Abstract
Chemical address tags can be defined as specific structural features shared by a set of bioimaging probes having a predictable influence on cell-associated visual signals obtained from these probes. Here, using a large image dataset acquired with a high content screening instrument, machine vision and cheminformatics analysis have been applied to reveal chemical address tags. With a combinatorial library of fluorescent molecules, fluorescence signal intensity, spectral, and spatial features characterizing each one of the probes' visual signals were extracted from images acquired with the three different excitation and emission channels of the imaging instrument. With multivariate regression, the additive contribution from each one of the different building blocks of the bioimaging probes towards each measured, cell-associated image-based feature was calculated. In this manner, variations in the chemical features of the molecules were associated with the resulting staining patterns, facilitating quantitative, objective analysis of chemical address tags. Hierarchical clustering and paired image-cheminformatics analysis revealed key structure-property relationships amongst many building blocks of the fluorescent molecules. The results point to different chemical modifications of the bioimaging probes that can exert similar (or different) effects on the probes' visual signals. Inspection of the clustered structures suggests intramolecular charge migration or partial charge distribution as potential mechanistic determinants of chemical address tag behavior.
Keywords: Cheminformatics, machine vision, bioimaging, fluorescence, high content screening, image cytometry, combinatorial chemistry
INTRODUCTION
Microscopic imaging studies examining the interaction of small fluorescent molecules with cells are challenging because cells are complex three-dimensional objects that may exist in many different structural and functional states(1–3). From highly branched neuronal cells to multinucleated myocytes, the morphological features of any particular cell type can be quite varied, and for any growing cell population, there is cell-to-cell variation in the structure, function and spatial relationships between the different cellular organelles. In addition, the quantum yield and spectral properties of fluorescent molecules can be affected by local microenvironments within cells, and by interaction with specific cellular components.
Nevertheless, to optimize the signal of a bioimaging probe for specific applications, the interactions between molecules and cells are generally studied directly based on the fluorescence staining phenotype apparent in images of cells incubated with the probes (4–6). These fluorescence staining phenotypes can be visualized using automated microscopes equipped with specialized optics and filter sets to excite the molecules with light and capture images recording the fluorescence emission patterns at specific wavelengths (7–10). Development of fluorescent, organelle-targeted probes has been driven by an interest in discovering new probes that excite and emit in the visible spectrum, and that possess specific subcellular localization features so they can be used as organelle markers or physiological biosensors (9,11–14).
Today, high content screening instruments can generate large image data sets with combinatorial libraries of fluorescent probes(15–18). Although bioimaging probe development has traditionally relied on visual inspection by human experts, new analysis strategies are being pursued to quantitatively assess relationships between the chemical structures of fluorescent small molecules and cell-associated visual signals (16,18,19). These strategies combine basic image feature extraction algorithms (20,21), machine vision techniques derived from the study of location proteomics (22–28) and multivariate analysis and cheminformatics techniques used to study the activity of compounds across multiple different assays (1,2,29–32). Ultimately, probe optimization studies could benefit from objective analysis of how the chemical features of a fluorescent probe are related to cell-associated, quantitative image features.
In the case of styryl molecules, their simple bipartite structure lends itself to analysis in terms of differential contribution of the two basic building blocks of the molecule to the spectral and subcellular localization properties of the molecules' fluorescence (17–19). Using a high content screening dataset, we previously determined that many styryl molecules exhibit idiosyncratic interactions with cells, leading to very different staining phenotypes even amongst closely related isomers (18). In spite of these idiosyncratic interactions, visualization of the images strongly suggested that much of the variation in localization could be related to the molecule's chemical structure. Thus, we hypothesized that the building blocks of the molecules could behave as “chemical address tags”, and proceeded to determine the extent to which cell-associated image-based features derived from the images could be linked to additive contributions of the chemical building blocks of the molecules.
METHODS
Data acquisition and preprocessing
Synthesis and screening of the styryl library, and image data acquisition and preprocessing steps have been previously described(6,18). Briefly, each styryl molecule was synthesized from a conjugation reaction between 168 aldehyde building blocks with 8 methyl pyridinium/quinolinium building blocks. For notation purposes, each aldehyde building block is referred to as a number from 1 to 168, and the each pyridinium/quinolinium building block is referred to as a letter (from A to H) (6, 18). To facilitate data acquisition and analysis, an orthogonal fluorescent dye (Hoechst™ 33342) was used to label the cell nucleus. After incubation of Hela cells with the individual probes in 96 well plates, images were a acquired at 20× magnification with a Cellomics™ Kineticscan high content screening instrument, using the standard XF93 filter set's Hoechst™, FITC, TRITC, Cy5 acquisition channels(18). Analysis of the styryl molecules' fluorescence signal was based on 1 sec exposure images acquired from the FITC, TRITC and Cy5 channels. Each set of images was computationally and manually filtered to remove images with saturation or showing extensive dye precipitates or crystals. Using the Hoechst™ channel image, nuclear pixels were automatically identified using a thresholding algorithm, and images were background subtracted, by subtracting the median of pixel intensities of the noncellular region (pixels more than 10 pixels distant from any nuclear pixel). All analyses were based on whole field image features (described below), calculated for the cellular region or total nuclear region in each image. The cellular region was defined as a 5 pixel dilation of the nuclear region so as to sample the signal from the cytoplasm.
Extraction of quantitative image features
From the images, we measured a set of image-based features capturing the intensity level and distribution of probe fluorescence in cells, within and across fluorescence channels. The “integrated intensity” over a region is defined as the sum of pixel intensities of every pixel encompassing that region. In turn, dividing the integrated intensity by the number of pixels in a region corresponds to the mean (or average) pixel intensity over that region. In this manner, the average pixel intensity for all pixels within the cellular region was calculated in FITC, TRITC and Cy5 channels (excluding the Hoechst™ channel). With a control set of background-subtracted images acquired from unlabeled cells, we determined that < 1% of the images showed mean cellular autofluorescence > 100 units/pixel, so this value was chosen as a threshold for distinguishing those styryl molecules that yielded a cell-associated fluorescence signal. The average pixel intensity for all pixels within the cellular regions of FITC, TRITC and Cy5 was then summed to create a total cellular intensity (TCI) feature image. In addition, the cellular intensities for the three fluorescence channels were normalized by their sum to produce three “channel proportion intensity” (CPI) features. These values were not computed for compounds where the sum of mean pixel intensities was < 100 units/pixel. Coefficient of variation (CV) of the cellular regions was calculated as the standard deviation of pixel intensities for pixels within cells divided by the mean pixel intensity. The CV value was not computed when the mean intensity in a given image was < 100 units/pixel. “Cytoplasm to nucleus ratio” (CNR) was calculated as the ratio of the mean intensity in the cytoring region (the cellular region excluding the nuclear region) to the mean intensity in the nuclear region for a given image. The CNR was not computed when the mean intensity in the nucleus was < 100 units/pixel. To quantify the partitioning of probe fluorescence inside the cell in relation to the background fluorescence intensity, a “cell associated fluorescence” feature was computed as the ratio of mean cellular fluorescence (after background subtraction) to the median background intensity (corresponding to the probes' fluorescence in solution, before background subtraction). The CV, CNR and cell associated fluorescence features were computed for each channel, and also for the sum of the pixel intensities from all channels.
Extraction of reference image features
Several image features were also computed as controls or reference features: The size and number of cells in an image, quantified as the total number of pixels in either cell nuclei, or in the cellular region. The background intensity (BM) of each of the images acquired corresponds to the median background pixel intensity as used in background subtraction. The distribution of cells in an image was described in terms of the proportion of nuclear pixels in each of the four image quadrants (NQ1–NQ4). We also considered three features that represent the orientations and shapes of the cell nuclei in an image. These features were computed by first identifying all connected regions in the nuclear region consisting of more than 100 pixels in area. The x,y coordinates of the pixels in each connected region were used to construct a 2 × 2 covariance matrix of the x,y coordinates. This covariance matrix was then decomposed according to its eigenvectors to identify the principal axis (the dominant eigenvector) and the reciprocal aspect ratio (the ratio of the larger to the smaller eigenvalue). The median of the reciprocal aspect ratios across all regions in the cell (AR) was used to describe the aspect ratios of nuclei in the image. The angle of the principal axis for each nuclear region was also computed. This angle was measured relative to the increasing horizonal axis, using an origin placed at the center of mass of each cell. Angles greater than π radians were converted to lie in the range (0, π) by subtracting π radians, since the principal axis has no specific orientation. We then calculated the angular mean (AM) of these angles over all nuclei in an image. The AM was calculated by considering the angles for all cells in the image as points on a common unit circle (scaling the angles by 2 to cover the full unit circle). The arithmetic centroid of these points was then calculated, and scaled to a unit vector. The angle of this vector relative to the increasing horizontal axis divided by 2 to yield an angle between 0 and π that we used as the AM. An AM of π /2 radians corresponds to no favored orientation of the cell nuclei. In addition, the distance from the centroid to the origin (which is the centroid of all pixels in a nucleus) was also computed and used as a measure of variability, called “angular variance.”
Statistical analysis of chemical address tags
Regression methods were used to assess the extent to which additive contributions associated with the aldehyde and pyridinium/quinolinium building blocks could be used to describe the variation in image features. We first asked whether image features were predictable from additive contributions of the aldehyde and pyridinium/quinolinium building blocks. Ridge regression with a nominal ridging parameter of 1, with additive factorial main effects for the aldehyde and pyridinium/quinolinium moieties, was used for prediction. Ridge regression was chosen over ordinary least squares due to its lower prediction variance when correlated variables are used for prediction (although the combinatorial library leads to a full factorial structure, a fraction of the images were excluded from analysis as noted in the Methods, so substantial correlation between the predictor variables was expected in the data). For analysis, the image features were centered by plate, subtracting the mean feature value for each plate while retaining the original scale. A ridge regression model was fit using indicator (dummy) variables for each aldehyde and pyridinium/quinolinium group. To assess predictive accuracy without bias, 100 rounds of cross-validation were performed, holding out 10% of the data for testing. The correlation between fitted and observed image features was used to assess predictivity. Next, we used partial R2 values to quantify the additive contributions of aldehyde and pyridinium/quinolinium groups to each feature. All models included an additive effect for plate to remove plate-associated effects (due to any differences in cell preparation or instrument operation from one plate to the other). For example, the pyridinium/quinolinium group contribution was quantified in terms of the fit of the model with aldehyde, pyridinium/quinolinium, and plate effects compared to the model with pyridinium/quinolinium and plate effects. Adjusted R2 estimates (first-order unbiased estimates of the population R2) were used in calculating the partial R2 values. Bootstrapping was used to estimate standard errors of the R2 values, and confidence intervals calculated as the point estimate +/− two standard errors.
Relating chemical structures of styryl molecule building blocks to their effect on image-based features
To compare variations in chemical structure and image features, we considered every pair of compounds with image feature data. For each image-compound pair, we compared the Tanimoto similarity T between the two compounds' structures to the absolute difference D between the image feature values. The Pearson correlation between T and D was used to quantify the relationship between chemical structure and image features, with a negative correlation reflecting the strongest consistency.
Comparing the effect of isomer variants of pyridinium/quinolinium vs. aldehyde groups
Isomeric pairs of pyridinium/quinolinium groups and of aldehyde building blocks as identified in our previous study(18) were used to assess whether the image feature values were more sensitive to isomeric changes in the position of functional groups in the pyridinium/quinolinium moiety or in the aldehyde moiety. First, relative difference of image-based feature values X (measured for styryl molecule A(i)) and Y(measured for styryl molecule B(i)) was defined as 2|X−Y|/(|X|+|Y|), to assess the lack of conformity between feature values X and Y. Then, for every isomeric pair of pyridinium/quinolinium building blocks (pairs A:B, C:G and D:E; see (18)), we considered each one of them conjugated to the same aldehyde group “i” (where i corresponds to aldehydes 1 through 168), and calculated the relative difference in each image-based feature for each styryl pair A(i) vs B(i); C(i) vs. G (i); and, D(i) vs. E(i). Conversely, the relative difference of feature values X and Y was determined for each isomeric pair of aldehyde building blocks conjugated to the same pyridinium/quinolinium groups.
Comparing the effect of related chemical variants of pyridinium/quinolinium vs. aldehyde groups
As in the case of isomer variants, the relative difference of the calculated image-based features between of styryl molecules containing a pyridinium (A, B, C, G and H) vs quinolinium (D, E, F) group conjugated to every possible aldehyde building block was compared to the difference in image-based features between all pairs of styryl molecules containing a phenyl (aldehyde building blocks 1, 11, 85, 90, 126) vs. naphthalene (aldehyde building blocks 3, 6, 18, 20, 26, 34, 51, 67, 118, 141, 143) conjugated to every possible pyridinium/quinolinium building block. Conversely, the relative difference of feature values X and Y was determined for aldehyde moieties containing a phenyl vs. a naphthalene group conjugated to the same pyridinium/quinolinium groups. The relative difference of feature values X (measured for styryl molecule A(i)) and Y(measured for styryl molecule B(i)) was defined as 2|X−Y|/(|X|+|Y|), to assess the lack of conformity between feature values X and Y.
Hierarchical cluster analysis of chemical address tags
The additive contributions of each aldehyde group and each pyridinium/quinolinium group as estimated in the fitted regression model were concatenated over all image features into a vector. Using these vectors, we calculated the L1 norm (sum of absolute differences) between the features for any two chemical groups (either A groups or P groups). These L1 dissimilarities were used to perform an average linkage hierarchical cluster analysis of the chemical groups. The regression coefficients (scaled to (0,1)) were then displayed in a heat map.
RESULTS
Measuring the contribution of aldehyde vs. pyridinium/quinolinium building blocks to the probes' visual signals
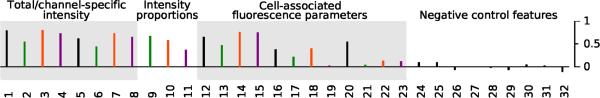
The styryl library is a 168 × 8 combinatorial library, so there are 1344 styryl structures. Excluding images with artifacts, extensive pixel saturation, dye precipitates, a total of 1291 images were used for chemical address tag analysis (including images lacking detectable styryl signal in the cellular region). A total of 23 specific probe-associated image-based features and 9 general control image-based features were extracted from the images (Table I). To address the extent to which global trends in probe behavior -apparent in the fluorescence images acquired in the FITC, TRITC and Cy5 acquisition channels of the imaging instrument- can be traced back to additive contributions from aldehyde and pyridinium/quinolinium building blocks of the styryl molecules, multivariate ridge regressions were performed, using each image feature as a response variable, and additive factorial effects for the two building blocks of the styryl molecule as predictors. The correlation coefficients between the observed and predicted image features were calculated, using cross-validation to provide unbiased estimates of the extent to which the image features can be predicted from additive contributions of the different building blocks (Figure 1).
Table 1.
List of quantitative, visual (image) features analyzed in this study.
|
Figure 1.
Correlation coefficients between actual and predicted values for the 32 image-based features analyzed in this study, using additive factors for the two styryl components as predictors. The bars represent the correlation value (estimated unbiasedly using cross-validation) for each specific feature.
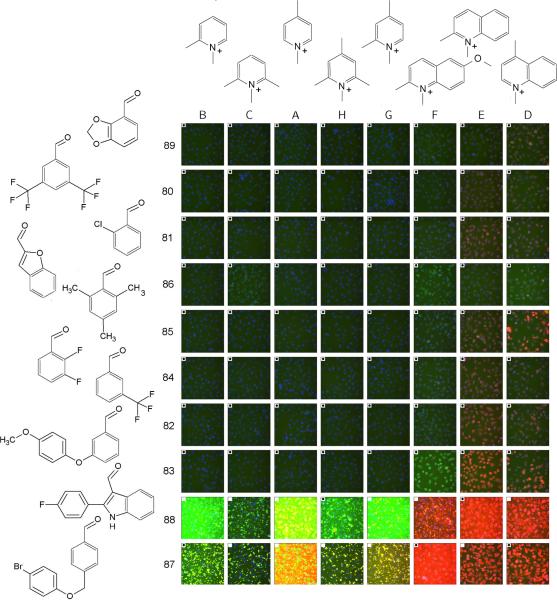
This quantitative analysis indicated that for many fluorescence intensity-related features, the correlation coefficient between predicted and measured values was strong and significant (ranging from 0.6 to 0.8) (Figure 1, features 1–11). In contrast, control features showed little correlation (correlation coefficient < 0.1) between predicted and measured values irrespective of wavelength (Figure 1, features 24–32). Visual inspection of arrays of images sorted according to the sign and magnitude of the regressed coefficients of the total intensity contribution from each aldehyde and pyridinium/quinolinium building blocks confirmed the expected trend: dark images in the top left of the array, with image brightness increasing towards the right and bottom (Figure 2). Furthermore, it is apparent that total intensity is linked with increased fluorescence in the TRITC channel, with the brightest images being the ones having most intense staining in the red (TRITC) channel, and images of intermediate brightness having most intense staining in the green (FITC) channel.
Figure 2.
Array of images acquired from styryl compounds, sorted based on the regression effects of different aldehyde building blocks (rows) and pyridinium/quinolinium building blocks (columns) to the total intensity feature of the styryl molecules. The effect increases from left to right for the pyridinium/quinolinium building blocks, and top to bottom for the aldehyde building blocks. Each image is a color composite of the signal obtained from the Hoechst™ channel (blue), FITC channel (green) and TRITC channel (red). For this figure, the images shown were acquired in the presence of the styryl molecules in the extracellular medium, so some of the images have high background fluorescence. A white, filled square in the upper left hand corner of an image indicates that the image was not used to train the regression model. For display purposes, the images were grouped into an array, and the array of images was then scaled (as a whole) to preserve the relative differences in the staining intensity of each probe.
For the spatial features, the relative accumulation of the fluorescence signal intensity in the cells relative to the background was moderately predictable from the regressed, additive contributions of the aldehyde and pyridinium/quinolinium building blocks (Figure 1, features 12–15). Based on the total intensity of the probe, the correlation coefficient for the CV (Figure 1, feature 16) and CNR (Figure 1, feature 20) of probe signal were 0.3 and 0.6 respectively However, in the individual wavelength acquisition channels, only the correlations of CV in the FITC and TRITC channel were as large (Figure 1, features 17, 18), and the correlation coefficient for the CNR feature was close to zero for each separate wavelength channel (it was positive only for the TRITC channel). To summarize, the total probe signals, several channel specific probe signals, the cellular accumulation of total probe signal relative to the background (for each separate acquisition channels and for the sum of the signal acquired in FITC, TRITC and Cy5 channels) were strongly predictable from additive effects of the molecules' basic building blocks (Figure 1). The CV and CNR features were moderately predictable based on the total sum of the signals acquired from FITC, TRITC and Cy5 channels, although they were not predictable based on the signal from the individual acquisition channels.
Elucidating chemical address tags with respect to fluorescence signal intensity, spectral and localization features
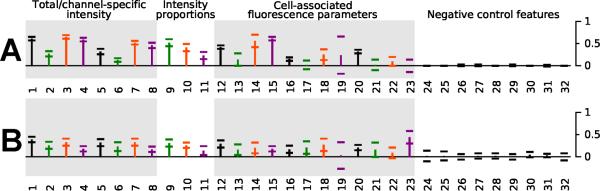
Next, based on the regression coefficients, the extent to which the different aldehyde and pyridinium/quinolinium building blocks differentially contributed to variations in the observed phenotype was established (Figure 3). The contribution of the aldehyde and pyridinium/quinolinium building blocks to signal in the total intensity and FITC channel tended to be equally strong (Figure 3, features 1, 2, 5, 6, 9, 11, 12, 13, 16, 18 and 20), indicating that the different building blocks both contributed as chemical address tags to determine the probes' visual signals in the FITC wavelength. Nevertheless, in the TRITC and Cy5 channel, the pyridinium/quinolinium group generally behaved as the determining chemical address tag relative to the aldehyde group, by showing substantially greater contribution to the image features (Figure 3, features 3, 4, 7, 8, 14, 15).
Figure 3.
The calculated, relative contribution of the pyridinium/quinolinium building blocks (A) and the aldehyde building blocks (B) towards the 32 image-based features analyzed in this study. The vertical bars represent the partial R2 value, capturing the additional information in A groups not contained in B groups, or vice-versa, based on the regression model. Error bars show the 95% confidence interval for the partial R2 value.
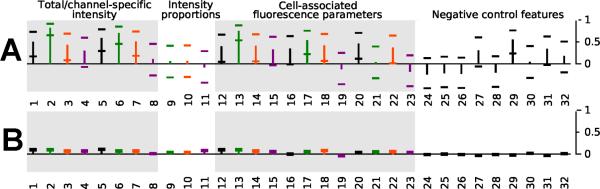
By relating the chemical features of the pyridinium/quinolinium building block to the variations spectral and localization properties, we established the extent to which chemical variations in the building blocks influenced spectral and localization features (Figure 4). For the pyridinium building block, variation in the chemical structure of the pyridinium/quinolinium group showed good correlation with variations in the image-based features (Figure 4A). In contrast in the case of the aldehyde building block, the relationship between the variation in chemical structure of the building block and variation in the image-based features was minimal (Figure 4B).
Figure 4.
Correlation coefficients between the degree of chemical structure variation in the building blocks of the styryl molecules and their contributions towards each image-based feature analyzed in this study. The bars represent the calculated correlation coefficient between Tanimoto structure similarities and the absolute differences in image feature regression coefficients over all pairs of structures, obtained while individually varying the pyridinium/quinolinium (A) or aldehyde building block (B), while keeping the other building block constant. Error bars show the 95% confidence interval for the partial R2 value.
Relating chemical structure variations to visual signal variations in the context of chemical address tags
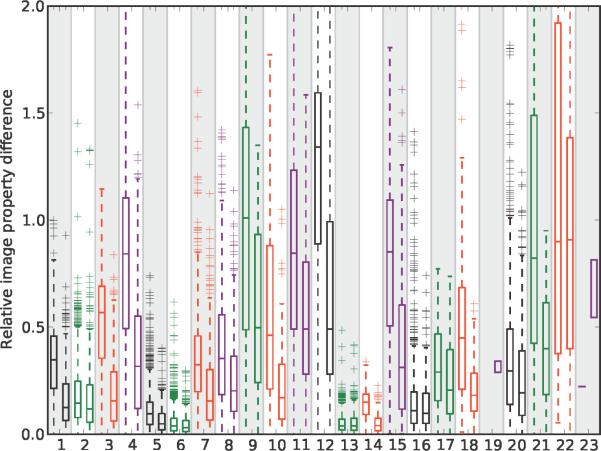
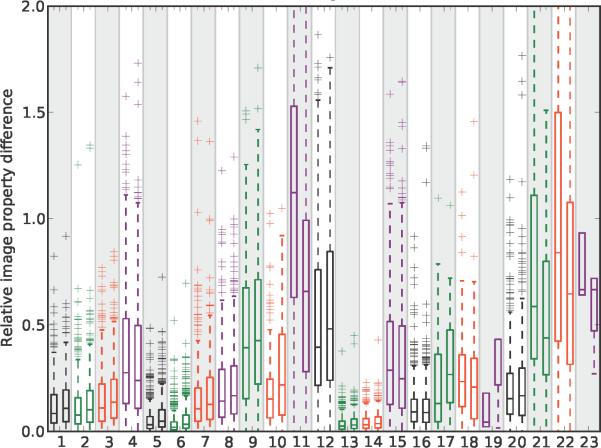
Probing how chemical variations in the pyridinium/quinolinium group affected the visual signal of the styryl molecules relative to similar variations in the aldehyde group, the results revealed that changing from a pyridinium to quinolinium exerted a major effect in relation to a phenyl-to-napthalene change in the aldehyde building block (Figure 5, features 1, 3, 4, 5, 7, 8, 12, 14, 15, 18). In comparison, isomers of pyridinium/quinolinium and aldehyde building blocks exerted comparable effects on the probe's visual signal (Figure 6). For the aldehyde building blocks, the magnitude of the effect of isomer variants (Figure 6) was similar to the magnitude of the effect of phenyl vs. naphthalene substitutions (Figure 5). For the pyridinium/quinolinium building blocks, the isomer effect (Figure 6) was generally less than the effect of substituting a quinolinium for pyridinium (Figure 5).
Figure 5.
The relative effect of a pyridinium vs. quinolinium building block (left box plot in each feature) and a phenyl vs. naphthalene aldehyde building block (right box plot in each feature) on each one of the 23 noncontrol image-based features analyzed in this study. For the left box plots, the difference in each image feature value was calculated for every possible pair of molecules possessing a pyridinium vs. quinolinium building block, with the aldehyde building block held constant. For the right box plots, the pyridinium/quinolinium building block was held constant, and the difference in each image feature value was calculated for all possible phenyl vs. naphthalene aldehyde building block pairs. The boxes extend from the 25th to the 75th percentiles of the data, with the central line denoting the median. The “whiskers” lie at the median plus and minus 1.5 times the median. All points outside the whiskers are plotted explicitly with '+' symbols.
Figure 6.
The relative effect of a pyridinium/quinolinium building block isomers (left box plot in each feature) and aldehyde building block isomers (right box plot in each feature) on each one of the 23 non-control image-based features analyzed in this study. For the left box plots, the difference in each image feature value was calculated for every pair of molecule possessing a pyridinium/quinolinium building block isomer, with the aldehyde building block held constant. For the right box plots, the pyridinium/quinolinium building block was held constant, and the difference in each image feature value was calculated for every aldehyde building block isomer pair. See the caption to figure 5 for a description of the boxplots.
Using cluster analysis to reveal relationships between chemical address tags
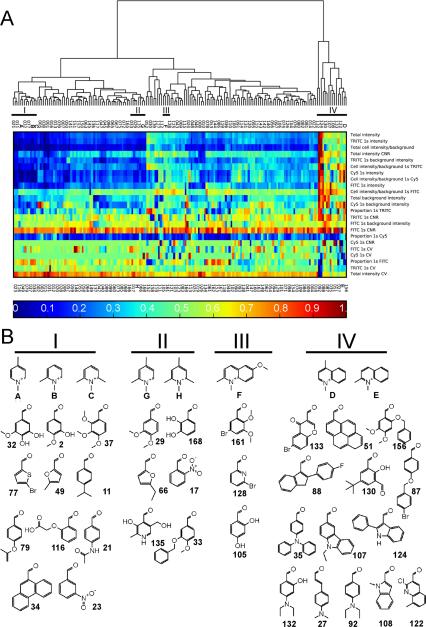
Based on hierarchical clustering (Figure 7), we analyzed how the quinolinium/pyridinium groups contributed to the image-based features of styryl molecules, in relation to the contribution of the aldehyde groups. A dendrogram (Figure 7A) revealed that the pyridinium/quinolinium groups formed distinct clusters with the different aldehyde groups (I, II, III, and IV). Note that the dendrogram divided the building blocks into two major clusters: one formed by group IV and the other one associated with groups I, II and III. Most aldehyde groups clustered with pyridinium/quinolinium groups A, B, C, F, G and H (Figure 7A, groups I, II and III). Nevertheless, a significant number of aldehydes formed a separate cluster with quinolinium groups D or E (Figure 7A, group IV). Visualizing the global pattern of regressed coefficients in a heat map (Figure 7A), group I and II appeared most similar to each other in terms of their contribution towards the staining patterns, with group IV being distinctively different.
Figure 7.
Hierarchical cluster analysis of the relationship between the regressed contribution of all the styryl building blocks to the 23 noncontrol image-based features analyzed in this study. A) Dendrogram and heat map visualization of the global relationship between different building blocks of the styryl molecules. The columns correspond to different aldehydes (numbers) or pyridinium/quinolinium (letters A–H) building blocks. Clusters I, II, III and IV are highlighted since they contain one or more pyridinium/quinolinium building block (from the dendrogram, the major branch point effectively separates cluster IV from the rest of the styryl building blocks). Colors of the heat map correspond to the regression coefficient values transformed to the unit interval, as indicated in the scale bar at the bottom of the heat map. B) Chemical structures of the selected pyridinium/quinolinium building blocks (letters) and surrounding aldehyde building blocks (numbers) associated with clusters I, II, III and IV.
Visual inspection of the building blocks in clusters I, II, III and IV (Figure 7B) indicated that half of the aldehyde building blocks that appeared closely related to pyridinium/quinolinium groups D or E in terms of their contribution to the styryl molecule's visual signals possessed a nitrogen as part of the conjugated structure (Figure 7B, group IV). As part of the conjugated structure, a nitrogen atom in the aldehyde building block can facilitate the migration of the molecule's positive charge across the central methine bridge of the styryl molecule, through resonance structures that would delocalize the positive charge normally associated with the imminium nitrogen on the pyridinium/quinolinium group.
In terms of the aldehyde groups that were most like pyridinium groups A, B, C, G, or H, many of them contained one or more hydroxyl, methoxy, or ether substituents (Figure 7B, groups I, II). For the aldheyde groups that were most like quinolinium group F (Figure7B, group III), two out of three were bromobenzene derivatives. Cluster IV also contained two aldehyde building blocks with bromine atoms, while clusters I and II contained none. These chemical functionalities that were prominent in several of the aldehyde groups in each of these clusters while being less represented in other clusters suggest that specific mechanisms can strongly influence image-based features across a large number of probes.
DISCUSSION
With a combinatorial library of bioimaging probes (6, 17–19), chemical address tags can be defined as a specific part of a molecule that contributes in an additive manner to a specific, quantitative image-based feature. Applying a statistical regression approach to a combinatorial library of styryl molecules (6), we demonstrated how the building blocks of the styryl library can be analyzed as chemical address tags with respect to cell-associated image-based features. Based on the results obtained, the behavior of chemical address tags is wavelength-dependent: chemical address tags were most prominent across all noncontrol features after summing the signals from the various acquisition channels, as compared to the individual FITC, TRITC or Cy5 acquisition channels.
Our results indicate that in the styryl library, building block isomers tend to behave similarly as chemical address tags, although isomer-specific phenotypic effects underlie many idiosyncratic interactions observed between cells and styryl molecules (18). In this study, we observed that isomer variants of the pyridinium/quinolinium or aldehyde building blocks were associated with relatively small variations in image-based features. Also, the results indicate that chemical variation in the pyridinium/quinolinium building block generally had the greatest effect on the probes' cell-associated image-based features.
A key additional finding explaining the behavior of chemical address tags comes from hierarchical cluster analysis: specific chemical variations in the aldehyde building blocks associated with the presence of a conjugated nitrogen atom can lead to a major effect on the image-based features, mimicking the behavior of quinolinium group D or E as chemical address tags. Notably, every styryl molecule in this library contains a positive charge because of the quaternary, imminium nitrogen in every pyridinium/quinolinium building block (6, 17, 19). Therefore electrostatics alone cannot explain chemical address tag behavior. Instead, our observations are consistent with chemical modifications affecting charge migration or the partial charge distribution of the styryl molecules being the major determinant of chemical address tag behavior. When the aldehyde building blocks contains a conjugated nitrogen atom, the free electrons of the nitrogen atom can pi-bond with the rest of the aromatic system, and the positive charge associated with the imminium nitrogen an become delocalized across the conjugated system, resonating with the nitrogen atom on the aldehyde group. Because of resonance effects, the positive charge of the molecule can shift from the pyridinium/quinolinium group to the nitrogen atom on the aldehyde group.
Demonstrably, in spite of complex interactions between individual styryl molecules and cellular components, quantitative analysis of the probes' visual signals can be used to study the effect of chemical structure on fluorescent probe behavior. For optimizing a probes' fluorescence and intracellular accumulation properties, elucidation and quantitative analysis of chemical address tags using simple linear regressions can be useful. For future work, the apparent association between different building blocks, as revealed by hierarchical clustering analysis of the regressed coefficients, points to specific mechanisms through which different chemical variations may lead to similar effects on the probes' phenotypic, image-based features. The importance of the nitrogen atom in the aromatic structure of the aldehyde group coupled to resonance effects constitutes a testable hypothesis, in terms of determining the behavior of chemical address tags through effects on charge migration or partial charge distribution.
To conclude, the development of organelle-targeting bioimaging probes has traditionally relied on qualitative, subjective criteria (i.e. visual inspection by experts). Therefore, the ability to apply automated, objective machine vision techniques and rigorous statistical analysis to biomaging probe development constitutes an important advance. Because the molecules of the styryl library differ in their fluorescence properties (17, 19), it is practically impossible to screen this library with filter sets tailored to the specific excitation and emission properties of each molecule. Nevertheless, the results of this study indicate that it is feasible to identify chemical address tags and analyze their behavior using the sum of the signals from the FITC, TRITC and Cy5 channels of the standard XF93 multipass filter set. Indeed, although the magnification of the image dataset analyzed in this study does not resolve specific organelles, the observed structure-property relationships reveal a potential mechanism underlying chemical address tag behavior. Paralleling advances in location proteomics (22–28), we envision using higher magnification 3D image data sets, together with orthogonal, organelle-specific markers and a more elaborate set of image features, to analyze chemical address tags responsible for fluorescence signal localization to specific organelles.
ACKNOWLEDGEMENTS
This work was funded by NIH grant RO1GM078200 to G.R.R. The authors thank Andrew Parth for assistance with the figures.
REFERENCES
- 1.Loo LH, Wu LF, Altschuler SJ. Image-based multivariate profiling of drug responses from single cells. Nat Methods. 2007;4(5):445–53. doi: 10.1038/nmeth1032. [DOI] [PubMed] [Google Scholar]
- 2.Perlman ZE, Slack MD, Feng Y, Mitchison TJ, Wu LF, Altschuler SJ. Multidimensional drug profiling by automated microscopy. Science. 2004;306(5699):1194–8. doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- 3.Slack MD, Martinez ED, Wu LF, Altschuler SJ. Characterizing heterogeneous cellular responses to perturbations. Proc Natl Acad Sci U S A. 2008;105(49):19306–11. doi: 10.1073/pnas.0807038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner BK, Carrinski HA, Ahn YH, Kim YK, Gilbert TJ, Fomina DA, Schreiber SL, Chang YT, Clemons PA. Small-molecule fluorophores to detect cell-state switching in the context of high-throughput screening. J Am Chem Soc. 2008;130(13):4208–9. doi: 10.1021/ja077656d. [DOI] [PubMed] [Google Scholar]
- 5.Lee JW, Jung M, Rosania GR, Chang YT. Development of novel cell-permeable DNA sensitive dyes using combinatorial synthesis and cell-based screening. Chem Commun (Camb) 2003;(15):1852–3. doi: 10.1039/b303960a. [DOI] [PubMed] [Google Scholar]
- 6.Li Q, Kim YK, Namm J, Kulkarni A, Rosania G, Ahn YH, Chang YT. RNA-selective, live cell imaging probes for studying nuclear structure and function. Chem. Biol. 2006 doi: 10.1016/j.chembiol.2006.04.007. in press. [DOI] [PubMed] [Google Scholar]
- 7.Abraham VC, Taylor DL, Haskins JR. High content screening applied to large-scale cell biology. Trends Biotechnol. 2004;22(1):15–22. doi: 10.1016/j.tibtech.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 8.Cervantes S, Prudhomme J, Carter D, Gopi KG, Li Q, Chang YT, Le Roch KG. High-content live cell imaging with RNA probes: advancements in high-throughput antimalarial drug discovery. BMC Cell Biol. 2009;10:45. doi: 10.1186/1471-2121-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giuliano KA. Optimizing the integration of immunoreagents and fluorescent probes for multiplexed high content screening assays. Methods Mol Biol. 2007;356:189–93. doi: 10.1385/1-59745-217-3:189. [DOI] [PubMed] [Google Scholar]
- 10.Mitchison TJ. Small-molecule screening and profiling by using automated microscopy. Chembiochem. 2005;6(1):33–9. doi: 10.1002/cbic.200400272. [DOI] [PubMed] [Google Scholar]
- 11.Giuliano KA, Post PL, Hahn KM, Taylor DL. Fluorescent protein biosensors: measurement of molecular dynamics in living cells. Annu Rev Biophys Biomol Struct. 1995;24:405–34. doi: 10.1146/annurev.bb.24.060195.002201. [DOI] [PubMed] [Google Scholar]
- 12.Giuliano KA, Taylor DL. Fluorescent-protein biosensors: new tools for drug discovery. Trends Biotechnol. 1998;16(3):135–40. doi: 10.1016/S0167-7799(97)01166-9. [DOI] [PubMed] [Google Scholar]
- 13.Giuliano KA, Taylor DL, Waggoner AS. Reagents to measure and manipulate cell functions. Methods Mol Biol. 2007;356:141–63. doi: 10.1385/1-59745-217-3:141. [DOI] [PubMed] [Google Scholar]
- 14.Taylor DL, DeBiasio R, LaRocca G, Pane D, Post P, Kolega J, Giuliano K, Burton K, Gough B, Dow A, et al. Potential of machine-vision light microscopy in toxicologic pathology. Toxicol Pathol. 1994;22(2):145–59. doi: 10.1177/019262339402200208. [DOI] [PubMed] [Google Scholar]
- 15.Ljosa V, Carpenter AE. High-throughput screens for fluorescent dye discovery. Trends Biotechnol. 2008;26(10):527–30. doi: 10.1016/j.tibtech.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Chen VY, Khersonsky SM, Shedden K, Chang YT, Rosania GR. System dynamics of subcellular transport. Mol Pharm. 2004;1(6):414–25. doi: 10.1021/mp049916t. [DOI] [PubMed] [Google Scholar]
- 17.Rosania GR, Lee JW, Ding L, Yoon HS, Chang YT. Combinatorial approach to organelle-targeted fluorescent library based on the styryl scaffold. J Am Chem Soc. 2003;125(5):1130–1. doi: 10.1021/ja027587x. [DOI] [PubMed] [Google Scholar]
- 18.Shedden K, Li Q, Liu F, Chang YT, Rosania GR. Machine vision-assisted analysis of structure-localization relationships in a combinatorial library of prospective bioimaging probes. Cytometry A. 2009;75(6):482–93. doi: 10.1002/cyto.a.20713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shedden K, Brumer J, Chang YT, Rosania GR. Chemoinformatic analysis of a supertargeted combinatorial library of styryl molecules. J Chem Inf Comput Sci. 2003;43(6):2068–80. doi: 10.1021/ci0341215. [DOI] [PubMed] [Google Scholar]
- 20.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones TR, Carpenter AE, Lamprecht MR, Moffat J, Silver SJ, Grenier JK, Castoreno AB, Eggert US, Root DE, Golland P, et al. Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning. Proc Natl Acad Sci U S A. 2009;106(6):1826–31. doi: 10.1073/pnas.0808843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boland MV, Murphy RF. A neural network classifier capable of recognizing the patterns of all major subcellular structures in fluorescence microscope images of HeLa cells. Bioinformatics. 2001;17(12):1213–23. doi: 10.1093/bioinformatics/17.12.1213. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Murphy RF. Objective clustering of proteins based on subcellular location patterns. J Biomed Biotechnol. 2005;2005(2):87–95. doi: 10.1155/JBB.2005.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Y, Murphy RF. Automated interpretation of subcellular patterns from immunofluorescence microscopy. J Immunol Methods. 2004;290(1–2):93–105. doi: 10.1016/j.jim.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 25.Huang K, Murphy RF. From quantitative microscopy to automated image understanding. J Biomed Opt. 2004;9(5):893–912. doi: 10.1117/1.1779233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang K, Murphy RF. Boosting accuracy of automated classification of fluorescence microscope images for location proteomics. BMC Bioinformatics. 2004;5:78. doi: 10.1186/1471-2105-5-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy RF. Cytomics and location proteomics: Automated interpretation of subcellular patterns in fluorescence microscope images. Cytometry A. 2005 doi: 10.1002/cyto.a.20179. [DOI] [PubMed] [Google Scholar]
- 28.Murphy RF. Location proteomics: a systems approach to subcellular location. Biochem Soc Trans. 2005;33(Pt 3):535–8. doi: 10.1042/BST0330535. [DOI] [PubMed] [Google Scholar]
- 29.Hansch C. The QSAR paradigm in the design of less toxic molecules. Drug Metab Rev. 1984;15(7):1279–94. doi: 10.3109/03602538409029960. [DOI] [PubMed] [Google Scholar]
- 30.Hansch C, Leo A. Exploring QSAR: fundamentals and applications in chemistry and biology. American Chemical Society; Washington DC: 1995. [Google Scholar]
- 31.Lee AC, Shedden K, Rosania GR, Crippen GM. Data mining the NCI60 to predict generalized cytotoxicity. J Chem Inf Model. 2008;48(7):1379–88. doi: 10.1021/ci800097k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shedden K, Rosania GR. Exploratory chemoinformatic analysis of cell type-selective anticancer drug targeting. Mol Pharm. 2004;1(4):267–80. doi: 10.1021/mp049953k. [DOI] [PubMed] [Google Scholar]