Abstract
The present study was conducted to evaluate the possible antibacterial activity of Anabaena extracts. Anabaena was isolated from a natural source and cultured in vitro. after suitable growth, cyanobacterial culture was harvested using different solvents. Extracts, thus prepared, were evaluated for their antibacterial potential by agar-well diffusion assay against bacterial species of clinical significance. MIC values were determined further to check the concentration ranges for significant inhibition. HPTLC analysis was done to separate the components of active crude extract in an attempt to identify the bio-active chemical entity. Methanol extract exhibited more potent activity than that of hexane and ethyl acetate extracts. No inhibitory effect was found against Pseudomonas aeruginosa, Salmonella typhi, and Klebsiella pneumoniae. Staphylococcus aureus required about 256 µg/ml of the crude methanol extract for effective inhibition. HPTLC evaluation at λ 254 nm was performed for the separation of a complex mixture of the methanol extract. The results provide evidence that Anabaena sp. extracts might indeed be potential sources of new antibacterial agents.
Keywords: Blue-green algae, drug-resistance, biologically active substances, bacterial species
Introduction
Cyanobacteria (also known as blue-green algae due to the presence of certain pigments) are among the oldest phototrophic procaryotic organisms. The search for cyanobacteria with antimicrobial activity has gained importance in recent years due to growing worldwide concern about an alarming increase in the emergence of antibiotic resistance and further increase in the rate of infection by these antibiotic-resistant microorganisms. Various strains of cyanobacteria are known to produce intracellular and extracellular metabolites with diverse biological activities such as antibacterial and antifungal.[1‐3] The aim of study reported here was to investigate the antibacterial activity of various organic extracts and spent medium of Anabaena sp. against the bacteria of clinical significance.
Materials and Methods
Isolation and Culturing of Cyanobacterium
Cyanobacterial mats collected from Naini-Lake, Nainital, India, were crushed with sterilized glass beads and diluted in presterilized double distilled water and were spread on solidified agar plates containing BG-11 medium and incubated at a light intensity of 3500 lux until visible colonies were appeared. Isolated cyanobacterium was identified by Prof. P. Kaushik at the Department of Botany and Microbiology, Gurukul Kangri University, Hardwar, India, as Anabaena species on the basis of its characteristic features. Scanning electron microscopy of isolated species was done for further confirmation on the basis of its morphological characteristics [Figure 1].
Figure 1.
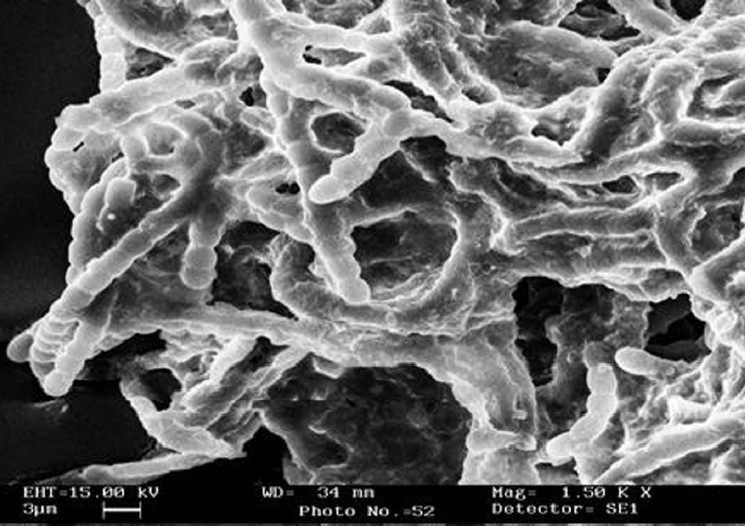
Scanning electron micrograph of Anabaena sp. isolated from Naini-Lake Nainital, India
Preparation of Extracts
At the stationary phase of growth (25 days), Anabaena culture was dried in a hot air oven at 60°C for 1 h. The dried biomass was extracted with the solvents viz. methanol, ethyl acetate, hexane, and dichloromethane (Qualigens, AR grade). The extracts were centrifuged at 4000 rpm for 10 min, and were further concentrated in vacuum under reduced pressure and dried in a desiccator. Dried extracts were stored in labeled sterile screw capped bottles in a refrigerator till further use.
Test Microorganisms
The bacterial cultures selected for the present study were Staphylococcus aureus MTCC-740, Bacillus subtilis MTCC-736, Bacillus cereus MTCC-430, Bacillus pumilus MTCC-1607, Escherichia coli MTCC-739, Pseudomonas aeruginosa MTCC-741, Salmonella typhi MTCC-733, and Klebsiella pneumoniae MTCC-139. All organisms were maintained on specific media slants at 4°C and revived prior to use. An inoculum size of 106 CFU/ml of bacteria, compared with 0.5 McFarland turbidity standards, was used.
Antibacterial Testing
Agar well diffusion assay
In vitro evaluation of antibacterial activity of different crude extracts of Anabaena sp. was done using the agar well diffusion assay.[4] Petri dishes (90 mm diameter) were poured with Mueller Hinton Agar media (HI-Media) and allowed to solidify to make a base layer. A seed layer was prepared by inoculating 100 μl of each test organism's suspension in 25 ml Mueller Hinton Agar. After complete solidification of media, wells (of 6 mm in diameter) were made in the agar medium with the help of a sterile steel borer. 100 μl of each extract (stock solution 10 mg/ml) was added aseptically in respective wells. All the plates were incubated at 37±1°C for 24 h. At the end of the period, the inhibition zone formed in the medium was measured in millimeters.
Microdilution assay
MIC values were determined for the sensitive bacteria to the methanol extract using a two-fold serial microdilution method.[5] The concentrations of the extract used in experiment ranged from 10 μg/ml to 512 μg/ml. The methanol extract was added to sterile microtiter plate containing Cation-Adjusted Mueller-Hinton Broth (CAMHB, Hi-Media). In each plate, 1 × 106 CFU/ml bacterial suspensions were added. The MIC value was taken as the lowest concentration of the extract in wells of the microtiter plate that showed no turbidity after 24 h of the incubation at 37°C. The turbidity of the wells in the microtiter plate was interpreted as visible growth of the microorganisms. Experiments were carried out in duplicate.
HPTLC analysis
HPTLC analysis was performed on a silica gel F-254 (E-Merck grade) pre-coated aluminum plate. A band of 7 mm was applied at a distance of 10 mm from the bottom of the plate. The methanol extract (10 μl) was applied on a chromatoplate (CAMAG Linomat-5) and run in the solvent system (chloroform and methanol, 7:3).The plate was developed up to 8 cm in a twin trough chamber previously equilibrated with mobile phase for 20 min. Densitometric evaluation of the plate was performed at λ 254 nm.
Results
The results shown in Table 1 indicate that the methanol extract revealed a prominent effect on various bacterial species. However, the hexane and ethyl acetate extract exhibited a moderate effect and DCM and spent medium did not exhibit any antibacterial activity. As shown in Table 2, MIC (minimum inhibitory concentrations) for the methanol extract was found to be >512 μg/ml for Bacillus pumilus and Bacillus cereus. On the other hand, Bacillus subtilis and Escherichia coli was inhibited at the dose of 512 μg/ml of the crude extract. While Staphylococcus aureus required about 256 μg/ml. Results of HPTLC analysis revealed that the methanol extract contained a mixture of different component that were eluted at RF = 0.19, RF = 0.30, RF = 0.33, RF = 0.42, RF = 0.54, RF = 0.66, RF = 0.73 as shown in Figure 2a and b.
Table 1.
Antimicrobial activity of various crude extracts of Anabaena sp.
| Test organisms |
Zone of inhibition (in mm) |
||||
|---|---|---|---|---|---|
| Hexane | Ethyl acetate | Methanol | Dichloromethane | Spent medium | |
| S. aureus | 10 | 12 | 18 | NZ | NZ |
| B. subtilis | NZ | NZ | 16 | NZ | NZ |
| B. pumilus | NZ | NZ | 14 | NZ | NZ |
| B. cereus | NZ | NZ | 12 | NZ | NZ |
| E. coli | 12 | NZ | 15 | NZ | NZ |
| P. aeruginosa | NZ | NZ | NZ | NZ | NZ |
| K. pneumoniae | NZ | NZ | NZ | NZ | NZ |
| S. typhi | NZ | NZ | NZ | NZ | NZ |
NZ- No zone of Inhibition
Table 2.
Minimum inhibitory concentration of active crude extracts of Anabaena sp.
| Test organisms |
Minimum inhibitory concentration (μg/ml) |
|
|---|---|---|
| Methanol extracts | Ciprofloxacin | |
| Staphylococcus aureus | 256 | <1 |
| Bacillus subtilis | 512 | <1 |
| Bacillus pumilus | >512 | <1 |
| Bacillus cereus | >512 | <1 |
| Escherichia coli | 512 | <1 |
Figure 2.
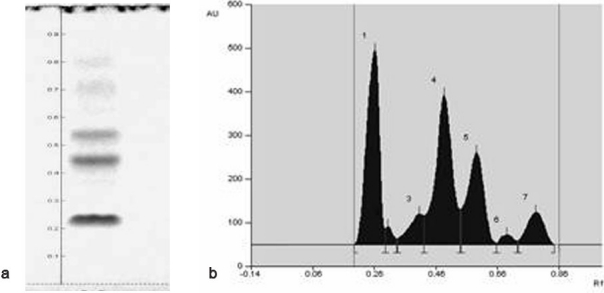
HPTLC profile of methanol extract of Anabaena sp. (a) HPTLC separation at UV-254 nm. (b) Densitometric chromatogram at wavelength 254 nm.
Discussion
There is a clear indication that Anabaena sp. extracted in different solvents has significant antibacterial effects on both Gram-positive and Gram-negative bacterial species. The current scenario of antibiotics is very threatening due to significant emergence of resistance among bacterial pathogens against available antibiotics.[6] The present study focused to the discovery of novel chemicals that can lead to the development of pharmaceuticals of cyanobacterial origin. The combination of a chromatographic technique with serial dilution in tubes was capable of identifying the active components of methanol extract responsible for this activity. This manifests the importance of using the correct combination of chemical screening and biological analysis.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared
References
- 1.Kreitlow S, Mundt S, Lindequist U. Cyanobacteria-a potential source of new biologically active substances. J Biotechnol. 1999;70:61–3. doi: 10.1016/s0168-1656(99)00058-9. [DOI] [PubMed] [Google Scholar]
- 2.Falch BS, Konig GM, Wright AD, Sticher O, Angerhofer CK, Pezzuto JM. Biological activities of cyanobacteria: Evaluation of extracts and pure compounds. Planta Med. 1995;61:321–8. doi: 10.1055/s-2006-958092. [DOI] [PubMed] [Google Scholar]
- 3.Mundt S, Kreitlow S, Nowotny A, Effmert U. Biological and pharmacological investigation of selected cyanobacteria. Int J Hyg Environ Health. 2001;203:327–34. doi: 10.1078/1438-4639-00045. [DOI] [PubMed] [Google Scholar]
- 4.Perez C, Pauli M, Bazerque P. An antibiotic assay by agar-well diffusion method. Acta Biol Med Exp. 1990;15:113–5. [Google Scholar]
- 5.NCCLS-National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, Approved Standard M7-A4, Wayne, Pa. 1997 [Google Scholar]
- 6.Kaushik P, Chauhan A, Chauhan G, Goyal P. Evaluation of Nostoc commune for potential antibacterial activity and UV-HPLC analysis of methanol extract. Int J Microbiol. 2008;5:1–5. [Google Scholar]


