Abstract
The enzyme UDP-GlcNAc:α3-D-mannoside β1,2-N-acetylglucosaminyltransferase I (GnT1, encoded by Mgat1) controls the synthesis of paucimannose N-glycans in Drosophila. We have previously reported that null mutations in Drosophila Mgat1 are viable but exhibit defects in locomotion, brain abnormalities, and a severely reduced life span. Here, we show that knockdown of Mgat1 in the central nervous system (CNS) of wild-type flies decreases locomotor activity and life span. This phenotype is similar to that observed in Drosophila Mgat11 null mutants, demonstrating that Mgat1 is required in the CNS. We also found that neuronal expression of a wild-type Mgat1 transgene rescued the shortened life span of Mgat11 null mutants and resulted in a dramatic 135% increase in mean life span relative to genetically identical controls. Neuronal expression of a wild-type Mgat1 transgene in wild-type flies resulted in a modest 9% increase in mean life span relative to genetically identical controls. In both Mgat11 null mutants and wild-type flies, neuronal expression of wild-type Mgat1 transgene resulted in a significant increase in GnT1 activity and resistance to oxidative stress. Whereas dietary restriction is not absolutely essential for the increased life span, it plays a role in the process. Interestingly, we observe a direct correlation between GnT1 activity and mean life span up to a maximum of ~136 days, showing that the ability of GnT1 activity to increase life span is limited. Altogether, these observations suggest that Mgat1-dependent N-glycosylation plays an important role in the control of Drosophila life span.
Keywords: locomotion, longevity, N-glycosylation, N-acetylglucosaminyltransferase, GnT1
N-glycans are large sugar molecules attached to proteins by a GlcNAcβ1-N-Asn linkage. This type of protein N-glycosylation is an ancient pathway present in both protozoan and metazoan eukaryotes (1). The enzyme UDP-GlcNAc:α3-D-mannoside β1,2-N-acetylglucosaminyltransferase I (GnT1 encoded by Mgat1) appeared at about the same time as metazoa and catalyzes the first essential step in the synthesis of paucimannose N-glycans (2, 3). These glycans are major components in Drosophila (4, 5) and other invertebrates but not in vertebrates. Although deletion of Mgat1 does not affect the viability of metazoan cells in culture, it has severe effects on animal development. For example, mice that are homozygous null for Mgat1 die at embryonic days 9.5–10.5 (6, 7). Caenorhabditis elegans Mgat1 mutants are viable but exhibit abnormal responses to bacterial infection (8). Similarly, Drosophila Mgat11 null mutants are also viable but exhibit pronounced locomotor defects, abnormal brain structures, and a severely reduced life span (5). The precise mechanisms by which mutations in Mgat1 affect these various processes remain unclear. Here we show that knockdown of Mgat1 in the central nervous system (CNS) can recapitulate the null mutant phenotype, demonstrating that Mgat1 is required in the CNS. We also show that neuronal expression of a wild-type Mgat1 transgene increases the mean and maximum life span of Mgat11 null mutants by 135% relative to genetically identical animals that do not express the transgene. Finally, we show that neuronal expression of a wild-type Mgat1 transgene in wild-type flies results in significant increases in GnT1 activity and resistance to oxidative stress and a modest increase in life span relative to genetically identical controls. These observations suggest that Mgat1-dependent N-glycosylation plays an important role in the control of Drosophila life span.
Results
Knockdown of Mgat1 in the CNS of Wild-Type Flies Gives Rise to Defects in Locomotion and Reduced Longevity.
Drosophila Mgat11 null mutants exhibit defects in locomotion, abnormal brain structures, and a severely reduced life span (5). To determine if the defects observed in Drosophila Mgat1 mutants are due to a specific requirement for Mgat1 in the CNS we performed tissue-specific knockdown experiments using the conditional pan-neuronal ELAV-GeneSwitch line to drive expression of a UAS-Mgat1-RNAi transgene. Briefly, the GeneSwitch system is based on the GAL4/UAS system (9, 10) and employs a hormone-inducible tissue-specific GAL4 driver (11, 12) that allows for expression of any transgene under the transcriptional control of the GAL4 upstream activating sequence (UAS). The major advantage of GeneSwitch over the traditional GAL4/UAS method is that it also allows for the comparison of genetically identical animals from one cohort, whereby one group receives a hormone (RU486) to activate the UAS transgene whereas the other does not. Addition of RU486 to flies carrying the ELAV-GeneSwitch and various other transgenes has no effect on life span (13–16).
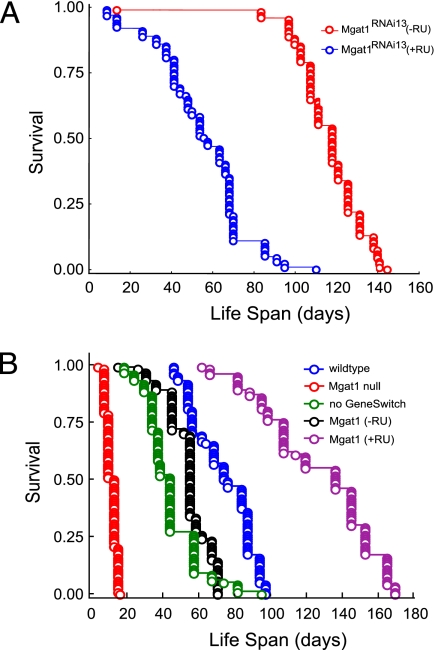
Two distinct Drosophila lines (Mgat1RNAi13 and Mgat1RNAi18) were maintained both in the presence (Table 1, lines B and D) and in the absence (Table 1, lines A and C) of RU486 to induce expression of the RNAi transgene. We found that knockdown of Mgat1 in the CNS of both lines resulted in RU486-dependent reductions of GnT1 activity in both heads and whole bodies of 41–56% (Table 1, lines B and D), confirming effective knockdown. We also found concomitant RU486-dependent reductions of locomotion (28%, 70%) and of mean and maximum life spans (17–53%) (Table 1, lines A–D; Fig. 1A). These observations are consistent with previous data on the effects of a null mutation of Mgat1 (compare Table 1, lines E and I). Because heterozygous Mgat11 null mutants also have ~50% reduction in GnT1 enzyme activity (5) but do not display any defects in locomotion or life span, the data suggest that GnT1 activity in the CNS must be reduced by >50% to cause abnormal locomotion and life span.
Table 1.
Properties of Drosophila lines used in this study
| GnT1 (pmol/h/mg), mean/SE (%Δ) |
Life span (days) |
|||||
| Genotype | RU486 | Body | Head | Locomotion, mean/SE | Mean/SE (%Δ) | Max (%Δ) |
| RNAi knockdown | ||||||
| A ELAV-GeneSwitch;UAS-Mgat1RNAi13 | − | 184/8.5 | 228/18 | 60/7.5 | 116/1.8 | 138 |
| B ELAV-GeneSwitch;UAS-Mgat1RNAi13 | + | 92/16 (−50) | 135/18 (−41) | 18/5.0 | 55.1/2.1 (−53) | 85 (−38) |
| C ELAV-GeneSwitch;UAS-Mgat1RNAi18 | − | 180/20 | 130/6.5 | 75/15 | 91.9/2.9 | 126 |
| D ELAV-GeneSwitch;UAS-Mgat1RNAi18 | + | 88/8.5 (−51) | 57/4.5 (−56) | 54/14 | 74.1/2.6 (−20) | 105 (−17) |
| Mgat1-null series | ||||||
| E w1118;Mgat11/Mgat11 (null mutant) | − | 0 | 0 | 1.4/0.04 | 10.7/0.3 | 15 |
| F w1118/UAS-Mgat1;Mgat11/Mgat11;+/ELAV-GeneSwitch | − | 24/2 | 143/20 | 34.7/12 | 54.1/1.2 | 70 |
| G w1118/UAS-Mgat1;Mgat11/Mgat11;+/ELAV-GeneSwitch | + | 118/3 (+392) | 202/6.5 (+41) | 39.7/8.5 | 127/3.0 (+135) | 165 (+136) |
| H w1118/UAS-Mgat1;Mgat11/Mgat11 (no GeneSwitch) | − | 61/16 | 71/15 | 12/7.0 | 43.8/1.5 | 57 |
| Wild-type series | ||||||
| I w1118;Mgat+9/Mgat+9 (wild type control) | − | 148/21 | 249/39 | 60/7.0 | 73/1.6 | 93 |
| J w1118/UAS-Mgat1;Mgat+9/Mgat+9;+/ELAV-GeneSwitch | − | 177/29 | nd | nd | 125/1.4 | 142 |
| K w1118/UAS-Mgat1;Mgat+9/Mgat+9;+/ELAV-GeneSwitch | + | 257/49 (+45) | nd | nd | 136/1.6 (+9) | 160 (+13) |
| L w1118/UAS-Mgat1;Mgat+9/Mgat+9 (no GeneSwitch) | − | nd | nd | nd | 103/1.4 | 118 |
| M w1118;Mgat+9/Mgat+9;+/ELAV-GeneSwitch (no transgene) | − | nd | nd | nd | 84.5/1.4 | 103 |
Targeted expression of Mgat1 RNAi and wild-type Mgat1 transgenes in the CNS is shown. The conditional ELAV-GeneSwitch line was used to knock down or overexpress Mgat1 in the CNS of Mgat11 null mutant and wild-type flies. Transgene expression from ELAV-GeneSwitch was induced in the presence of the drug RU486. GnT1 enzyme activity (5) was measured in duplicate in two independent experiments from whole-body extracts or isolated adult heads. The locomotory activity of adult male and female flies was measured using a modified open field test (32). Individual flies from each genotype were placed in a covered Petri dish (15 mm) and allowed to adapt to their environment for 5 min. The number of times the fly crossed four lines drawn on the Petri dish over a period of 3 min was measured. Twenty-five flies of each genotype were used for each experiment. Life span determinations of flies were performed as previously described (5). The mean and maximum (90%) life span for each genotype is shown. Significant differences between each of these groups (P < 0.001) were determined using the Cox–Mantel log-rank test. %Δ refers to the percentage of differences in GnT1 activities and mean and maximum life spans between “with and without RU486” pairs with the same genotype (A/B, C/D, F/G, and J/K). SE represents standard error. nd, not done. Two independent UAS-RNAi lines and one UAS-Mgat1 transgenic line were analyzed.
Fig. 1.
Targeted gene expression. (A) Targeted RNAi knockdown of Mgat1 in the wild-type CNS affects Drosophila life span. The conditional ELAV-GeneSwitch line was used to knock down Mgat1 in the CNS of wild-type flies. Transgene expression from ELAV-GeneSwitch was induced in the presence of the drug RU486 to down-regulate an Mgat1 RNAi line (no. 13) and life span was measured as previously described (5). The complete genotype and life span of the line are indicated in Table 1. (B) Targeted expression of Mgat1 in the CNS of Mgat11 null Drosophila affects life span. Mgat11 null mutants have a maximum life span of 15 days whereas wild-type flies live to 93 days. Targeted expression of Mgat1 to the CNS of Mgat11 null mutants [Mgat1 (+RU)] increases the mean and maximum life spans by 73 and 77%, respectively, relative to wild-type flies and by 135% relative to Mgat11 null mutant flies that were not treated by drug [Mgat1 (−RU)]. All life span plots were derived from a single population of flies.
Neuronal Expression of Mgat1 in Mgat11 Null Flies Does Not Rescue Locomotion but Dramatically Extends Life Span.
We used the ELAV-GeneSwitch line to drive expression of a wild-type Mgat1 transgene in the CNS of Mgat11 null mutants to determine whether neuron-specific expression of Mgat1 is sufficient to rescue the locomotory defects and shortened life span of the mutants. Of note, we detect GnT1 activity in Mgat11 null mutant flies that contain the UAS-Mgat1 transgene in the absence of RU486 (Table 1, line F), suggesting that the transgene gives rise to nonspecific (“leaky”) expression of Mgat1 not only in neurons but also in other tissues. Transgene leakage independent of the GeneSwitch has been reported previously (13). The increased locomotory activity and life span due to leaky Mgat1 expression (compare Table 1, lines E and F) are probably due primarily to the increased GnT1 activity but may also be due, at least partly, to the different genotypes of the two fly lines. Addition of RU486 to Mgat11 null flies with the Mgat1 transgene and ELAV-GeneSwitch does not result in a significant increase in locomotory activity (compare Table 1, lines F and G) and the locomotory activities of these flies are significantly lower than those of wild-type flies (compare Table 1, lines G and I), suggesting that optimum locomotion requires GnT1 activity not only in the CNS but also in other tissues such as muscle. Targeted expression of a wild-type Mgat1 transgene in the CNS of Mgat11 null mutants increases life span and rescues the shortened life span of the mutants (compare Table 1, lines F and G; Fig. 1B). As in the case of the RNAi knockdown experiments, we observed a correlation between RU486-dependent changes in GnT1 activity (increases of 392 and 41% in whole bodies and heads, respectively) and mean and maximum life spans (increases of 135 and 136%) (Table 1, line G).
Increased Longevity in Rescued Mgat11 Null Mutant Flies Is Associated with Increased Resistance to Oxidative Stress and Is Influenced by Caloric Intake.
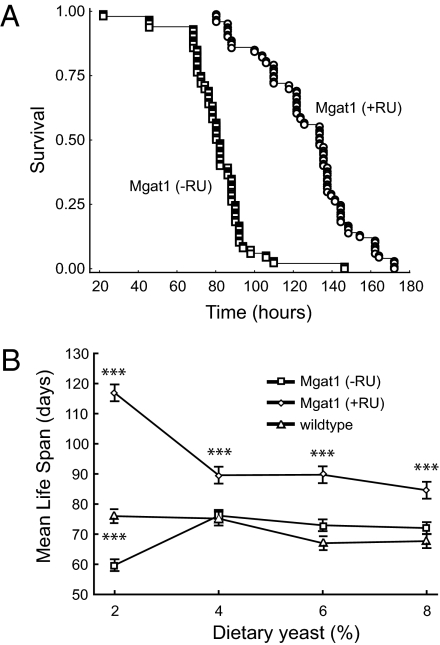
Increased longevity in organisms as diverse as worms, flies, and mice has been associated with increased resistance to oxidative stress. To determine if the increase in life span we observe in the Mgat1 rescued flies is also associated with resistance to oxidative stress, we examined the life span of the control (−RU486) and rescued flies (+RU486) in the presence of 3% hydrogen peroxide. Targeted expression of Mgat1 in the CNS of Mgat11 null mutants rendered them significantly more resistant to hydrogen peroxide than control flies (Fig. 2A). Specifically, the rescued flies had a mean life span of 127.9 ± 2.4 h compared with controls whose mean life span was 80.8 ± 1.7 h, demonstrating that the increase in longevity is also associated with an increase in oxidative stress resistance.
Fig. 2.
Life span studies on Mgat1 rescued Mgat11 null flies. (A) The effect of oxidative stress on the life span of Mgat1 rescued flies. Resistance to oxidative stress was measured in adult Mgat11 null mutant flies that expressed a wild-type Mgat1 transgene in the CNS [Mgat1 (+RU)] compared with genetically matched controls that did not express Mgat1 [Mgat1 (−RU)]. Briefly, flies were fed normal food containing 3% hydrogen peroxide and their life span was measured over a 180-h period. The Mgat1 rescued flies [Mgat1 (+RU)] lived significantly longer under conditions of oxidative stress (mean life span = 127.9 ± 2.4 h) than control flies (mean life span = 80.8 ± 1.7 h). (B) The effect of percentage of yeast in the diet on the life span of Mgat1 rescued flies. The effect of dietary restriction on the life span of Mgat1 rescued flies [Mgat1 (+RU)] compared with Mgat11 null mutant flies that were not treated by drug [Mgat1 (−RU)] and to wild-type controls was determined by varying the amount of dietary yeast from 2 to 8% as described (34). Shown is mean life span with SE bars. Asterisks above the line for Mgat1 rescued flies [Mgat1 (+RU)] indicate that survival differed significantly between the Mgat1 rescued flies and controls that were not treated by drug [Mgat1 (−RU)] at yeast concentrations from 2 to 8% using the log-rank rest (P < 0.001). Asterisks between the lines for controls that were not treated by drug [Mgat1 (−RU)] and wild-type flies indicate that survival differed significantly between Mgat1 rescued flies [Mgat1 (+RU)] and wild-type flies only at 2% yeast, using the log-rank rest (P < 0.001). All life span plots were derived from a single population of flies. The standard media used for the data shown in Table 1 contained 2% yeast.
We also examined whether dietary restriction had an effect on the increased longevity conferred by neuronal expression of Mgat1. Initially, we measured the amount of food intake in our control and long-lived flies using a diet that contained food dye and measured the amount of dye ingested in a given period. We found that the long-lived Mgat1 flies ate the same amount of food as controls that were not treated with RU486, indicating that caloric intake is not a factor in the increased life span (Table S1).
It has been shown that flies fed a low-yeast diet are long lived and consume about half the calories of flies on high-yeast diets, regardless of the energetic content of the diet itself (13, 17). We therefore measured the survival of flies maintained on several diets in which all of the ingredients were held constant except for dietary yeast, as previously described (17). At concentrations of dietary yeast between 2 and 8% (wt/vol), Mgat11 null mutants that contain the UAS-Mgat1 transgene and the ELAV-GeneSwitch driver in the presence of RU486 show a significant increase in mean life span relative to genetically identical controls that were maintained in the absence of RU486 and also relative to wild-type flies (Fig.2B and Table S2).
However, the increase in life span observed in Mgat11 null mutants that contain the UAS-Mgat1 transgene and the ELAV-GeneSwitch driver in the presence of RU486 is relatively modest at concentrations of yeast between 4 and 8% compared with controls. In contrast, when the same flies are maintained on a diet of 2% yeast, we observe a dramatic increase in mean life span compared with control flies maintained in the absence of RU486 and, as previously reported (13, 17), compared with wild-type flies (Fig.2B and Table S2). We therefore conclude that although dietary restriction is not essential for the increased life span of these flies, it plays a role in the process.
Effect of Neuronal Expression of Mgat1 in Wild-Type Flies.
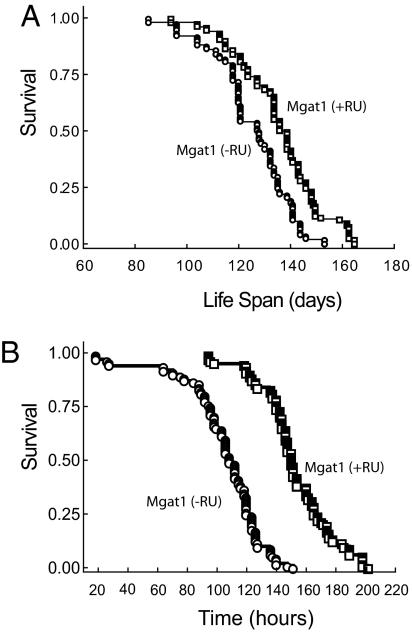
We used the ELAV-GeneSwitch line to drive expression of a wild-type Mgat1 transgene in the CNS of wild-type flies to determine whether we can also extend the life span of these flies. Similar to our observations on Mgat11 null flies, we detect increased GnT1 activity (relative to wild-type flies) in wild-type flies with the UAS-Mgat1 transgene in the absence of RU486 (compare Table 1, lines I and J), once again demonstrating that the transgene gives rise to leaky expression of Mgat1. Life span is significantly increased in association with leaky expression of the Mgat1 transgene in wild-type flies (compare Table 1, lines I, J, and L). Although this increase may be due to increased GnT1 activity, it may also be due, at least in part, to the different genotypes of the fly lines. We also show that RU486-dependent neuron-specific overexpression of the wild-type Mgat1 transgene in wild-type flies results in a 45% increase in GnT1 activity (Table 1, line K) but, in marked contrast to our observations on the Mgat11 null fly, the increase in GnT1 activity increases mean and maximum life spans by only 9 and 13%, respectively (Table 1, line K; Fig. 3A). The data suggest a correlation between GnT1 activity and mean and maximum life spans up to maximum levels of ~136 and 160 days, respectively. Importantly, the 45% increase in GnT1 activity shows that failure of the GeneSwitch cannot explain this upper limit to life span.
Fig. 3.
Life span studies on wild-type flies with neuron-specific overexpression of Mgat1. (A) Targeted expression of Mgat1 in the CNS of wild-type flies increases Drosophila life span by a relatively small amount. Targeted expression of Mgat1 to the CNS of wild-type flies increases the mean and maximum life spans by 8.8 and 12.7%, respectively, relative to wild-type flies. The genotypes, life spans, and GnT1 activities are shown in Table 1. (B) Wild-type flies overexpressing Mgat1 are resistant to oxidative stress. Resistance to oxidative stress was measured in adult wild-type flies that express a wild-type Mgat1 transgene in the CNS [Mgat1 (+RU)] compared with genetically matched controls that do not express Mgat1 [Mgat1 (−RU)]. Flies were fed normal food containing 3% hydrogen peroxide and their life span was measured over a 180-h period. The Mgat1 expressing flies [Mgat1 (+RU)] lived significantly longer under conditions of oxidative stress than control flies. All life span plots were derived from a single population of flies.
Oxidative Stress and Dietary Restriction in Wild-Type Flies Overexpressing Mgat1.
To determine if Mgat1-dependent increases in the life span of wild-type flies were also associated with resistance to oxidative stress, we examined the effect of 3% hydrogen peroxide on wild-type flies that contain the UAS-Mgat1 transgene and the ELAV-GeneSwitch driver in the absence (Table 1, line J) and presence (Table 1, line K) of RU486, as previously described. Targeted expression of Mgat1 in the CNS of these flies renders them significantly more resistant to hydrogen peroxide than control flies (Fig. 3B). Specifically, the mean and maximum life spans of wild-type flies (151.4 ± 2.4 and 187 h) in the presence of RU486 are significantly higher than the respective life spans (103.7 ± 2.7 and 131 h) in the absence of RU486. The increased longevity observed in both wild-type and null mutants expressing Mgat1 is therefore dependent on neuron-specific expression of Mgat1 and is associated with increased resistance to oxidative stress.
Finally, we also measured the effect of dietary restriction on the life span of wild-type flies overexpressing Mgat1 in the CNS. At all concentrations of dietary yeast tested (2–12% wt/vol), wild-type flies that carry the Mgat1 transgene either in the presence or in the absence of RU486 show the same mean life spans (123–136 days) (Table S2). Our inability to increase mean life span beyond ~136 days may be due to interference by other rate-limiting pathways.
Discussion
We find that knockdown of Mgat1 in the CNS of wild-type flies gives rise to defects in locomotion and life span similar to those in Mgat11 null mutants (compare lines A–D in Table 1). However, although Mgat1 may be required in the CNS for proper locomotor activity, expression in other tissues (such as muscle) may also be essential because we did not observe a significant increase in locomotion on neuron-specific Mgat1 transgene expression in Mgat11 null flies (compare lines F and G in Table 1).
We examined the effects of neuron-specific expression of Mgat1 in Mgat11 null and wild-type flies on locomotor activity and life span. Although Mgat1 transgene leakage in Mgat11 null flies results in increases of 43 and 55 days in mean and maximum life span, respectively (compare lines E and F in Table 1), it is important to note that neuron-specific Mgat1 transgene expression results in further increases of 73 days (135%) and 95 days (136%) in mean and maximum life span, respectively (compare lines F and G in Table 1). Therefore neuronal expression of Mgat1 dramatically increases the life span of Mgat11 null flies relative to genetically identical animals that do not express the transgene. Mgat1 transgene leakage in wild-type flies results in increases of 52 and 49 days in mean and maximum life span, respectively (compare lines I and J in Table 1), but neuron-specific Mgat1 transgene expression results in further increases of only 11 days (9%) and 18 days (13%) in mean and maximum life span, respectively (compare lines J and K in Table 1). We have not been able to increase mean and maximum life span of either Mgat11 null or wild-type flies beyond 136 and 165 days, respectively (Table 1 and Table S2), although the corresponding GnT1 activities are significantly increased (lines G and K in Table 1). Importantly, in the presence of 3% hydrogen peroxide, neuron-specific expression of the Mgat1 transgene in wild-type flies results in 45 and 43% increases in mean and maximum life span, respectively (Fig. 3B). Life spans are much shorter in the presence of hydrogen peroxide (hours instead of days), suggesting that the limitations to increasing life span do not apply under these conditions. We suggest that increased life span is due to increases in normal and/or unusual N-glycans and that our inability to increase mean life span beyond 136 days is due to rate-limiting interference of the GnT1 and/or other pathways. These questions require further investigation.
Interestingly, we find that although we can observe an increase in the life span of our long-lived, rescued Mgat1 null mutants at all concentrations of dietary yeast, the effects are most dramatic in flies maintained on a 2% yeast diet (Table S2), suggesting that although dietary restriction in not absolutely required to extend the life span of the Mgat1 rescued flies, it does play a role. Although the precise mechanisms by which dietary restriction extends life span have not been fully elucidated, there have been links to oxidative stress and various metabolic pathways such as insulin signaling. Further studies will be required to investigate if alterations in any of these pathways can modulate the increased life span conferred by neuronal expression of Mgat1.
Several other studies have previously identified long-lived strains of Drosophila that are associated either with loss-of-function mutations in specific genes including I'm not dead yet (Indy) (18), methuselah (19), components of the insulin signaling pathway (20, 21), and β-integrin (22) or with overexpression of specific genes such as superoxide dismutase (17). For the most part, these manipulations give rise to a 20–50% increase in mean or maximum life span (Table S3). Others have used the same ELAV-GeneSwitch line described here, to examine the effects of neuronal expression of various transgenes on Drosophila life span. For example, neuronal expression of dSir2 extended the mean life span by 52% in females and 20% in males (23). Our finding that neuronal expression of Mgat1 increases Drosophila life span suggests a unique role for Mgat1 and N-glycosylation in the regulation of organismal life span.
Protein N-glycosylation has been implicated in numerous biological processes including cell adhesion, control of the immune system, and embryonic development (24, 25). The common factor responsible for these diverse functions appears to be the ability of cell surface N-glycans to interact in a specific manner with a variety of ligands. In mammals, the biosynthesis of complex N-glycans has been shown to regulate the cell surface residency of glycoprotein receptors and their respective downstream signaling pathways (26–28). Signaling pathways in Drosophila may be controlled by an analogous mechanism involving paucimannose instead of complex N-glycans. Such a mechanism may explain the global metabolic changes that occur in our Mgat1-dependent long-lived flies. Although GnT1 action is specific for the N-glycan synthetic pathway, overexpression of Mgat1 may lower the level of the UDP-GlcNAc pool and affect UDP-GlcNAc-dependent enzymes required for the synthesis of other glycans (29, 30).
An antibody raised against horseradish peroxidase (HRP) has been used for many years to stain neural tissues in D. melanogaster (5, 31). The epitopes recognized by anti-HRP are Mgat1-dependent N-glycans carrying α1,3-fucose residues (5, 31). Drosophila proteins carrying the HRP epitope have been identified, e.g., cell adhesion and signal transduction proteins like fasciclins I and II, neurotactin, neuroglian, and receptor-linked protein tyrosine phosphatases (31). An exciting aspect of our findings is that mass spectrometry can be used to identify Mgat1 target proteins (such as those that carry the HRP epitope) in the CNS of the long-lived flies, offering a feasible approach to understanding some of the mechanisms involved in Drosophila longevity.
Materials and Methods
Food Medium and Rearing Conditions and GeneSwitch Induction.
Adults were collected from larvae grown on a diet of cornmeal (6.4%), molasses (8.5% vol/vol), yeast (2%), agar (0.79%), and 0.2% Tegosep (methyl 4-hydroxybenzoate; Sigma). Adults were maintained on this diet supplemented with yeast (2, 4, 6, and 8% wt/vol, as specified in each experiment). Induction of UAS transgenes using the conditional ELAV-GeneSwitch line was achieved using RU486 (mifepristone; Sigma). RU486 was dissolved in ethanol and added to the media at a concentration of 0.2 mM; ethanol alone was added to the media of the control experiments. First instar larvae were raised on the media in the presence or absence of RU486 until they reached adulthood. Adult flies were then transferred to fresh vials to initiate life span studies. All life span studies were carried out at 2% yeast unless otherwise mentioned.
Drosophila Stocks.
Mgat11 null mutants were generated by imprecise excision of a nearby P-element insertion and have previously been described (5). Mgat+9 is a wild-type control line that was generated at the same time as Mgat11 and is a precise excision of the initial P-element insertion. Neuronal expression of Mgat1 in Mgat11 null mutants was achieved using a conditional ELAV-GeneSwitch line in Drosophila with the following genotype: w1118/UAS-Mgat1; Mgat11/Mgat11; +/ELAV-GeneSwitch.
Locomotor Activity and Life Span Studies.
The locomotor activity of adult male and female flies was measured using a slightly modified open field test (32). Briefly, individual flies from each genotype were placed in a covered Petri dish (15 mm diameter) and allowed to adapt to their environment for 5 min. The number of times a fly crossed four lines drawn on the Petri dish over a period of 3 min was measured. Twenty-five flies of each genotype were used for each experiment. Life span determinations of flies with different genotypes were performed as previously described (5). The life span of w1118/UAS-Mgat1; Mgat11/Mgat11; +/ELAV-GeneSwitch was performed in triplicate; all other life span studies were performed in duplicate.
Enzyme Activity.
The activity of GnT1 was measured in duplicate as previously described (5).
Oxidative Stress Resistance.
The effect of experimentally induced oxidative stress on the survival of flies was measured by administration of hydrogen peroxide (33). Medium was freshly prepared in a 50-mL vial containing 1.5% agar, 2% sucrose, and 3% hydrogen peroxide (added to the melted medium at 45 °C to avoid loss of oxidative activity). Eight-day-old female flies were placed in vials in groups of 10, and the dead flies were counted every 2 h until the end of the experiment. Experiments were always carried out simultaneously with control flies and flies of various genotypes to prevent unavoidable variations in viability between different experiments that may arise from slight differences in experimental conditions.
Meal Volume Assay and Dietary Restriction.
The exact volume of a single meal of adult flies was determined for D. melanogaster, using a diet that contains a food dye (Brilliant Blue FCF or Blue No. 1; MD McCormick). After feeding, the flies were homogenized and the amount of dye ingested was assayed with a spectrophotometer. Blue No. 1 absorbs light strongly in a narrow band at a wavelength of 625 nm. This dye remains within the digestive tract and passes out of the fly unaffected by gut pH and enzymes. Groups of 30 female flies, previously food deprived for 24 h (starved) or nonfood deprived (fed), were fed for 30 or 60 min on 2 mL of diet containing 0.5% (wt/vol) Blue No. 1. After feeding for the required time, the flies were anesthetized with carbon dioxide and homogenized in 1 mL PBS (pH 7.4) and centrifuged for 5 min. The supernatant was removed, recentrifuged for 5 min, and transferred to cuvettetes. The homogenization and centrifugation steps were performed as rapidly as possible to prevent eye pigments from leaching into the solution and interfering with the absorbance of the dye. To correct for any background absorbance of the flies, the absorbance of the supernatant from flies that were not fed dye was measured at 625 nm and subtracted from the absorbance of the supernatant from the dye-fed fly. The net absorbance was used to calculate the amount of food ingested. Dietary restriction was achieved by rearing flies on a standard cornmeal diet and varying the yeast concentrations between 2 and 12% as described (34). Proportional hazard (Cox) regression analysis was used to determine if life span was modified by dietary restriction.
Generation of RNAi and UAS Constructs.
pWIZ is a pUAST transformation vector containing the 74-bp second intron of the white gene flanked by several unique restriction sites to facilitate cloning. The RNAi construct was generated in the vector by the following procedure (35): A 700-bp cDNA fragment (36) was amplified by PCR. The restriction site XbaI, compatible with AvrII and NheI restriction sites, was present at the 5′ end of each primer. The PCR product was subcloned into the TOPO vector (Invitrogen) by TA cloning. The PCR fragment was cut from the TOPO vector by the XbaI restriction enzyme and subcloned into the NheI site of the pWIZ vector. A second ligation of the same PCR product into the AvrII site of the pWIZ vector created a “tail-to-tail” inverted repeat of the RNAi construct. The clone with the desired orientation of the insert was selected by sequencing. The PCR product described above was directly subcloned into the EcoRI site of the pUAST vector. The clone containing the insert in the reverse orientation was selected and the same PCR product was subcloned into this plasmid in the sense orientation, generating a “head-to-head” inverted repeat of the RNAi construct with a 29-bp spacer between the two repeated sequences.
Drosophila Mgat1 cDNA (36) was used as a template for PCR amplification of the entire coding region of the gene including the transmembrane domain segment. The PCR products were subcloned into the pUAST expression vector using NotI and KpnI restriction sites introduced by the PCR primers to yield pUAST-Mgat1 plasmids. Plasmid DNA sequences were verified by sequencing.
Supplementary Material
Acknowledgments
We thank Drs. W. S. Trimble, J. Dennis, and M. Tatar for critically reading the manuscript and members of the Boulianne Laboratory for helpful discussions. This work was supported by grants from the Canadian Institutes for Health Research (MOP 67130 to G.L.B. and MOP 68811 to H.S.). G.L.B. is the recipient of a Tier I Canada Research Chair in Molecular and Developmental Neurobiology.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004431107/-/DCSupplemental.
References
- 1.Henrissat B, Surolia A, Stanley P. A genomic view of glycobiology. In: Varki A, et al., editors. Essentials of Glycobiology. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2008. pp. 89–99. [Google Scholar]
- 2.Schachter H. The ‘yellow brick road’ to branched complex N-glycans. Glycobiology. 1991;1:453–461. doi: 10.1093/glycob/1.5.453. [DOI] [PubMed] [Google Scholar]
- 3.Schachter H. The joys of HexNAc. The synthesis and function of N- and O-glycan branches. Glycoconj J. 2000;17:465–483. doi: 10.1023/a:1011010206774. [DOI] [PubMed] [Google Scholar]
- 4.Stanley P, Schachter H, Taniguchi N. N-glycans. In: Varki A, et al., editors. Essentials of Glycobiology. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2008. pp. 101–114. [Google Scholar]
- 5.Sarkar M, et al. Null mutations in Drosophila N-acetylglucosaminyltransferase I produce defects in locomotion and a reduced life span. J Biol Chem. 2006;281:12776–12785. doi: 10.1074/jbc.M512769200. [DOI] [PubMed] [Google Scholar]
- 6.Ioffe E, Stanley P. Mice lacking N-acetylglucosaminyltransferase I activity die at mid-gestation, revealing an essential role for complex or hybrid N-linked carbohydrates. Proc Natl Acad Sci USA. 1994;91:728–732. doi: 10.1073/pnas.91.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Metzler M, et al. Complex asparagine-linked oligosaccharides are required for morphogenic events during post-implantation development. EMBO J. 1994;13:2056–2065. doi: 10.1002/j.1460-2075.1994.tb06480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi H, Tan J, Schachter H. N-glycans are involved in the response of Caenorhabditis elegans to bacterial pathogens. Methods Enzymol. 2006;417:359–389. doi: 10.1016/S0076-6879(06)17022-6. [DOI] [PubMed] [Google Scholar]
- 9.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 10.Brand AH, Dormand EL. The GAL4 system as a tool for unravelling the mysteries of the Drosophila nervous system. Curr Opin Neurobiol. 1995;5:572–578. doi: 10.1016/0959-4388(95)80061-1. [DOI] [PubMed] [Google Scholar]
- 11.Roman G, Endo K, Zong L, Davis RL. P[Switch], a system for spatial and temporal control of gene expression in Drosophila melanogaster. Proc Natl Acad Sci USA. 2001;98:12602–12607. doi: 10.1073/pnas.221303998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osterwalder T, Yoon KS, White BH, Keshishian H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc Natl Acad Sci USA. 2001;98:12596–12601. doi: 10.1073/pnas.221303298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ford D, et al. Alteration of Drosophila life span using conditional, tissue-specific expression of transgenes triggered by doxycyline or RU486/Mifepristone. Exp Gerontol. 2007;42:483–497. doi: 10.1016/j.exger.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giannakou ME, et al. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science. 2004;305:361. doi: 10.1126/science.1098219. [DOI] [PubMed] [Google Scholar]
- 15.Hwangbo DS, et al. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 16.Lee KS, et al. JNK/FOXO-mediated neuronal expression of fly homologue of peroxiredoxin II reduces oxidative stress and extends life span. J Biol Chem. 2009;284:29454–29461. doi: 10.1074/jbc.M109.028027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parkes TL, et al. Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nat Genet. 1998;19:171–174. doi: 10.1038/534. [DOI] [PubMed] [Google Scholar]
- 18.Rogina B, Reenan RA, Nilsen SP, Helfand SL. Extended life-span conferred by cotransporter gene mutations in Drosophila. Science. 2000;290:2137–2140. doi: 10.1126/science.290.5499.2137. [DOI] [PubMed] [Google Scholar]
- 19.Lin YJ, Seroude L, Benzer S. Extended life-span and stress resistance in the Drosophila mutant methuselah. Science. 1998;282:943–946. doi: 10.1126/science.282.5390.943. [DOI] [PubMed] [Google Scholar]
- 20.Clancy DJ, et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- 21.Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- 22.Goddeeris MM, et al. Delayed behavioural aging and altered mortality in Drosophila beta integrin mutants. Aging Cell. 2003;2:257–264. doi: 10.1046/j.1474-9728.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 23.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varki A, Lowe JB. Biological roles of glycans. In: Varki A, et al., editors. Essentials of Glycobiology. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2008. pp. 75–88. [Google Scholar]
- 25.Freeze HH. Genetic defects in the human glycome. Nat Rev Genet. 2006;7:537–551. doi: 10.1038/nrg1894. [DOI] [PubMed] [Google Scholar]
- 26.Lau KS, et al. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129:123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 27.Partridge EA, Cheung P, Dennis JW. Cytokine sensitivity and N-glycan processing mutations. Methods Enzymol. 2006;417:3–11. doi: 10.1016/S0076-6879(06)17001-9. [DOI] [PubMed] [Google Scholar]
- 28.Partridge EA, et al. Regulation of cytokine receptors by Golgi N-glycan processing and endocytosis. Science. 2004;306:120–124. doi: 10.1126/science.1102109. [DOI] [PubMed] [Google Scholar]
- 29.Dennis JW, Lau KS, Demetriou M, Nabi IR. Adaptive regulation at the cell surface by N-glycosylation. Traffic. 2009;10:1569–1578. doi: 10.1111/j.1600-0854.2009.00981.x. [DOI] [PubMed] [Google Scholar]
- 30.Dennis JW, Nabi IR, Demetriou M. Metabolism, cell surface organization, and disease. Cell. 2009;139:1229–1241. doi: 10.1016/j.cell.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paschinger K, Rendić D, Wilson IB. Revealing the anti-HRP epitope in Drosophila and Caenorhabditis. Glycoconj J. 2009;26:385–395. doi: 10.1007/s10719-008-9155-3. [DOI] [PubMed] [Google Scholar]
- 32.Iliadi KG, et al. Sexual differences for emigration behavior in natural populations of Drosophila melanogaster. Behav Genet. 2002;32:173–180. doi: 10.1023/a:1016017028041. [DOI] [PubMed] [Google Scholar]
- 33.Monnier V, Girardot F, Audin W, Tricoire H. Control of oxidative stress resistance by IP3 kinase in Drosophila melanogaster. Free Radic Biol Med. 2002;33:1250–1259. doi: 10.1016/s0891-5849(02)01019-5. [DOI] [PubMed] [Google Scholar]
- 34.Min KJ, Yamamoto R, Buch S, Pankratz M, Tatar M. Drosophila lifespan control by dietary restriction independent of insulin-like signaling. Aging Cell. 2008;7:199–206. doi: 10.1111/j.1474-9726.2008.00373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee YS, Carthew RW. Making a better RNAi vector for Drosophila: Use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 36.Sarkar M, Schachter H. Cloning and expression of Drosophila melanogaster UDP-GlcNAc:alpha-3-D-mannoside beta1,2-N-acetylglucosaminyltransferase I. Biol Chem. 2001;382:209–217. doi: 10.1515/BC.2001.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.