 2010;192:29–37. doi: 10.1128/JB.01139-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith JN, et al. SdiA, an N-acylhomoserine lactone receptor, becomes active during the transit of Salmonella enterica through the gastrointestinal tract of turtles. PLoS ONE. 2008;3:e2826. doi: 10.1371/journal.pone.0002826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmer BM. Cell-to-cell signalling in Escherichia coli and Salmonella enterica. Mol Microbiol. 2004;52:933–945. doi: 10.1111/j.1365-2958.2004.04054.x. [DOI] [PubMed] [Google Scholar]
- 12.Michael B, Smith JN, Swift S, Heffron F, Ahmer BM. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J Bacteriol. 2001;183:5733–5742. doi: 10.1128/JB.183.19.5733-5742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu J, Winans SC. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc Natl Acad Sci USA. 1999;96:4832–4837. doi: 10.1073/pnas.96.9.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu J, Winans SC. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc Natl Acad Sci USA. 2001;98:1507–1512. doi: 10.1073/pnas.98.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang RG, et al. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature. 2002;417:971–974. doi: 10.1038/nature00833. [DOI] [PubMed] [Google Scholar]
- 16.Yao Y, et al. Structure of the Escherichia coli quorum sensing protein SdiA: Activation of the folding switch by acyl homoserine lactones. J Mol Biol. 2006;355:262–273. doi: 10.1016/j.jmb.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 17.Huisman GW, Kolter R. Sensing starvation: A homoserine lactone—dependent signaling pathway in Escherichia coli. Science. 1994;265:537–539. doi: 10.1126/science.7545940. [DOI] [PubMed] [Google Scholar]
- 18.Rasko DA, et al. Targeting QseC signaling and virulence for antibiotic development. Science. 2008;321:1078–1080. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price SB, Wright JC, DeGraves FJ, Castanie-Cornet MP, Foster JW. Acid resistance systems required for survival of Escherichia coli O157:H7 in the bovine gastrointestinal tract and in apple cider are different. Appl Environ Microbiol. 2004;70:4792–4799. doi: 10.1128/AEM.70.8.4792-4799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dziva F, van Diemen PM, Stevens MP, Smith AJ, Wallis TS. Identification of Escherichia coli O157:H7 genes influencing colonization of the bovine gastrointestinal tract using signature-tagged mutagenesis. Microbiology. 2004;150:3631–3645. doi: 10.1099/mic.0.27448-0. [DOI] [PubMed] [Google Scholar]
- 21.Kanamaru K, Kanamaru K, Tatsuno I, Tobe T, Sasakawa C. SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol. 2000;38:805–816. doi: 10.1046/j.1365-2958.2000.02171.x. [DOI] [PubMed] [Google Scholar]
- 22.Mellies JL, Elliott SJ, Sperandio V, Donnenberg MS, Kaper JB. The Per regulon of enteropathogenic Escherichia coli: Identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler) Mol Microbiol. 1999;33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- 23.Erickson DL, et al. Evidence of quorum sensing in the rumen ecosystem: Detection of N-acyl homoserine lactone autoinducers in ruminal contents. Can J Microbiol. 2002;48:374–378. doi: 10.1139/w02-022. [DOI] [PubMed] [Google Scholar]
- 24.Edrington TS, et al. Acyl-homoserine-lactone autoinducer in the gastrointesinal tract of feedlot cattle and correlation to season, E. Coli O157:H7 prevalence, and diet. Curr Microbiol. 2008;58:227–232. doi: 10.1007/s00284-008-9312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuqua C, Winans SC. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J Bacteriol. 1996;178:435–440. doi: 10.1128/jb.178.2.435-440.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunny GM, Winans SC. Cell-Cell Signaling in Bacteria. Washington, DC: American Society for Microbiology; 1999. [Google Scholar]
- 27.de Sablet T, et al. Human microbiota-secreted factors inhibit shiga toxin synthesis by enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 2009;77:783–790. doi: 10.1128/IAI.01048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong YH, Xu JL, Li XZ, Zhang LH. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc Natl Acad Sci USA. 2000;97:3526–3531. doi: 10.1073/pnas.060023897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brulc JM, et al. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci USA. 2009;106:1948–1953. doi: 10.1073/pnas.0806191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chao A. Non-parametric estimation of the number of classes in a population. Scand J Stat. 1984; 11:265–270. [Google Scholar]
- 31.Chao A. Estimating the population size for capture-recapture data with unequal catchability. Biometrics. 1987;43:783–791. [PubMed] [Google Scholar]
- 32.Shannon CE, Weaver W. The Mathematical Theory of Communication. Urbana, IL: University of Illinois Press; 1949. [Google Scholar]
- 33.Romero M, Avendano-Herrera R, Magarinos B, Camara M, Otero A. Acylhomoserine lactone production and degradation by the fish pathogen Tenacibaculum maritimum, a member of the Cytophaga-Flavobacterium-Bacteroides (CFB) group. FEMS Microbiol Lett. 2010;304:131–139. doi: 10.1111/j.1574-6968.2009.01889.x. [DOI] [PubMed] [Google Scholar]
- 34.Rasko DA, et al. Targeting QseC signaling and virulence for antibiotic development. Science. 2008;321:1078–1080. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.