Abstract
We analyzed the nuclear import and regulation of the yeast histone variant Htz1 (H2A.Z), and the role of histone chaperones Nap1 and Chz1 in this process. Co-purification suggested that Htz1 and H2B dimerized in the cytoplasm prior to import. Like H2B, Htz1 contained an NLS in its N-terminus that is recognized by multiple karyopherins (also called importins), indicating multiple transport pathways into the nucleus. However, Kap114 and Kap123 appeared to play the major role in Htz1 import. We also identified a role for Nap1 in the import of Htz1/H2B heterodimers, and Nap1 formed a Ran-GTP insensitive import complex with Htz1/H2B and Kap114. Nap1 was necessary for maintaining a soluble pool of Htz1, indicating that its chaperone function may be important for the dynamic exchange of histones within nucleosomes. In contrast, Chz1 was imported by a distinct import pathway, and Chz1 did not appear to interact with Htz1 the cytoplasm. Genetic analysis indicated that NAP1 has a function in the absence of HTZ1 that is not shared with CHZ1. This provides further evidence that the histone chaperones Nap1 and Chz1 have separate Htz1-dependent and -independent functions.
Introduction
Histones comprise the bulk protein component of chromatin in eukaryotic cells(1, 2). Changes to the chromatin can be facilitated by the action of chromatin remodeling complexes, through posttranslational modification of the histones, and by replacement of canonical histones with nonallelic histone variants. Yeast histone Htz1 is the evolutionarily conserved H2A.Z variant and its incorporation into chromatin has been linked to various cellular processes including transcription, heterochromatin silencing, and chromosome transmission (3–5). Genome wide localization studies have indicated that Htz1 is often deposited at one or two nucleosomes proximal to the transcriptional start site (6–10). Htz1 plays a role in transcription acting in parallel with chromatin remodelers and histone modifying enzymes (11). Htz1 itself is acetylated on four lysine residues in its N-terminus, and acetylation of K14 has been shown to be enriched at transcriptionally active promoters (8, 12, 13).
Non-nucleosomal histones are found in complex with one of several histone chaperones (14–17). One of the primary functions of the chaperones is to shield the highly basic histones from aggregating and forming non-specific interactions with DNA, and as histones and most histone chaperones are conserved from yeast to humans, this function is likely conserved in all eukaryotic cells (14–18). Nap1 and Chz1 are chaperones for Htz1, and in vitro both deliver Htz1/H2B heterodimers to the SWR1 complex (19). The SWR1 complex catalyzes the exchange of H2A for Htz1 within a nucleosome (20, 21). Chz1 preferentially interacts with Htz1/H2B, whereas Nap1 is an H2A/H2B and Htz1/H2B chaperone and interacts with either histone dimer (19–23). Immunoprecipitation of Htz1 from whole cell extracts demonstrated that Chz1 and Nap1 form histone – chaperone complexes in the cell, suggesting functional redundancy (19). Recently, Nap1 was found to be a phospho-protein and associated with chromatin by chromatin immunoprecipitation (ChIP) analysis, and is enriched at the 3' end of ACT1 and ADH1 (24, 25). Nap1 is involved in transcript elongation and may promote the reassembly of nucleosomes following the progression of RNA polymerase II (25). Nap1 was also found in association with the promoter region of the inducible genes PHO5 and GAL1, suggesting a role in Htz1 assembly or disassembly (25).
The transport of most proteins into the nucleus is facilitated by soluble nuclear transport factors referred to as karyopherins (Kaps) or importins (26). Kaps interact directly with the nuclear localization signal (NLS) of a target protein to form an import complex and transport the protein into the nucleus through the nuclear pore complex (NPC) (27–29). Once inside the nucleus, Ran-GTP binds to the Kap to stimulate the release of the cargo protein (28, 30). The Kap – Ran-GTP complex then exits the nucleus through the NPC back into the cytoplasm to begin another round of transport. The nuclear transport pathways of the core histone proteins in budding yeast have been described (31, 32). Histones H3 and H4 are primarily imported by Kap123 and histones H2A and H2B are primarily imported by Kap114, but other Kaps, especially Kap121, are involved. Also, histone chaperones can play a role in histone import exemplified by Nap1 (22). We previously showed that Nap1 interacts directly with the NLS domains of H2A and H2B, and in the absence of NAP1 a decrease in the import of H2A and H2B NLS GFP reporters was observed (22). Nap1 also binds directly to Kap114, which results in an increased affinity of Kap114 for H2A/H2B (22). Conversely, the association of other Kaps to H2A/H2B is significantly inhibited in the presence of Nap1. This suggests that Nap1 serves as an import cofactor involved in the nuclear transport of H2A/H2B, and as a karyopherin specificity determinant for H2A/H2B. The association of histone chaperones with the import complex provides a link between nuclear transport and nucleosome assembly. In addition, Htz1 has been linked to NPC-mediated transcriptional control and heterochromatin boundary activity, suggesting that Htz1 may serve as a target for the transport machinery to specific regions of DNA (33).
In this report, we analyze the nuclear import of Htz1 and the role of Htz1 chaperones in this process. We describe Htz1 nuclear import pathways and identify an Htz1 NLS in the N-terminus. We also identify a role for Nap1 in Htz1 import. Surprisingly, we do not detect Chz1 associated with Htz1 in cytosol. We show that Chz1 contains a classical NLS suggesting it utilizes distinct import pathways from Htz1. Although both are histone chaperones, the function of Nap1 and Chz1 are seemingly different. We show that Nap1 is important for maintaining a soluble pool of Htz1. This may be important for the dynamic exchange of this histone within nucleosomes. In addition, we uncovered genetic interactions not shared between NAP1 and CHZ1 further suggesting their distinct functions.
Results
Htz1 interacts with Nap1 and multiple karyopherins in cytosol
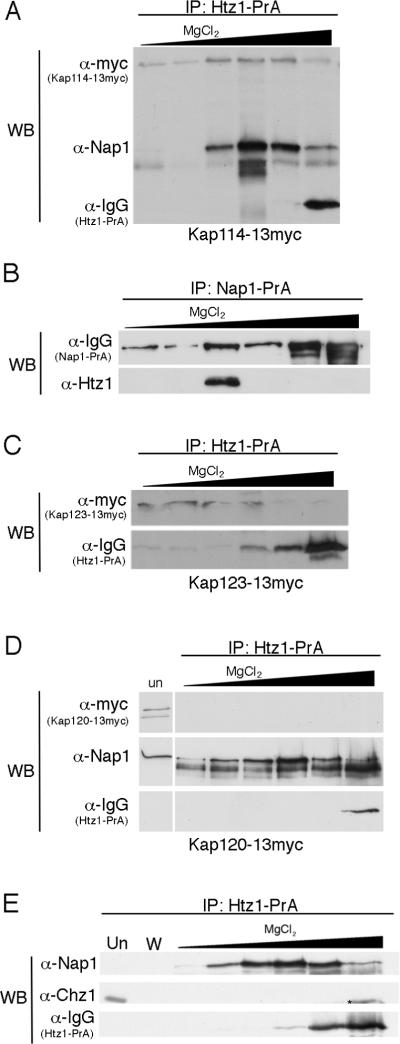
It is generally assumed that Htz1 forms an obligate heterodimer with H2B shortly after synthesis. As Htz1 synthesis occurs throughout the cell cycle, this suggests a model where Htz1/H2B is imported separately, both spatially and temporally, from H2A/H2B, demonstrating that the import of the two heterodimers is likely distinctly regulated. To understand how the nuclear transport of histone variant Htz1 is coordinated with its deposition into chromatin, we immunopurified Htz1 from yeast cytosol to identify interacting proteins. A strain containing an allele of HTZ1 with a C-terminal protein A (PrA) tag was grown to logarithmic phase and Htz1-PrA and associated proteins were purified on IgG sepharose from the post-ribosomal cytosolic fraction. Bound protein was eluted with 1 M MgCl2, enzymatically digested, and the complex mixture was submitted for analysis by liquid chromatography/tandem mass spectrometry. Among the interacting proteins, four karyopherins (Kaps) were identified; Kap114, Kap123, Kap95, and Kap108 (Sxm1). These Kaps were previously shown to transport canonical H2A and H2B into the nucleus, with a specific role identified for Kap114. Histone chaperone Nap1 was also identified. Nap1 acts as an import cofactor for H2A and H2B and has been shown to be a chaperone for Htz1. Chz1 is also a histone chaperone specific for Htz1, but surprisingly we did not find any in our cytosolic interacting protein fraction.
To verify the Kap interactions by Western blot, we immunoprecipitated Htz1-PrA from a strains expressing Kap114-Myc, Kap123-Myc, Kap95-Myc and Kap108-Myc. Bound proteins were eluted with a step gradient of MgCl2 and the Kaps were detected with α-Myc antibodies, confirming the mass spectrometry results (Figure 1A, B and data not shown). As a negative control we determined that Kap120-Myc did not co-elute with Htz1-PrA (Figure 1C). We also tested whether Nap1 was co-precipitated with Htz1-PrA, and determined that it eluted in the same fractions as Kap114 (Figure 1A). In the reciprocal experiment, Htz1 was co-immunoprecipitated from cytosol with Nap1-PrA (Figure 1D). Additionally, we tested for the presence of Chz1 co-eluting with Htz1-PrA using anti-Chz1 antibodies. We were unable to detect any co-eluting Chz1 by Western blot although we did detect Chz1 in the unbound fraction (Figure 1E). In contrast, Nap1 co-elution was clearly evident in the same experiment. These results suggested that Htz1 interacted with several Kaps representing multiple import pathways, and that Nap1 and Htz1 associated in the cytoplasm prior to nuclear import. The Kaps we observed were similar to those we reported for H2A and H2B and raised the possibility that H2B mediated the import of the Htz1/H2B heterodimer.
Figure 1.
Htz1 associates with Kap114, Kap123, and Nap1 in yeast cytosol. (A) Htz1-PrA and associated proteins were purified from cytosol prepared from a strain expressing Kap114-13myc, eluted with a MgCl2 step gradient, separated by SDS-PAGE and analyzed by Western blot (WB) using anti-Myc and anti-Nap1 antibodies. Htz1-PrA is recognized by IgG. (B) Nap1-PrA and associated proteins were purified from cytosol as above and probed with anti-Htz1 antibodies. (C) Htz1-PrA was purified from a strain expressing Kap123-13myc or (D) Kap120-13myc and analyzed as in (A). `Un' lane contains 0.02% of depleted post-bead starting cytosol. (E) Htz1-PrA was purified as in (A) and probed with anti-Chz1 and anti-Nap1 antibodies. W lane contains the last wash fraction. * indicates a proteolytic product of Htz1-PrA.
The N-terminus of Htz1 contains an NLS
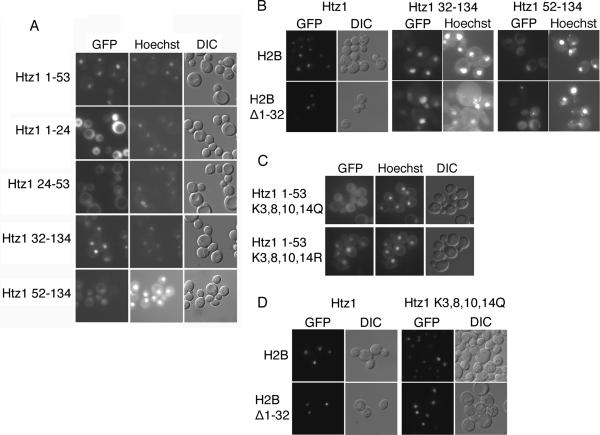
We therefore addressed whether Htz1 contains its own active NLS. All histones contain a histone fold domain that consists of three α-helices (α1, α2 and α3), as well as the unstructured N terminal domain. In addition some histones contain additional alpha helical domains N-terminal to α1 (αN), and C-terminal to α-3 (α-C). All four core histones, H2A, H2B, H3 and H4, have been shown to contain an NLS in their N-terminus (31, 32). To test the Htz1 N-terminus for NLS activity, we expressed Htz1 residues 1–53 fused to two GFP moieties (Htz11–53-GFP2) in wild type cells. This fragment of Htz1 corresponds to the fragment of H2A that contains full NLS activity, and encodes the flexible N terminus, αN and α1 domains (32). Upon induction of GFP reporter expression, the Htz11–53-GFP2 fusion protein was localized to the nucleus, indicating that Htz1 contained an NLS in its N-terminus (Figure 2A). We then expressed Htz1 residues 1–24 (containing the unstructured N terminus) or 24–53 (containing the αN and α1 domain) fused to GFP2 to determine if the entire 1–53 segment is necessary to direct the reporter into the nucleus. The Htz11–24-GFP2 fusion was distributed throughout the nucleus and cytoplasm, indicating the extreme N-terminus was not sufficient for NLS activity (Figure 2A). Htz124–53-GFP2 was localized to the nucleus and cytoplasm, but the nucleus was discernable in most cells, indicating that Htz1 residues 24–53 contained partial NLS activity. Our results indicated that Htz1 contains an NLS in its N-terminus, and that the entire 1–53 sequence is required for full NLS activity. We wanted to determine whether Htz1 contained an NLS in the C terminal part of the protein, therefore we expressed Htz132–134-GFP2, (deleting the unstructured N terminus and the αN domain) and Htz152–134-GFP2. (deleting the unstructured N terminus, the αN domain and the α1 domain). The Htz132–134-GFP2 was nuclear, and Htz152–134-GFP2 was expressed very poorly, but in expressing cells was visible in both nuclear and cytoplasmic compartments, with some nuclear acccumulation. This result suggested that there maybe second weaker NLS in Htz1, or that some nuclear import was mediated by the H2B NLS, via dimerization with H2B.
Figure 2.
Htz1 contains an NLS in its amino terminus (A) Wild type (WT) yeast containing the indicated plasmid, were induced to express Htz1 residues 1–53, 1–24, 24–53, 32–134 or 52–134 fused to GFP2 and visualized by fluorescent microscopy. Coincident Hoechst staining and DIC is shown. Htz1 32–134 or 52–134 GFP2 expressing strains were induced for an additional 2 hours. (B) JHY200 strains deleted for HTZ1, and expressing wild type H2B or H2BΔ1–32, were induced to express the Htz1- GFP2 fusions indicated. Htz1 32–134 GFP2 and Htz1 52–134 GFP2 expressing strains were induced for an additional 5 hours, and only a small fraction of cells expressed Htz1 52–134 GFP2. Coincident Hoechst staining or DIC is shown. (C) Wild type yeast expressing mutant NLS reporters htz11–53K3,8,10,14Q or htz11–53K3,8,10,14R-GFP2 were visualized by fluorescent microscopy as above. (D) JHY200 strains deleted for HTZ1, and expressing wild type H2B or H2BΔ1–32, were induced to express the Htz1-GFP2 or htz1K3,8,10,14R-GFP2 fusions as above.
To test this we expressed our GFP constructs in a strain (JHY200) that bears deletions of both copies of each of the four core histones, and growth is supported by plasmid pQQ18 that encodes one copy of each of the core histones. The genomic copy of HTZ1 was deleted in this strain and Htz1-GFP2, Htz132–134-GFP2, and Htz152–134-GFP2, were expressed from plasmids. A mutation was created in HTB1, the gene encoding H2B, whereby amino acids 1–32, containing the H2B NLS, were deleted in the context of pQQ18. Htz1-GFP2 and Htz132–134-GFP2 were nuclear in strains with and without the H2B NLS (Figure 2B). Htz152–134-GFP2 was expressed at very low levels in this strain background. In cells with intact H2B, Htz152–134-GFP2 appeared to be both cytoplasmic and nuclear, however, in a subset of cells (<10%) distinct nuclear accumulation of GFP was observed (Figure 2B). In contrast, nuclear accumulation was not apparent in cells lacking the H2B NLS, and GFP was equilibrated through the cell (Figure 2B). However, Htz152–134-GFP2 was very poorly expressed in these cells, and cytoplasmic and nuclear aggregates were also visible. These data are consistent with the finding that the major Htz1 NLS is located in the N terminal half of the protein and the α1 domain (between residue 32 and 52) is particularly important for this activity.
Acetylation of Htz1 is not necessary for import or growth
We have shown that mutation of key acetylation sites in H3 and H4 perturbs NLS function, and loss of positive charge, mimicking acetylation, reduces nuclear import (34). Htz 1 can be acetylated on four lysine residues located in the N-terminus (8, 12, 13). We mutated the four acetylated lysines within the Htz1 NLS to glutamine to mimic acetylation, and to arginine, which mimics the positively charged unacetylated lysine. We expressed the Htz1 NLS mutants in the context of our Htz11–53-GFP2 reporter and observed the localization of GFP. The Htz11–53K3,8,10,14Q-GFP2 reporter was localized throughout the cell while the Htz11–53K3,8,10,14R-GFP2 reporter was localized primarily to the nucleus similar to wild type (Figure 2C). These results suggested that positive charge rather than acetylation of these lysines promotes import. In the context of full length Htz1-GFP, mutation of these residues had no effect on localization (data not shown). To prevent co-import via the H2B NLS we expressed full length htz1 K3,8,10,14Q-GFP2 in strains expressing H2B Δ1–32. The observed nuclear signal demonstrated that H2B Δ1–32 /Htz1 K3,8,10,14Q dimers were able to effectively access the nucleus (Figure 2D). This suggests that in the context of the full length protein the htz1 K3,8,10,14Q mutation does not abrogate NLS activity.
The Htz1 NLS directly interacts with several karyopherins
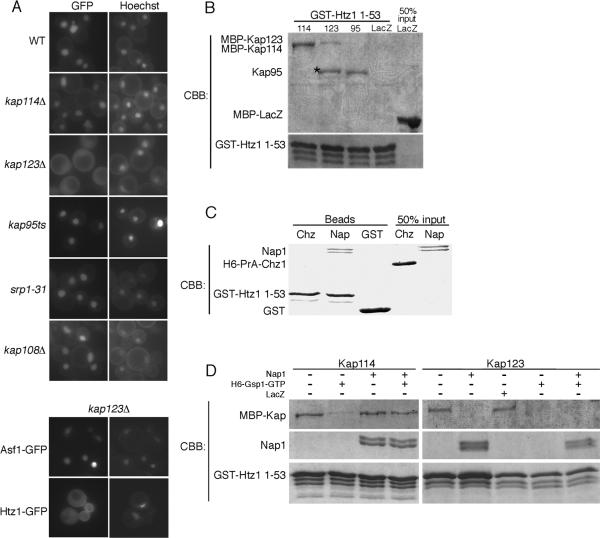
An Htz1/H2B dimer contains two distinct NLSs, and therefore the associated Kaps identified by mass spectrometry may associate with either the Htz1 or H2B NLS. To test which Kaps were important for Htz1 mediated import, we performed an import assay using the Htz11–53GFP2 reporter, which should not dimerize with endogenous H2B. We expressed Htz11–53GFP2 in strains containing a deletion or mutation of one of the Kap genes. Deletion of KAP123 resulted in strong mislocalization of the Htz11–53GFP2 reporter to the cytoplasm, but did not cause mislocalization of an Asf1-GFP2 reporter (Figure 3A). Deletion of KAP114 or KAP108/SXM1 had little affect on Htz11–53GFP2 reporter localization when compared to wild type (Figure 3A). Strains containing a mutant allele of KAP95(kap95ts) or SRP1 (srp1–31), also showed no significant mislocalization of the GFP reporter at the restrictive temperature (Figure 3A). As mislocalization of the NLS reporter was not observed in most of the single Kap deletion strains, even those Kaps that co-purified with Htz1-PrA, it suggested that Htz1 import was mediated by several Kaps. The mislocalization observed in the kap123Δ strain suggested that Kap123 is a major import karyopherin for Htz1.
Figure 3.
Htz1, Nap1, Kap114 form a co-complex. (A) The Htz1 NLS (residues 1–53) or Asf1 GFP2 reporter was expressed in yeast strains containing mutations of individual KAP genes as indicated and localized by fluorescent microscopy. Coincident Hoechst staining is shown. (B) Recombinant GST-Htz11–53 (250 nM) was immobilized on glutathione sepharose beads and incubated with the indicated recombinant Kap protein (MBP-Kap114 100 nM, MBP-Kap123 500 nM, Kap95 500 nM). MBP-LacZ (500 nM) was utilized as a negative control. After extensive washing, bound protein was analyzed by SDS-PAGE and Coomassie blue staining (CBB). A 50% input of MBP-LacZ is shown. * indicates a proteolytic product of MBP-Kap123. (C) Recombinant GST-Htz11–53 (400 nM) was immobilized on glutathione sepharose beads and incubated with either recombinant Nap1 (250 nM) or H6-PrA-Chz1 (500 nM). Similarly, GST (500 nM) was incubated with Nap1 (250 nM) as a control. After washing, protein was eluted from the beads and analyzed by SDS-PAGE and Coomassie stain. 50% input of Nap1 and H6-PrA-Chz1 are indicated. (D) Recombinant GST-Htz11–53 (400 nM) was immobilized on glutathione sepharose in the presence or absence of Nap1 (250 nM). MBP-Kap114 (100 nM) or MBP-Kap123 (300 nM) was pre-incubated with or without H6-Gsp1Q71L-GTP (50 μM) and/or MBP-LacZ (400nM) and then added to the binding reaction. After extensive washing, bound protein was analyzed by SDS-PAGE and Coomassie stain.
To determine whether the interaction between Htz1 and the Kaps detected in cytosol was direct or mediated through histone H2B, we tested these interactions using recombinant proteins. Htz1 residues 1–53 was expressed with a n N-terminal GST tag and immobilized on glutathione Sepharose beads. The beads were incubated with recombinant MBP tagged Kap114, Kap123, or untagged Kap95, and bound protein was analyzed by SDS-PAGE and Coomassie blue staining. Unlike the MBP-LacZ control protein, each of these Kaps was able to bind directly to the Htz1 NLS (Figure 3B), further supporting their proposed role in Htz1 transport. This suggests that Htz1 binds these Kaps directly.
Htz1, Nap1, and Kap114 form an import co-complex
Previously, we have demonstrated that Nap1 plays a role in H2A/H2B import and promotes association of the histones with Kap114 in the cytoplasm (22). In this study we used a RanGTP dissociation assay to demonstrate for the existence of an H2A-Nap1-Kap114p co-complex (22). In the absence of Nap1, RanGTP readily dissociates H2A from Kap114. However, the Nap1-Kap114p complex is insensitive to RanGTP, and we showed that addition of Nap1 to the Kap114-H2A complex, rendered this complex insensitive to RanGTP dissociation. This suggested that Nap1 was able to bridge the histone-Kap interaction (22). We used this assay again to determine whether Htz1, Kap114 and Nap1 formed a co-complex. Nap1 interacts with the NLS domain of H2A and we tested whether Nap1 directly associated with an Htz1-NLS fusion protein. Recombinant Nap1, but not Chz1, directly interacted with the GST-Htz11–53 (Figure 3C). Recombinant GST-Htz11–53 fusion protein was incubated with or without Nap1. MBP-Kap114 was then added with or without pre-incubation with Gsp1Q71L-GTP, a GTPase deficient yeast Ran mutant. As seen in Figure 3D, the interaction between GST-Htz11–53 and MBP-Kap114 was sensitive to Gsp1Q71L-GTP. In contrast, in the presence of Nap1, the GST-Htz11–53, MBP-Kap114 interaction was insensitive to Gsp1Q71L-GTP (Figure 3D). This result suggests that Nap1 can serve as a bridge between the Htz1 NLS and Kap114 in an import complex. In a parallel experiment, we showed that the GST-Htz11–53, MBP-Kap123 interaction was also sensitive to Gsp1Q71L-GTP (Figure 3D). However, in this case addition of Nap1 did not render the GST-Htz11–53, MBP-Kap123 complex insensitive to Gsp1Q71L-GTP. In fact, Nap1 appeared to compete with MBP-Kap123 for binding to GST-Htz11–53 (Figure 3D). In a control experiment we showed that the complex remained intact when incubated with the nonspecific protein MBP-LacZ (Figure 3D). This suggests that Kap123, Nap1 and Htz1 do not form a co-complex, and similarly to its role with H2A, Nap1 may promote binding of Htz1/H2B to Kap114 (22).
Chz1 contains an NLS and interacts with Kap95 in cytosol
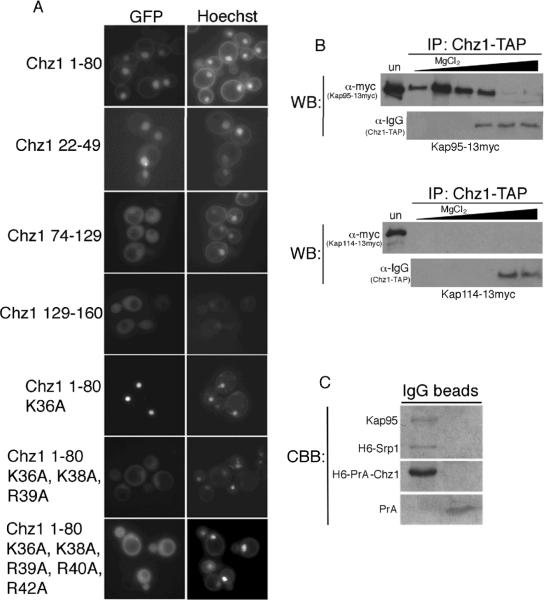
It has been argued that Nap1 and Chz1 are redundant chaperones for Htz1. Our Htz1 immunoprecipitation experiments suggest Chz1 did not interact with Htz1 in cytosol. As Chz1 is nuclear we wanted to determine whether Chz1 did indeed enter the nucleus independently of Htz1 and contained its own NLS (19). Chz1 was identified in a bioinformatics search for yeast nuclear proteins with consensus classical NLS (cNLS) sequences, although the putative Chz1 NLS was not examined experimentally (27). We tested whether the putative cNLS of Chz1 was functional using a GFP reporter assay. An N-terminal fragment of Chz1 (amino acids 1–80) was fused to GFP2 and expressed in wild type cells. The GFP signal was localized to the nucleus, suggesting that the N-terminus of Chz1 does contain NLS activity (Figure 4A). We then expressed Chz1 residues 22–49, which contain the predicted cNLS (36KPKR39), fused to GFP2 and expressed in wild type cells. The GFP signal was predominately localized to the nucleus, indicating that the cNLS is indeed functional (Figure 4A). Chz1 74–128-GFP2, which contains the Htz1/H2B-interacting “CHZ” domain, was localized throughout the cell (19) (Figure 4A). We also tested the C-terminus of Chz1 (residues 129–160) for NLS activity, but this reporter was also localized throughout the cell (Figure 4A).
Figure 4.
Chz1 contains an NLS and associates directly with Srp1/Kap95 in cytosol. (A) Fragments of Chz1, bearing the indicated mutations, fused to GFP2 were expressed in wild type yeast as indicated and visualized by fluorescence microscopy. Coincident Hoechst staining is shown. (B) Chz1-TAP was immunoprecipitated from cytosol prepared from strains expressing also Kap95-Myc or Kap114-Myc. Bound protein was eluted with MgCl2, precipitated, and analyzed by SDS-PAGE and Western blotting (WB) using anti-Myc antibodies. Chz1-TAP is recognized by IgG. `Un' lane contains 0.02% of depleted post-bead starting cytosol. (C) Recombinant H6-PrA-Chz1 (250 nM) was immobilized on IgG sepharose and incubated with H6-Srp1 (500 nM) and Kap95 (500 nM). zz protein (~250 nM) was used as a negative control. Bound protein was eluted and analyzed by SDS-PAGE and Coomassie blue staining (CBB).
To further delineate the NLS we made a series of lysine and arginine to alanine mutations in the context of Chz11–80GFP2. The mutant Chz11–80K36A, K38A, R39A, R40A, R42A-GFP2 was mislocalized to the cytoplasm (Figure 4A). We tested a mutant with fewer changes and showed that Chz11–80K36A, K38A, R39A-GFP2 was also mislocalized suggesting that these are key amino acids in the NLS (Figure 4A). In contrast mutation of one lysine was not sufficient as Chz11–80K36A GFP2 was localized to the nucleus (Figure 4A). These data suggest that Chz1 contains a cNLS in its N-terminus and that amino acids K36, K38 and K39 are critical residues.
The presence of a cNLS within Chz1 suggested import by the classical import pathway, which is mediated by the Srp1(Kap60)/Kap95 heterodimer. To test this we tagged KAP95 with a 13-Myc epitope in the CHZ1-TAP strain. If Chz1 was co-imported with Htz1, we would expect an interaction with Htz1-interacting Kaps, so we also tested Kap114. Chz1-TAP was immunopurified from cytosol and bound protein was analyzed by Western blotting. Kap95-Myc eluted in the 50mM to 1M MgCl2 fractions, consistent with Chz1 being an import cargo of Srp1/Kap95 (Figure 4B). No Kap114-Myc was detected in association with Chz1, suggesting that Chz1 does not use Kap114 for import (Figure 4B). We next tested if the Srp1/Kap95, Chz1 association was direct and we could assemble a Chz1-Srp1/Kap95 import complex in vitro. Recombinant H6-PrA-Chz1 was immobilized and incubated with H6-Srp1 and untagged Kap95. Analysis of bound protein by Coomassie blue staining showed that similar amounts of H6-Srp1 and Kap95 bound to H6-PrA-Chz1 (Figure 4C). Neither Srp1 nor Kap95 bound PrA alone. These results suggest that while Nap1 and Htz1 are co-imported, Chz1 is imported independently by the Srp1/Kap95 pathway.
We tested whether Chz1 (amino acids1–80) was mislocalized in strains bearing mutant alleles of srp1 and kap95 grown at the nonpermissive temperature. We did not see any obvious mislocalization in these strains suggesting that Chz1 may also have additional, secondary Kap-mediated pathways into the nucleus (data not shown).
Nap1, but not Chz1, maintains the soluble pool of Htz1
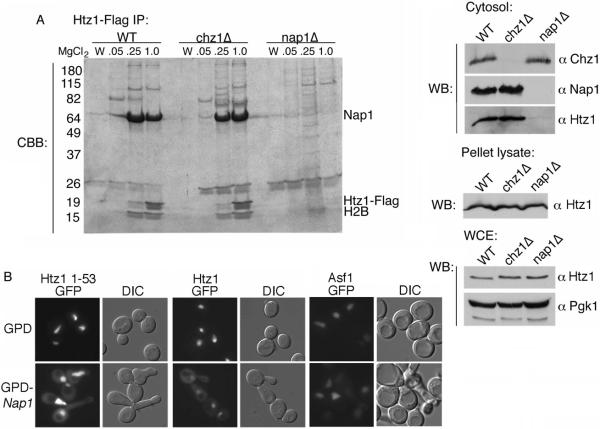
Our results predict that Nap1, but not Chz1, is an import cofactor for Htz1. This finding is consistent with the fact that we did not identify Chz1 in the Htz1 interacting proteins from cytosol and Chz1 does not interact with Kap114. As NAP1 is a non-essential gene, we wanted to determine whether in the absence of Nap1, Chz1 could serve as import cofactor and interact with Htz1 in cytosol. We generated cytosol from yeast strains expressing Htz1-FLAG in wild type, nap1Δ, or chz1Δ genetic backgrounds. Htz1-FLAG was immunoprecipitated using anti-FLAG agarose beads and associated proteins were eluted with MgCl2. Purified proteins were separated by SDS-PAGE and visualized with Coomassie blue stain. In the wild type strain, Htz1-FLAG was associated with Nap1, as well as H2B, and other copurifying proteins (Figure 5A). Surprisingly, in the nap1Δ strain we were not able to detect any soluble Htz1-FLAG by Coomassie stain and consequently no other major interacting proteins, including Chz1, were evident (Figure 5A). Parallel experiments using mutant strain chz1Δ were similar to wild type (Figure 5A). To verify that all our strains expressed Htz1-FLAG, we analyzed the cytosolic unbound fractions, the nuclei enriched pellet fractions, and whole cell lysates from each strain for the presence of Htz1-FLAG. In the absence of NAP1, Htz1-FLAG was not visible in the unbound cytosolic fraction as determined by Western blot, in contrast, Chz1 was apparent (Figure 5A). This could suggest that in the nap1Δ strain, cytoplasmic Htz1 is shifted to the nucleus. In the nuclei enriched pellet and whole cell lysate fractions, seemingly equivalent amounts of Htz1-FLAG were visible from all strains suggesting that the total cellular levels were unchanged (Figure 5A). This leaves the cytoplasmic fraction of Htz1 unaccounted for, however, as there is far more Htz1 in the nucleus, complete loss of Htz1 from the cytoplasm would be expected to only lead to a very modest increase in the nucleus which may be difficult to detect.
Figure 5.
Nap1 is important to maintain soluble pool of Htz1. (A) Htz1-FLAG was immunopurified from yeast cytosol from the indicated strain backgrounds. Bound protein was eluted with the indicated concentration of MgCl2 and analyzed by SDS-PAGE and Coomassie stain (CBB). The last wash fraction before elution is also shown (W). Indicated proteins are labeled according to molecular weight. Different cellular fractions were also analyzed for the presence of Htz1 as indicated. Cytosol: Proteins from the depleted, post-bead cytosol fractions (0.02% from above) were separated by SDS-PAGE and analyzed by Western blot (WB). Htz1 was detected via the Flag antibody. Pellet lysate: Nuclei enriched pellet fractions were analyzed by Western blot for the presence of Htz1-FLAG. WCE: Whole cell extracts of starting yeast cultures were analyzed by Western blot for the presence of Htz1-FLAG. Pgk1 levels were used as a loading control. (B) Htz11–53GFP2, Htz1-GFP2, or Asf1-GFP2 was expressed in wild type yeast containing pRS415GPD-NAP1 or empty vector (pRS415GPD). Localization of GFP was visualized by fluorescence and DIC microscopy.
This result further suggested that Chz1 does not associate with Htz1 in cytosol, whilst Nap1 appeared to be important for the presence of a cytoplasmic pool of Htz1. In this case, we predicted that overexpression of Nap1 would increase the cytosolic pool of Htz1 and lead to relocalization of the Htz11–53GFP2 reporter. To test this, NAP1 was overexpressed under the control of the yeast GPD1 promoter. Overexpression was confirmed by the presence of long budded cells, a phenotype indicative that Nap1 overexpression (Figure 5B) (35). In this strain the Htz11–53GFP2 reporter was partially relocalized to the cytoplasm, suggesting the excess Nap1 was increasing the amount of Htz11–53GFP2 in the cytoplasm at steady state (Figure 5B). We observed a similar result when we expressed full length Htz1-GFP2 (Figure 5B). These results were consistent with a role of Nap1 in maintaining a soluble pool of Htz1. To ensure that excess Nap1 did not result in a general transport defect, we also expressed Asf1-GFP2 in this strain and did not observe relocalization of the reporter to the cytoplasm (Figure 5B).
CHZ1 and NAP1 are not functionally redundant
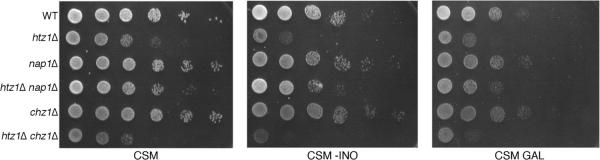
Our data suggests that Nap1 is a transport cofactor for Htz1/H2B and helps maintain a soluble pool of Htz1, and Chz1 does not appear to share these functions. However, both Nap1 and Chz1 have been described to deliver Htz1/H2B heterodimers to the SWR1 complex, indicating functional redundancy (19). We next wanted to explore if NAP1 and CHZ1 display similar genetic interactions indicative of the extent of their functional overlap. Deletion of NAP1 or CHZ1 did not lead to reduced fitness on YPD, while deletion of HTZ1 does lead to reduced fitness as exhibited by slow growth on YPD (11). If HTZ1 and its chaperones belong in a linear functional pathway, then a double mutant strain should have a similar growth phenotype as the htz1Δ null. We deleted NAP1 or CHZ1 in an htz1Δ null strain and observed the fitness of the double mutants on complete synthetic media (CSM). The htz1Δ mutant grew more slowly than wild type whereas both the nap1Δ and chz1Δ single mutants grew similarly to wild type (Figure 6). The chz1Δ htz1Δ double mutant grew similarly to htz1Δ (Figure 6). Surprisingly, the nap1Δ htz1Δ double mutant grew better than htz1Δ, indicating a positive genetic interaction between NAP1 and HTZ1 (Figure 6). htz1Δ null strains are sensitive to growth on media lacking inositol and media containing galactose as the carbon source, suggesting defects in transcription (11, 36). We tested nap1Δ htz1Δ and chz1Δ htz1Δ double mutants for growth defects on CSM lacking inositol (-INO) and CSM galactose (GAL). Yeast containing htz1Δ or chz1Δ htz1Δ grew more slowly on CSM GAL and CSM -INO than wild type, nap1Δ, or chz1Δ single mutants (Figure 6). The nap1Δ htz1Δ double mutant strain grew slightly less well than wild type but better than htz1Δ (Figure 6). These results suggest that deletion of NAP1 relieves the apparent transcriptional defects observed in an htz1Δ strain, a function that is not shared with CHZ1. This gives further evidence that the histone chaperones Nap1 and Chz1 have separate Htz1-dependent and -independent functions.
Figure 6.
NAP1, SWR1, and SWC2, but not CHZ1, exhibit genetic interactions with HTZ1. Strains of the indicated genotype were grown to logarithmic phase, equalized, and spotted as 10-fold serial dilutions onto CSM, CSM lacking inositol (CSM –INO), or CSM containing 2% galactose as the sole carbon source (CSM GAL) and grown at 30°C for 3 days.
Discussion
Transport of the histone variant Htz1 into the nucleus precedes its assembly into chromatin, and we wanted to determine whether common factors, such as histone chaperones, link the regulation of import with chromatin assembly. Here we show that the histone chaperone Nap1 functions in the nuclear import of Htz1. Surprisingly, we did not find any of the Htz1 chaperone Chz1 associated with Htz1 in cytosol, suggesting that Chz1 is not co-imported with Htz1, and that Nap1 and Chz1 play different roles in the biogenesis of Htz1-containing nucleosomes.
Histones contain N terminal targeting signals
Although imported as a dimer with H2B, our data indicated that Htz1, like H2B and the other core histones, contains an N terminal NLS. Similarly to H2A, the entire N terminus was required to specify full targeting of GFP, however in the context of the full protein we showed that the α1 helix contains an essential domain. The Htz1 N-terminus can be acetylated, and some studies have indicated that Htz1 is acetylated after nucleosome deposition, while another suggested that acetylation occurs prior to deposition suggesting a role in nuclear import, (8, 12, 13). Our NLS-GFP data suggests that the basic charge of lysine is an important NLS determinant, arguing against a role for acetylation in nuclear import. Surprisingly, in the context of the full-length protein, Htz1 K3,8,10,14Q appeared to be able to access the nucleus in the absence of the H2B NLS, suggesting that this mutation does not ablate NLS activity in vivo. It is not clear why we see a stronger defect with the acetylation site mutant in the context of the NLS fusion than the full-length protein, it is possible that while they are both imported at decreased rates, the full length protein is more efficiently retained in the nucleus. It is also possible that there is a second NLS in Htz1 C-terminus, although this would be inconsistent with the observed loss of nuclear localization of Htz1-52–134GFP in the absence of the H2B NLS. However, as GFP-fusions with C terminal fragments of Htz1 were poorly expressed, future experiments are needed to clarify this point.
Htz1 is imported by more than one karyopherin
We identified the Kaps responsible for Htz1/H2B import from cytosol and observed the most unique peptides from Kap114, followed by Kap123, suggesting that import occurs by more than one pathway. Previous purifications of Htz1 binding partners from whole cell lysates also identified Kap114 and Nap1, but surprisingly not Kap123 (19, 20). The direct binding between Htz1 and Kap114, and Kap123, and observation of a Kap114-Nap1-H2B-Htz1 co-complex, was also consistent with a role for Kap114 and Kap123 in Htz1 import. Unexpectedly, the Htz1 NLS reporter was strongly mislocalized in the absence of Kap123 but not in kap114Δ strain. Kap123, although not essential, is the most abundant karyopherin, and this result is comparable to that observed with H3-NLS-GFP2, which is also mislocalized in the kap123Δ genetic background (31). As Kap123 was not isolated previously from whole cell lysate experiments, but from cytosol as shown here, it is possible that the interaction between Kap123 and Htz1/H2B is short-lived and only stable in the cytoplasm(19, 20). This also correlates with the finding that this interaction is sensitive to RanGTP, and suggests that Kap123 plays a distinct role in Htz1 import.
Nap1 promotes Htz1 import
We showed Nap1 was in a complex with Htz1/H2B and Kap114, and similar to its role with H2A/H2B, we propose that Nap1 serves as a transport co-factor for Htz1/H2B import by promoting association of the dimer with Kap114 in the cytoplasm(22). Nap1 would likely only promote interaction of Htz1 with Kap114, as Nap1 and Kap123 do not form a co-complex with Htz1. We tested whether the import of an Htz1-NLS GFP reporter was reduced in the absence of Nap1. A small but statistically significant decrease was observed, where the nuclear to cytoplasmic ratio was 91 % of that observed in wild type cells (data not shown). The decrease is small because, as is shown here, Htz1 likely has several redundant import pathways. However, co-import of Nap1 and Htz1/H2B would provide an efficient mechanism to deliver the histones to the SWR1 complex for assembly. As Nap1 and Kap123 compete for Htz1 NLS binding, it suggests a model whereby Kap123 would deliver Htz1/H2B dimers to Nap1 or Chz1, once inside the nucleus.
Nap1 and Chz1 have distinct functions as Htz1 chaperones
Both Nap1 and Chz1 can deliver Htz1/H2B dimers to the SWR1 complex for histone exchange onto chromatin in vitro, and similar amounts of both proteins are found associated with Htz1 in whole cell lysates, leading to the suggestion that they have redundant functions in Htz1 assembly (19, 21). Unlike Nap1, which shuttles between the nucleus and cytoplasm, Chz1 appears to be a strictly nuclear protein and our data indicates that Chz1 contains a classical basic NLS and is a cargo of the classical import pathway. Consequently, we propose that the histone chaperone functions of Chz1 all occur within the nucleus, in coordination with the SWR1 complex. In contrast, Nap1 is necessary for maintaining the soluble pool of Htz1 in the cytoplasm, and we propose that Nap1 maintains a dynamic pool of histones in the cell. These data are in agreement with Luk et al, who showed that there was virtually no free Htz1 in the cell and that in the absence of Nap1, the fractionation pattern of total cellular Htz1 by glycerol gradient centrifugation was altered, although they did not carry out subcellular fractionation (19). This suggests that Nap1 is a bona fide chaperone for Htz1, whereas Chz1 may play a more specialized role in chromatin assembly. It is possible that Nap1 has a function in buffering the amount of free soluble Htz1/H2B in the cell that is competent for nucleosome assembly, similar to the function described for Asf1 and H3/H4 dimers (37).
Chaperones have specialized and redundant functions
Previous analysis of nap1Δchz1Δ double mutant did not uncover any genetic interactions (19). The authors also showed that recruitment of Htz1 to two promoters appeared normal in the absence of both chaperones, although a more recent paper analyzing Chz1, has shown a decrease in Htz1 promoter recruitment at specific genes in the absence of Chz1(19, 38). Our genetic analysis of htz1Δ double mutants indicated that concomitant loss of NAP1, but not CHZ1, partially rescued the growth defect observed with the htz1Δ deletion. This was most apparent when plated on media that put the cells under transcriptional stress. Why would loss of NAP1 result in better cell growth? It is possible that in the absence of HTZ1, Nap1 may sequester H2A, or load H2A in the place of Htz1 leading to H2A containing nucleosomes in the promoter. These aberrant nucleosomes would be deleterious for cell growth if they lead to defects in transcription, possibly due to reduced nucleosome mobility or impaired activation and histone eviction. In this case, the simultaneous loss of NAP1 would reduce H2A misincorporation. The lack of a genetic interaction between CHZ1 and HTZ1 gives more evidence that Chz1 and Nap1 play distinct roles, and indicates that Chz1 function may be restricted Htz1 assembly/disassembly. Chz1 has recently been shown to interact with nucleosomal Htz1 (39). It is possible that although Chz1 acts as a donor and acceptor for Htz1, its function is spatially restricted to the surface of chromatin, in contrast to the more dynamic function of the nucleo-cytoplasmic shuttling protein Nap1.
It is interesting that most histone chaperones, such as Nap1 and Chz1, are not encoded by essential genes, and many seem to have overlapping functions. This suggests that there is a high degree of functional redundancy, however, as discussed here, it has been shown that each chaperone most likely does have some degree of functional specialization(14, 16). It has been speculated that the diversification of histone chaperones may have evolved in parallel to the diversification of histones themselves, this is particularly apparent in higher eukaryotes, where many histone variants have been identified(40, 41). Interestingly the karyopherin family is also very redundant, and although specific cargoes have been identified for most karyopherins, how cargoes are recognized and imported by several different karyopherins at the molecular level is not well understood.
Is the function of Nap1 and Chz1 conserved in human cells?
These results suggest a model whereby Htz1 and H2B are made in the cytoplasm and immediately bound by the chaperone Nap1. As Htz1 is made throughout the cell cycle, but H2B is made mainly in S phase, we speculate that Nap1 interacts with either histone in the cytoplasm, until the dimer partner is available. In this way Nap1 may play an important role in maintaining a soluble pool of histone dimers in the cytoplasm. We propose that Nap1 then targets the dimer to Kap114, in addition, other dimers interact directly with Kap123. The histone dimers are subsequently imported into the nucleus. Acetylation likely takes place once in the nucleus, which may help trigger the Ran-mediated release of the Kaps. Nap1-bound dimers can be targeted to chromatin and assembled by the Swr1 complex, and dimers not bound by Nap1 may be passed directly to the other chaperone Chz1. How the chaperones are targeted to the Swr1 complex, and how the complex is targeted to the correct promoters is not well understood and it is possible that chaperones themselves recognize different chromatin domains or differently modified histones. Nap1 and Chz1 are both associated with chromatin and it is likely that a pool of these chaperones also act there as histone acceptors and donors during transcription.
These findings are likely to be relevant to H2A.Z import and assembly in human cells. In human cells there are orthologs of Kap114 and Kap123, which have been shown to import core histones although, a direct role in H2A.Z import has yet to be investigated(18, 42). Nap1 has several conserved orthologs in human cells, which have been shown to function as a histone chaperones(43). There is not a direct human homolog of Chz1, although interestingly the nuclear protein HIRIP3 shares homology with Chz1 in the histone binding domain, and has been shown to interact with histones and the human HIRA histone chaperone(19). The Swr1 complex is also conserved in human cells, and future experiments are needed to determine whether human Nap1 and HIRIP3 play a role in the import of H2A.Z, and as histone chaperones for H2A.Z nucleosome incorporation.
In summary, here we have demonstrated here that the yeast histone chaperones Nap1 and Chz1 play different roles in the import and biogenesis of Htz1- containing nucleosomes.
Materials and Methods
Yeast strains and plasmids
Yeast strains containing HTZ1-PrA and NAP1-PrA were created in DF5 as previously described (44, 45). Strain BY4741 CHZ1-TAP was obtained commercially (Open Biosystems). Strains containing KAP114-13MYC-kanMX4 or KAP95-13MYC-kanMX4 were derived from DF5 HTZ1-PrA∷HIS3∷URA3 and BY4741 CHZ1-TAP∷kanMX4 by targeted integration of the Myc tag to the C-terminus of each ORF as described (46). Strains containing KAP123-13MYC-kanMX4, SXM1-13MYC-kanMX4, or KAP120-13MYC-kanMX4 were derived from DF5 HTZ1-PrA∷HIS3∷URA3 as described above. Strain JHY200, which contains genomic deletions of each core histone gene, was previously described (34). Mutant strain JHY200 htz1Δ∷TRP1 was created by targeted integration of TRP1 to replace the HTZ1 ORF. Strains containing deletions or mutations of Kap genes (kap114Δ, kap123Δ, kap108Δ, kap95ts, and srp1-31) have been described elsewhere (47–51). Strains expressing Htz1-FLAG (MBY137, HTZ1-3FLAG-kanMX; yEL011, HTZ1-3FLAG-kanMX chz1Δ∷hphMX4; yEL012, HTZ1-3FLAG-kanMX nap1Δ∷natMX4; yEL013, HTZ1-3FLAG-kanMX chz1Δ∷hphMX4 nap1Δ∷natMX4) were generously provided by C. Wu and previously described (19). BY4741 strains containing htz1Δ∷kanMX4 or swc2Δ∷kanMX4 were obtained commercially (Open Biosystems). Strain Y2454 nap1Δ∷natMX4 was previously described (25). The htz1Δ∷kanMX4 nap1Δ∷natMX4 double mutant strain was created by mating BY4741 htz1Δ∷kanMX4 with Y2454 nap1Δ∷natMX4 and dissecting tetrads. The mutant strains chz1Δ∷HIS3 and swr1Δ∷HIS3 were created by replacing the respective ORFs with HIS3 in BY4741. The double mutant strains htz1Δ∷kanMX4 chz1Δ∷HIS3 and htz1Δ∷kanMX4 swr1Δ∷HIS3 were derived from BY4741 htz1Δ∷kanMX4 by replacing the respective ORFs with HIS3. The double mutant strain htz1Δ∷HIS3 swc2Δ∷kanMX4 was derived from BY4741 swc2Δ∷kanMX4 by replacing the HTZ1 ORF with HIS3.
Htz1-GFP2 reporter plasmids were created by inserting full length HTZ1 or DNA fragments encoding residues 1–53, 1–24, 24–53, 32–134, or 52–134 into plasmid pGFP2-C-FUS previously described (32). phtz1K3,8,10,14Q-GFP2, phtz11–53K3,8,10,14Q-GFP2 and phtz11–53K3,8,10,14R-GFP2 plasmids were created by inserting PCR fragments generated from plasmids pCM369 (HA-htz1K3,8,10,14Q-PTEFKanMX6-TTEF) or pCM368 (HA-htz1K3,8,10,14R-PTEF-KanMX6-TTEF), generously provided by M. Grunstein (8), into pGFP2-C-FUS. Chz1-GFP2 reporter plasmids were created by inserting CHZ1 DNA fragments encoding residues 1–80, 22–49, 74–128, or 129–160 into plasmid pGFP2-C-FUS. The Asf1-GFP2 plasmid was created by inserting the entire ASF1 orf into pGFP2-C-FUS. Yeast expression plasmids pRS416ADH-HTZ1 and pRS416ADH-Htz1K3,8,10,14Q were created by inserting the corresponding sequence of HTZ1 into pRS416 under control of the ADH promoter. Equal expression was checked by western blotting for Htz1 (data not shown). The pQQ18 plasmid containing htb1Δ1–32 was created by single strand mutagenesis of the wild type pQQ18 plasmid previously described (34). The NAP1 overexpression plasmid pRS425GPD-NAP1 was previously described (25). Bacterial expression plasmid pGEX4T1-Htz11–53 was created by inserting a DNA fragment encoding HTZ1 residues 1–53 into pGEX-4T1. Expression plasmids pGEX-4T1-NAP1, pMAL-KAP114, pMAL-KAP123, pGEX-2T-KAP95, and pQE9-GSP1-Q71L have been described elsewhere (22, 52, 53). Plasmid pQE-SRP1 was kindly provided by M. Floer. Expression plasmid pET28a+-CHZ1 (pEL0210) was provided by C. Wu (19).
Microscopy
Yeast expressing GFP reporters were subjected to fluorescence microscopy using a Nikon Microphot-SA microscope equipped with a X100 objective. GFP expression from the MET25 promoter was induced by growing cells in medium lacking methionine for 4hrs, except where extended incubation times are noted. GFP images were captured with OpenLab software using 500ms (Fig. 2A and Fig 2D) or 400ms (Htz1-52–134 in Fig. 2A, Fig. 2B and Fig 2C) exposure settings and each GFP figure part manipulated identically using Adobe Photoshop.
Purification of recombinant proteins and in vitro binding assays
Bacterially expressed recombinant GST tagged Htz11–53 and MBP tagged Kap114, Kap123 and LacZ were purified with glutathione Sepharose according to the manufacturer's instructions (NEB and GE Healthcare). Untagged recombinant Nap1 and Kap95 were generated by cleaving the GST tag from the fusion proteins using thrombin protease (Sigma). H6-PrA-Chz1 was purified under denaturing conditions as described (19). H6-Gsp1Q71L-GTP was purified from E. coli grown in LB containing 0.05 μg/mL kanamycin and 0.1 μg/mL ampicillin at 37°C until an OD A600 of 1. Protein expression was induced with 0.3 mM IPTG and grown at 37°C for 5 hours. Cells were collected by centrifugation and lysed in Lysis buffer (50 mM NaH2PO4 pH 8, 300 mM NaCl, 10 mM imidazole, 2 mM MgOAc, 0.2 mM GTP, 20% glycerol, 1 μg/mL pepstatin, and 0.1 mg/mL PMSF) using a French press. After centrifugation for 15 min at 30,000 Xg, 0.1% Tween-20 and 0.1 mM ATP were added. The lysate was incubated with Ni-NTA beads for 1 hr at 4°C. Beads were washed, in batch, with Lysis buffer followed by Wash buffer (50 mM NaH2PO4 pH 8, 300 mM NaCl, 20 mM imidazole, 2 mM MgOAc, 0.2 mM GTP, 20% glycerol, 0.1% Tween-20) plus 0.1 mM ATP, then Wash buffer plus 0.5% Tween-20, and finally with Wash buffer. The beads were loaded onto a mini-column and protein was eluted with Elution buffer (50 mM NaH2PO4 pH 8, 300 mM NaCl, 250 mM imidazole, 2 mM MgOAc, 0.2 mM GTP, 20% glycerol, 0.1% Tween-20). 2 mM GTP and 20 mM EDTA pH 8 were added to the eluted protein to generate H6-Gsp1Q71L-GTP. For in vitro binding assays, stated amounts of recombinant protein were added to a binding reaction with a total volume of 100 μL and incubated at 4°C with agitation. Transport buffer (TB: 20 mM Hepes, pH7.5, 110 mM KOAc, 2 mM MgCl2, 1 mM DTT, 0.1% Tween-20) containing 15% glycerol was used for binding reactions. Beads were pre-blocked in TB containing 10% BSA with a final concentration of 1.5% BSA per reaction. After incubation, beads were washed with TB containing 400 mM KOAc (except for Figure 4C where 110 mM KOAc was used). Bound protein was eluted with 4X sample buffer and analyzed by SDS-PAGE.
Immunoprecipitations
Cytosol fractions were prepared from 1 L of exponentially growing yeast as previously described (54). Briefly, cells were washed, spheroplasted with 10% glusulase in 1 M sorbitol, and lysed using a Polytron in 8% polyvinylchloride (PVP) buffer containing protease inhibitors. Centrifugation over a 0.3 M sucrose cushion yielded a crude cytosol fraction and nuclei enriched pellet. The cytosolic fraction was clarified by ultracentrifugation. Cytosol from cells expressing Htz1-PrA, Nap1-PrA, or Chz1-TAP was incubated with IgG Sepharose to purify the tagged protein. Cytosol from Htz1-FLAG expressing cells was incubated with anti-FLAG agarose (Sigma). Bound proteins were precipitated with MgCl2 and analyzed by SDS-PAGE. For the cells expressing Htz1-FLAG, the nuclei enriched pellet was washed with 8% PVP solution and centrifuged again over a 0.3 M sucrose cushion. The pellet was resuspended in 18.5 mL of TB and 5 mL of 8% PVP with protease inhibitors. 100 μL of material from the resuspended pellet was lysed by sonication in 4X sample buffer. Proteins were visualized by Coomassie brilliant blue (CBB) stain or by Western blot (WB) using specific antibodies. The anti-Chz1 antibody was a gift from C. Wu.
Nano-HPLC Microelectrospray Ionization Mass Spectrometry Analysis and Database Searching
Proteins co-purifying with Htz1-PrA isolated from 1 L of exponentially growing cells were submitted for analysis by Nano-HPLC Microelectrospray Ionization Mass Spectrometry as previously described (32). A list of interacting proteins was generated using SEQUEST as previously described (32).
Acknowledgements
We thank Drs. Don Hunt and Yurong Guo for Mass Spectrometry data, Dr. Carl Wu for anti Chz1 antibody, we thank Sarah Wilkinson and Dr. Nima Mosammaparast for their contributions to the initial stages of the project, and Drs M. Mitchell Smith and Meredith Calvert for reading the original version of the manuscript.
The project was funded by research grant NIH GM065385 to LFP.
References
- 1.Luger K. Structure and dynamic behavior of nucleosomes. Current opinion in genetics & development. 2003;13(2):127–135. doi: 10.1016/s0959-437x(03)00026-1. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Guillemette B, Gaudreau L. Reuniting the contrasting functions of H2A.Z. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2006;84(4):528–535. doi: 10.1139/o06-077. [DOI] [PubMed] [Google Scholar]
- 4.Raisner RM, Madhani HD. Patterning chromatin: form and function for H2A.Z variant nucleosomes. Current opinion in genetics & development. 2006;16(2):119–124. doi: 10.1016/j.gde.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Zlatanova J, Thakar A. H2A.Z: view from the top. Structure. 2008;16(2):166–179. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS biology. 2005;3(12):e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li B, Pattenden SG, Lee D, Gutierrez J, Chen J, Seidel C, Gerton J, Workman JL. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(51):18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes & development. 2006;20(6):711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5' ends of both active and inactive genes in euchromatin. Cell. 2005;123(2):233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123(2):219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santisteban MS, Kalashnikova T, Smith MM. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000;103(3):411–422. doi: 10.1016/s0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- 12.Babiarz JE, Halley JE, Rine J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes & development. 2006;20(6):700–710. doi: 10.1101/gad.1386306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keogh MC, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, Buratowski S. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes & development. 2006;20(6):660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eitoku M, Sato L, Senda T, Horikoshi M. Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell Mol Life Sci. 2008;65(3):414–444. doi: 10.1007/s00018-007-7305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loyola A, Almouzni G. Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta. 2004;1677(1–3):3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 16.Rocha W, Verreault A. Clothing up DNA for all seasons: Histone chaperones and nucleosome assembly pathways. FEBS letters. 2008 doi: 10.1016/j.febslet.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 17.De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: an escort network regulating histone traffic. Nature structural & molecular biology. 2007;14(11):997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- 18.Jakel S, Mingot JM, Schwarzmaier P, Hartmann E, Gorlich D. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. The EMBO journal. 2002;21(3):377–386. doi: 10.1093/emboj/21.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luk E, Vu ND, Patteson K, Mizuguchi G, Wu WH, Ranjan A, Backus J, Sen S, Lewis M, Bai Y, Wu C. Chz1, a nuclear chaperone for histone H2AZ. Molecular cell. 2007;25(3):357–368. doi: 10.1016/j.molcel.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 20.Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS biology. 2004;2(5):E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science (New York, NY. 2004;303(5656):343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 22.Mosammaparast N, Ewart CS, Pemberton LF. A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. The EMBO journal. 2002;21(23):6527–6538. doi: 10.1093/emboj/cdf647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park YJ, Chodaparambil JV, Bao Y, McBryant SJ, Luger K. Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J Biol Chem. 2005;280(3):1817–1825. doi: 10.1074/jbc.M411347200. [DOI] [PubMed] [Google Scholar]
- 24.Calvert ME, Keck KM, Ptak C, Shabanowitz J, Hunt DF, Pemberton LF. Phosphorylation by casein kinase 2 regulates Nap1 localization and function. Molecular and cellular biology. 2008;28(4):1313–1325. doi: 10.1128/MCB.01035-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Del Rosario BC, Pemberton LF. Nap1 links transcription elongation, chromatin assembly, and messenger RNP complex biogenesis. Molecular and cellular biology. 2008;28(7):2113–2124. doi: 10.1128/MCB.02136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mosammaparast N, Pemberton LF. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends in cell biology. 2004;14(10):547–556. doi: 10.1016/j.tcb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 27.Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282(8):5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nature reviews. 2007;8(3):195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 29.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annual review of biochemistry. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 30.Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic (Copenhagen, Denmark) 2005;6(3):187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- 31.Mosammaparast N, Guo Y, Shabanowitz J, Hunt DF, Pemberton LF. Pathways mediating the nuclear import of histones H3 and H4 in yeast. J Biol Chem. 2002;277(1):862–868. doi: 10.1074/jbc.M106845200. [DOI] [PubMed] [Google Scholar]
- 32.Mosammaparast N, Jackson KR, Guo Y, Brame CJ, Shabanowitz J, Hunt DF, Pemberton LF. Nuclear import of histone H2A and H2B is mediated by a network of karyopherins. The Journal of cell biology. 2001;153(2):251–262. doi: 10.1083/jcb.153.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dilworth DJ, Tackett AJ, Rogers RS, Yi EC, Christmas RH, Smith JJ, Siegel AF, Chait BT, Wozniak RW, Aitchison JD. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. The Journal of cell biology. 2005;171(6):955–965. doi: 10.1083/jcb.200509061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blackwell JS, Jr., Wilkinson ST, Mosammaparast N, Pemberton LF. Mutational analysis of H3 and H4 N termini reveals distinct roles in nuclear import. J Biol Chem. 2007;282(28):20142–20150. doi: 10.1074/jbc.M701989200. [DOI] [PubMed] [Google Scholar]
- 35.Calvert ME, Lannigan JA, Pemberton LF. Optimization of yeast cell cycle analysis and morphological characterization by multispectral imaging flow cytometry. Cytometry A. 2008;73(9):825–833. doi: 10.1002/cyto.a.20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adam M, Robert F, Larochelle M, Gaudreau L. H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Molecular and cellular biology. 2001;21(18):6270–6279. doi: 10.1128/MCB.21.18.6270-6279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Groth A, Ray-Gallet D, Quivy JP, Lukas J, Bartek J, Almouzni G. Human Asf1 regulates the flow of S phase histones during replicational stress. Molecular cell. 2005;17(2):301–311. doi: 10.1016/j.molcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 38.Wan Y, Saleem RA, Ratushny AV, Roda O, Smith JJ, Lin CH, Chiang JH, Aitchison JD. Role of the histone variant H2A.Z/Htz1p in TBP recruitment, chromatin dynamics, and regulated expression of oleate-responsive genes. Molecular and cellular biology. 2009;29(9):2346–2358. doi: 10.1128/MCB.01233-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Z, Feng H, Hansen DF, Kato H, Luk E, Freedberg DI, Kay LE, Wu C, Bai Y. NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B. Nature structural & molecular biology. 2008;15(8):868–869. doi: 10.1038/nsmb.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eirin-Lopez JM, Gonzalez-Romero R, Dryhurst D, Ishibashi T, Ausio J. The evolutionary differentiation of two histone H2A.Z variants in chordates (H2A.Z-1 and H2A.Z-2) is mediated by a stepwise mutation process that affects three amino acid residues. BMC Evol Biol. 2009;9:31. doi: 10.1186/1471-2148-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frehlick LJ, Eirin-Lopez JM, Ausio J. New insights into the nucleophosmin/nucleoplasmin family of nuclear chaperones. Bioessays. 2007;29(1):49–59. doi: 10.1002/bies.20512. [DOI] [PubMed] [Google Scholar]
- 42.Muhlhausser P, Muller EC, Otto A, Kutay U. Multiple pathways contribute to nuclear import of core histones. EMBO Rep. 2001;2(8):690–696. doi: 10.1093/embo-reports/kve168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zlatanova J, Seebart C, Tomschik M. Nap1: taking a closer look at a juggler protein of extraordinary skills. FASEB J. 2007;21(7):1294–1310. doi: 10.1096/fj.06-7199rev. [DOI] [PubMed] [Google Scholar]
- 44.Finley D, Ozkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48(6):1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- 45.Aitchison JD, Rout MP, Marelli M, Blobel G, Wozniak RW. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. The Journal of cell biology. 1995;131(5):1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mosammaparast N, Del Rosario BC, Pemberton LF. Modulation of histone deposition by the karyopherin kap114. Molecular and cellular biology. 2005;25(5):1764–1778. doi: 10.1128/MCB.25.5.1764-1778.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koepp DM, Wong DH, Corbett AH, Silver PA. Dynamic localization of the nuclear import receptor and its interactions with transport factors. The Journal of cell biology. 1996;133(6):1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loeb JD, Schlenstedt G, Pellman D, Kornitzer D, Silver PA, Fink GR. The yeast nuclear import receptor is required for mitosis. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(17):7647–7651. doi: 10.1073/pnas.92.17.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pemberton LF, Rosenblum JS, Blobel G. Nuclear import of the TATA-binding protein: mediation by the karyopherin Kap114p and a possible mechanism for intranuclear targeting. The Journal of cell biology. 1999;145(799315910):1407–1417. doi: 10.1083/jcb.145.7.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenblum JS, Pemberton LF, Blobel G. A nuclear import pathway for a protein involved in tRNA maturation. The Journal of cell biology. 1997;139(798075079):1655–1661. doi: 10.1083/jcb.139.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rout MP, Blobel G, Aitchison JD. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89(5):715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- 52.Maurer P, Redd M, Solsbacher J, Bischoff FR, Greiner M, Podtelejnikov AV, Mann M, Stade K, Weis K, Schlenstedt G. The nuclear export receptor Xpo1p forms distinct complexes with NES transport substrates and the yeast Ran binding protein 1 (Yrb1p) Molecular biology of the cell. 2001;12(3):539–549. doi: 10.1091/mbc.12.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83(5):683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- 54.Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science (New York, NY. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]