Abstract
The vacuolar (H+)-ATPases are ATP-dependent proton pumps that function to acidify intracellular compartments and, in some cases, transport protons across the plasma membrane of eukaryotic cells. Intracellular V-ATPases play an important role in such normal physiological processes as receptor-mediated endocytosis, intracellular membrane traffic, pro-hormone processing, protein degradation and the coupled uptake of small molecules, such as neurotransmitters. They also function in the entry of various pathogenic agents, including many envelope viruses, like influenza virus, and toxins, like anthrax toxin. Plasma membrane V-ATPases function in renal pH homeostasis, bone resorption and sperm maturation, as well as in various disease processes, including renal tubular acidosis, osteopetrosis and tumor metastasis. V-ATPases are composed of a peripheral V1 domain containing eight different subunits that is responsible for ATP hydrolysis and an integral V0 domain containing six different subunits that translocates protons. In mammalian cells most of the V-ATPase subunits exist in multiple isoforms which are often expressed in a tissue specific manner. Isoforms of one of the V0 subunits (subunit a) have been shown to possess information that targets the V-ATPase to distinct cellular destinations. Mutations in isoforms of subunit a lead to the human diseases osteopetrosis and renal tubular acidosis. A number of mechanisms are employed to regulate V-ATPase activity in vivo, including reversible dissociation of the V1 and V0 domains, control of the tightness of coupling of proton transport and ATP hydrolysis and selective targeting of V-ATPases to distinct cellular membranes. Isoforms of subunit a are involved in regulation both by control of coupling and by selective targeting. This review will begin with a brief introduction to the function, structure and mechanism of the V-ATPases followed by a discussion of the role of V-ATPase subunit isoforms and the mechanisms involved in regulation of V-ATPase activity.
V-ATPase Function
The vacuolar (H+)-ATPases (V-ATPases) are a family of ATP-dependent proton pumps localized to a variety of cellular membranes of eukaryotic cells, including endosomes, lysosomes, Golgi-derived vesicles, secretory vesicles and, for some cells, the plasma membrane (1-3). V-ATPases within endosomes function to dissociate internalized ligands, such as low density lipoprotein (LDL), from their receptors, thus facilitating recycling of the receptors to the plasma membrane (4). They are also required for budding of endosomal carrier vesicles that carry internalized ligands from early to late endosomes (5) and for dissociation of lysosomal enzymes from the mannose-6-phosphate receptors that target them from the Golgi to the lysosome (6). Acidic endosomal compartments also provide the entry point for the cytotoxic portions of many envelope viruses, including influenza and Ebola virus, and toxins, such as diphtheria and anthrax toxin (7). The ability of these pathogenic agents to infect or kill cells is thus critically dependent upon V-ATPase activity. Lysosomal acidification by V-ATPases is required for the activity of degradative enzymes located within the lysosomal lumen and for the export of degradation products via coupled transporters. V-ATPases within secretory vesicles create both the low pH required for processing of pro-hormones to their mature forms and the pH gradient and membrane potential used to drive the uptake of small molecules, such as neurotransmitters (1).
Plasma membrane V-ATPases predominantly play cell-type specific roles. Thus, in renal intercalated cells of the late distal tubule and collecting duct, V-ATPases located in the apical membrane function to secrete acid into the urine (3). A defect in isoforms of V-ATPase subunits that are selectively expressed in the kidney lead to the genetic disorder distal renal tubule acidosis, in which patients are unable to excrete sufficient acid in the urine (8). Plasma membrane V-ATPases in osteoclasts play a critical role in bone resorption by acidifying the space between the cell and the bone, thus dissolving the bone matrix (9). Defects in the osteoclast V-ATPase lead to the disease osteopetrosis, characterized by developmental defects resulting from the inability to degrade and remodel bone (10). V-ATPases localized to the apical membrane of clear cells in the epididymus maintain the semenal fluid at a low pH, a property crucial for the normal maturation and storage of sperm (11). Finally, plasma membrane V-ATPases have also been implicated in the metastasis of tumor cells (12, 13). In this context, plasma membrane V-ATPases may aid in tumor cell invasion by providing an acidic extracellular environment required for the activity of secreted cathepsins, proteases that have been shown to function in metastasis by many tumor cells (14). Because of their role in bone resorption and tumor invasion, V-ATPases are attractive targets in the development of drugs for the treatment of osteoporosis (characterized by excessive bone resorption) and cancer metastasis.
V-ATPase Structure and Mechanism
V-ATPases are large, multi-subunit complexes organized into two domains (Fig. 1, Table 1). The peripheral V1 domain is composed of eight different subunits (A-H) and functions to hydrolyze ATP (1, 2). The integral V0 domain is composed of six different subunits (a,c,c″,d,e and Ac45 in mammals and a,c,c′,c″,d and e in yeast) and functions to translocate protons across the membrane (1, 2). ATP hydrolysis occurs at catalytic sites located at the interface of the A and B subunits (15, 16), which are each present in three copies per complex and are arranged in alternating fashion in a ring. Most of the catalytic site residues are contributed by the A subunit (17). A second set of nucleotide binding sites is located at the other A/B subunit interface (termed “noncatalytic” sites), which are composed primarily of B subunit residues and may function to regulate activity (17, 18). Within the V0 domain, the proteolipid subunits (c, c′ and c″) are also organized into a ring containing single copies of subunits c′ and c″ and multiple copies of subunit c (1, 19). Analysis of chimeric constructs indicates that the proteolipid subunits adopt a well defined arrangement in the proteolipid ring (20). The proteolipid subunits are highly hydrophobic proteins composed of four (c and c′) or five (c″) transmembrane helices (TMs) (21) and each subunit contains a single essential buried acidic residue that undergoes reversible protonation during proton transport (22). Interestingly TM1 of subunit c″ appears to be dispensable for function (21, 23). Subunit a of V0 is thought to provide access channels (hemi-channels) that allow protons to reach and leave these buried acidic residues on the proteolipid ring (1).
Fig. 1. Structure and mechanism of the V-ATPase.
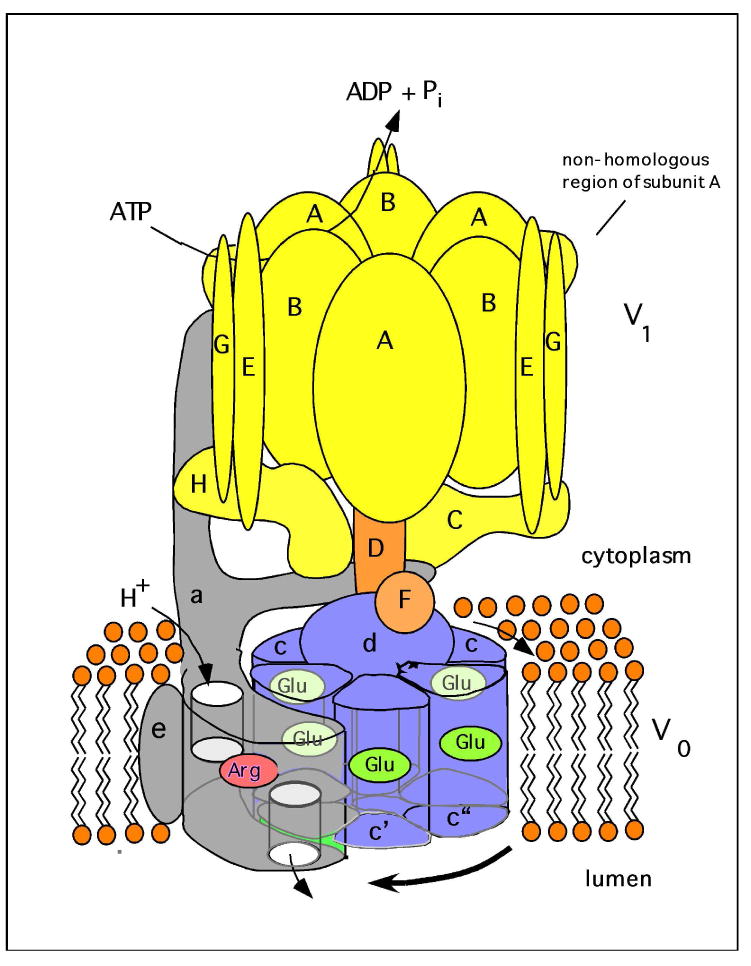
The V-ATPase is composed of two domains, V1 and V0. The peripheral V1 domain is composed of eight different subunits (A-H, shown in yellow and orange) and is responsible for ATP hydrolysis, whereas the integral V0 domain is composed of six subunits (in yeast these are subunits a, c, c′, c″, d and e, shown in blue and grey), and is involved in proton translocation across the membrane. ATP hydrolysis drives the rotation of a central rotor, which is composed of the D, F, d and proteolipid (c, c′, c″) subunits. Subunit a possesses two hemi-channels and a crucial arginine residue (shown in red), which are required for proton translocation. The hemi-channels allow protons to reach buried glutamic acid residues (shown in green) on the proteolipid ring from the cytoplasmic side of the membrane and to leave from these sites to the luminal side of the membrane following interaction of the glutamate residues with the a subunit arginine residue. The V1 and V0 domain are connected by a central stalk, which is composed of subunits D, F and d, and three peripheral stalks, which are composed of subunits C, E, G and the N-terminal cytoplasmic domain of subunit a. The peripheral stalks hold the A3B3 hexamer stationary with respect to subunit a. The non-homologous region of subunit A, which is absent from the F1FO ATP synthases, is involved in reversible dissociation (see text).
Tab.1. Characteristics of V-ATPase subunits.
| Subunit | Molecular Mass (kDa) | Yeast gene | Mammalian Isoforms | Subunit function | References | |
|---|---|---|---|---|---|---|
| V1 domain | tissue/cell | Alternative splicing | ||||
| A | 70 | VMA1 | ATP hydrolytic site, regulation via non-homologous domain, | (75, 76, 91) | ||
| B | 60 | VMA2 | B1 (renal, epididymis, olfactory epithelium); B2 (ubiquitous) |
Non-catalytic ATP site, binds actin and aldolase, | (11, 51, 54, 91-93) | |
| C | 40 | VMA5 | C1 (ubiquitous); C2 (lung, renal, epididymus) |
C2a (lung) C2b (renal) |
Regulatory, stator subunit, binds actin | (11, 30, 48, 55, 94-97) |
| D | 34 | VMA8 | Rotary subunit | (26, 31) | ||
| E | 33 | VMA4 | E1 (testis, olfactory epithelium); E2 (ubiquitous) |
Stator subunit, binds RAVE and aldolase | (26, 49, 55, 70, 92) | |
| F | 14 | VMA7 | Rotary subunit | (31, 98) | ||
| G | 13 | VMA10 | G1 (ubiquitous); G2 (neural); G3 (renal, epidiymus) |
Stator subunit, binds RAVE | (11, 32, 48, 55, 70, 99) | |
| H | 50 | VMA13 | SFD-α SFD-β |
Regulatory, stator subunit, binds NEF | (28, 100-103) | |
| V0 domain | ||||||
| a | 100 | VPH1 (vacuole) STV1 (Golgi) |
a1 (neural); a2 (endothelial, neurons); a3 (osteoclasts, pancreatic β-cells, premature melanosomes); a4 (renal, epididymis) |
a1-I (brain, neurons), a1-II (ubiquitous), a1-III (ubiquitous), a1-IV (neurons); a3-I (ubiquitous), a3-II (ubiquitous), a3-III (heart, lung), a4-I (kidney, heart, lung, skeletal muscle, testis), a4-II (kidney, lung, liver, testis) |
H+ transport, targeting, binds aldolase, stator subunit | (3, 8, 9, 11, 33, 38, 39, 41-44, 46, 91, 104-106) |
| d | 38 | VMA6 | d1 (ubiquitous) d2 (renal, epididymis, osteoclast, dendritic cells) |
Coupling, rotary subunit | (11, 27, 48, 55, 56, 107-109) | |
| e | 9 | VMA9 | Unknown | (110) | ||
| c | 17 | VMA3 | H+ transport, rotary subunit | (32, 91) | ||
| c′ | 17 | VMA11 | No mammalian gene | H+ transport, binds Vma21 assembly factor, rotary subunit | (91, 111) | |
| c″ | 21 | VMA16 | H+ transport, rotary subunit | (91) | ||
| Ac45 | 45 | no yeast gene | Ac45 Ac45LP (Xenopus) |
unknown | (112, 113) | |
Abbreviations: Ac45, accessory subunit-45; Ac45LP, Ac45-like protein; NEF, negative replication factor; RAVE, regulator of the ATPase of vacuolar and endosomal membranes; SFD, sub-fifty-eight-kDa Dimer; VMA, vacuolar membrane ATPase activity.
The V1 and V0 domains are connected by multiple stalks (24, 25). A central stalk (composed of subunits D, F and d (26, 27)) extends from the proteolipid ring up through the center of the A3B3 hexameric head whereas three peripheral stalks (composed of subunits C, E, G and H (25, 26, 28-30)) attach the A3B3 hexamer to the N-terminal hydrophilic domain subunit a of V0 (Fig. 1). The V-ATPases operate by a rotary mechanism in which ATP hydrolysis at the catalytic sites in V1 drives rotation of the central stalk and the attached ring of proteolipid subunits relative to subunit a (31, 32). The peripheral stalks thus serve as stators that prevent rotation of subunit a relative to the A3B3 hexamer. As the buried acidic residues on the proteolipid ring come into contact with subunit a they sequentially pick up protons from the hemi-channel in contact with the cytoplasm and, following rotation in the plane of the membrane, deliver those protons to the hemi-channel in contact with the luminal side of the membrane (1). A buried arginine residue in subunit a interacts with the buried acidic residues on the proteolipid ring, stabilizing them in their negatively charged, deprotonated form, thus facilitating release of protons into the luminal hemi-channel (33). In this way rotary motion driven by ATP hydrolysis in V1 drives unidirectional transport of protons across the membrane through V0. It should be noted that the V-ATPases are related to both the F1FO ATP-synthase (34) and the A1A0-ATPases (35) in both structure and mechanism.
Isoforms of Subunit a of the V-ATPase
Isoforms of many of the V-ATPase subunits exist in mammalian cells, but the most extensive information exists on isoforms of subunit a (Table 1). In yeast, all the V-ATPase subunits are encoded by single genes with the exception of subunit a, which is encoded by two genes (VPH1 and STV1). Vph1p and Stv1p are 54% identical at the amino acid level, with the C-terminal hydrophobic domain showing somewhat higher sequence identity than the N-terminal cytoplasmic domain (36). Vph1p targets the V-ATPase to vacuoles whereas Stv1p targets the V-ATPase to the Golgi (36, 37). Studies of chimeric proteins containing different parts of either Vph1p or Stv1p have demonstrated that the targeting information exists in the cytoplasmic N-terminal domain of subunit a (37). The nature of the specific targeting signal in the N-terminus of subunit a has not been determined. Differences in the degree of assembly between V1 and V0 and in the tightness of coupling between proton transport and ATP hydrolysis have been identified for complexes containing either Vph1p or Stv1p (37, 38), and these differences likely help to explain the lower pH of the vacuolar compartment relative to the Golgi (see section on Regulation of V-ATPases below).
In mammalian cells the a subunit exists in four isoforms (a1, a2, a3, and a4) which also contain targeting information (1). In humans, the four a subunit isoforms display 47-61% identity at the amino acid level (3). The a3 isoform is heavily expressed in osteoclasts and is responsible for targeting of the V-ATPase to the plasma membrane of these cells, where it is involved in bone resorption. In pre-osteoclast cells a3 is localized to lysosomes and appears to undergo movement to the plasma membrane upon differentiation of these cells into mature osteoclasts (9). Mutations in a3 lead to the defective bone resorption phenotype associated with osteopetrosis (10). Drugs which target a3-containing V-ATPase complexes could potentially be useful in the treatment of osteoporosis, where excessive bone resorption occurs. The a3 isoform has also been localized to insulin-containing secretory vesicles in pancreatic islet cells, where it likely contributes to the acidic environment required for proteolytic processing of insulin. Interestingly, mice bearing a knock-out in the a3 gene are still able to process insulin but show a defect in insulin secretion (39). Immunolocalization studies of islet cells from these mice suggest that the a2 isoform may compensate for the loss of a3 in acidifying the insulin-containing compartment, but a2 appears unable to replace a3 in its role in insulin secretion. The a3 isoform is also expressed in the adrenal, parathyroid, thyroid and pituitary glands (40), and in immature melanosomes, where it functions to keep this organelle acidic (41).
The a4 isoform is expressed almost exclusively in renal cells and is responsible for targeting of V-ATPases to the apical membrane of alpha-intercalated cells of the late distal tubule and collecting duct (3). Mutations in a4 cause the defective urinary acid secretion associated with renal tubular acidosis (8). Interestingly, a4 is also involved in targeting of V-ATPases to the apical membrane of epididymal clear cells, which are responsible for acidification of the epididymal lumen (11). Its role in sperm maturation and storage makes the a4 isoform a potential target in the development of male contraceptives. There exist two splice variants of the mouse a4 subunit, a4-I and a4-II, which show distinct tissue and developmental expression patterns. Besides strong expression in kidney, a4-I is detected in heart, lung, skeletal muscle and testis, whereas a4-II is detected in lung, liver, and testis. During development, a4-I is expressed beginning with the early embryonic stage, whereas a4-II is detected from day17 (42). In humans, similar splice variants of the a4 subunit exist as well (8). Splice variants have also been identified for a number of other V-ATPase subunits (see Table I).
The a1 isoform has been shown to localize to both the presynaptic membrane and synaptic vesicles present at nerve terminals (43). V-ATPase complexes containing a1 likely function to acidify synaptic vesicles, but V0 complexes containing a1 present in both the synaptic vesicle and the synaptic plasma membrane have been suggested to play a more direct role in fusion between these membranes during neurotransmitter release (44). The a1 subunit has also been implicated in fusion between phagosomes and lysosomes during phagocytosis in the brain (45), suggesting a role in the endocytic pathway for a1.
The a2 isoform has been identified in apical endosomes of proximal tubule cells of the kidney, where it functions to provide the low pH necessary to release endocytosed peptides from the receptors megalin and cubulin that are involved in their absorptive uptake from the renal fluid (46).
Despite the specific localizations noted above, it is likely that many cells express multiple a subunit isoforms that target V-ATPases to different cellular membranes. The highly invasive MB231 human breast tumor cell line has been shown to express much higher levels of both the a3 and a4 isoforms than the poorly invasive MCF7 cell line, and knock-down of either isoform significantly inhibits the invasiveness of MB231 cells (13). Nevertheless, knock-down of a3 appears to affect invasion by inhibiting V-ATPase activity in the endosome/lysosome compartment whereas knock-down of a4 may inhibit invasion by reducing plasma membrane V-ATPase activity.
Other V-ATPase Subunit Isoforms
Besides subunit a, at least six of the remaining fourteen V-ATPase subunits exist in multiple isoforms in mammalian cells (Table 1). In each case, one isoform is ubiquitously expressed while other isoforms are often expressed in a specific tissue. The sequence identities between human isoforms are as follows: subunit B (83%) (47), subunit C (62%) (48), subunit E (77%) (49), subunit G (53-64%) (48) and subunit d (68%) (48). We will begin with a discussion of the function and distribution profile of subunit B. The B1 isoform is expressed not only in renal and epididymal cells but also in hair cells of the inner ear (50). Mutations in B1 have thus been shown to lead not only to renal tubular acidosis but also to deafness. B1 is localized to the apical membrane of intercalated cells and is involved in proton secretion into the urine. In contrast, B2 is ubiquitously expressed and is predominantly localized to intracellular compartments, suggesting that these two isoforms have different functions (51). However, recent studies showed that B2 is able to compensate for at least some of the functional consequences of a loss of B1 in knock out mice (52, 53). V-ATPases have been identified in the mouse olfactory epithelium where the B1 isoform localizes to the apical membrane and subapical region of olfactory cells and B2 is cytoplasmic. In B1 knock-out mice, the B2 isoform is upregulated but does not appear in the apical membrane (54).
Other isoforms, including C2, G3, and d2, have also been shown to be shared between renal and epididymal cells (11, 48), suggesting the formation of V-ATPase complexes containing unique combinations of isoforms in these, and perhaps other cell types. In human and mouse, d1 is ubiquitously expressed, whereas d2 is expressed in osteoclasts in addition to the kidney and epdidiymus (48, 55, 56). It has been reported that d2 knock-out mice develop osteopetrosis due to defects in the formation of multinucleated osteoclasts (57). This suggests that the d2 isoform plays a specific role in osteoclast fusion. Recently it has been demonstrated that both d1 and d2 directly associate with the central stalk subunits D and F, supporting a role for subunit d as part of the central rotor of the V-ATPase (58). The existence of complexes containing unique combinations of V-ATPase isoforms raises the possibility of the development of drugs (or panels of drugs) that are highly specific for particular combinations of isoforms that could be used for the treatment of diseases (such as cancer) in which these complexes participate. For example, based upon recent studies of subunit a isoform expression in breast tumor cells (13), inhibitors specific for V-ATPase complexes containing a3 or a4 might inhibit breast cancer metastasis.
Regulation of V-ATPase Activity
Cells are able to regulate pH both spatially and temporally. Thus the luminal pH of compartments along the endocytic pathway decreases as one moves from early endosomes through late endosomes to lysosomes (1). This gradient of pH is essential for the correct timing of many essential steps in the endocytic process (4), including uncoupling of ligands from their receptors in early endosomes, the budding of endocytic carrier vesicles that move ligands from early to late endosomes and the degradation of endocytosed macromolecules in lysosomes. Similarly, precise regulation of vesicular pH along the pathway from the Golgi to lysosomes facilitates dissociation of lysosomal enzymes from the mannose-6-phosphate receptor and recycling of this receptor to the trans-Golgi (6). These represent examples of spatial regulation of the pH of intracellular compartments, where the pH of a given compartment is maintained within narrow limits. There are also many examples of temporal changes in V-ATPase activity in response to specific stimuli. For example, a variety of stimuli have been shown to lead to changes in assembly of the V-ATPase complex, which represents one of the principal mechanisms of controlling V-ATPase activity in cells. Changes in pump density through reversible fusion of V-ATPase-containing vesicles, most often with the plasma membrane, represents a second important temporal mechanism of regulating proton flux via the V-ATPase. We will discuss these and other mechanisms in the following section.
Regulation of V-ATPase Activity by Reversible Dissociation
Perhaps the best characterized mechanism of regulating V-ATPase activity is the reversible dissociation of the complex into its component V1 and V0 domains (Fig. 2). This process was first characterized in yeast and insect cells (2, 59), but occurs in mammalian cells as well. V-ATPase dissociation occurs in yeast in response to glucose depletion from the media and in insects during molting, most likely as a means of conserving cellular stores of ATP. V-ATPase assembly in renal cells is also responsive to glucose levels, although the physiological significance of these changes is less clear (60). Activation of dendritic cells to process antigen is accompanied by increased assembly of the V-ATPase on the lysosomal membrane, as acidification of this compartment is required for normal antigen processing (61). It is likely that reversible dissociation will be shown to regulate V-ATPase activity in other systems as well.
Fig. 2. Regulation of V-ATPase activity by reversible dissociation.
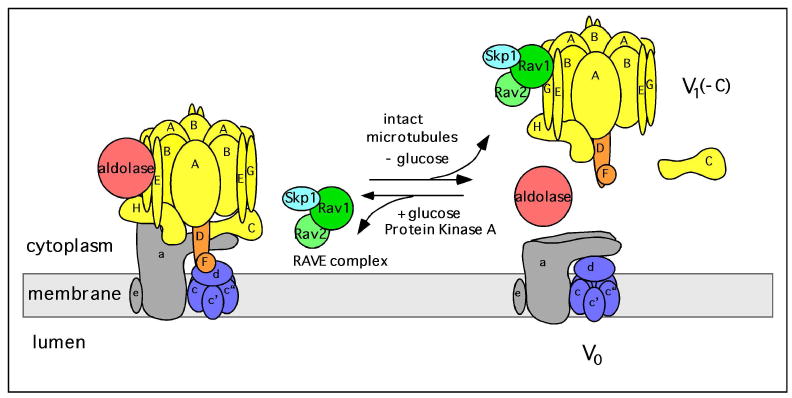
In yeast, glucose depletion initiates the reversible dissociation of the V-ATPase into a V1 complex minus C, free subunit C and V0. Dissociation requires an intact microtubule network whereas reassembly of V1 back onto V0 is mediated by the RAVE complex (composed of Skp1, Rav1 and Rav2) and aldolase. Dissociation may also be stimulated by altered interactions of the non-homologous region of subunit A with the V0 domain. Glucose-dependent assembly of the V-ATPase is driven by activation of the Ras/cAMP/protein kinase A (PKA) pathway. The free forms of V1 and V0 are silenced with respect to ATP hydrolysis and passive proton translocation.
Reversible dissociation of the V-ATPase has been most thoroughly studied in yeast (1, 2). In this system dissociation occurs rapidly, reversibly and does not require new protein synthesis, although some level of catalytic activity is generally required (62-64). The separated V1 and V0 domains are silenced with respect to ATP hydrolysis and passive proton translocation following dissociation, as expected given the deleterious effects of releasing an uncoupled ATP hydrolytic domain into the cytosol or creating an unregulated passive proton leak in cellular membranes. ATP hydrolysis by the free V1 domain is silenced by subunit H (65), either through direct bridging of the central and peripheral stalks (66) or as a result of a conformational change induced upon dissociation (67). Dissociation and reassembly appear to be independently controlled processes. Thus dissociation, but not reassembly, requires an intact microtubular network (68) whereas reassembly, but not dissociation, requires a novel protein complex termed RAVE (69). RAVE is composed of three proteins (Rav1p, Rav2p and Skp1, a ubiquitin ligase component) and appears to bind to subunits E and G of V1 as well as subunit C (a V1 subunit that dissociates from both V1 and V0 upon disassembly) (70, 71). RAVE appears to stabilize the dissociated V1 complex in an assembly-competent form and mediates assembly in both the normal biosynthetic pathway and the glucose-regulated process (70). Interaction with the glycolytic enzyme aldolase also appears to be important in glucose-mediated assembly as mutations in aldolase that disrupt this interaction (but leave the catalytic activity of aldolase intact) prevent assembly of the V-ATPase in vivo (72).
The signaling pathways that regulate reversible dissociation of the V-ATPase have begun to be unraveled. In yeast glucose causes assembly of the V-ATPase by activating the Ras/cAMP/protein kinase A pathway (73). Elevated glucose inhibits the activity of the Ras GAPs Ira1p and Ira2p which leads to an increase in the amount of GTP-bound Ras and hence increased Ras activity. Activated Ras in turn activates adenylate cyclase leading to increased cellular levels of cAMP. Elevated cAMP causes dissociation of the regulatory subunits of protein kinase A and hence activation of its catalytic subunits. Thus under conditions of glucose depletion the Ras/cAMP/PKA pathway would be suppressed leading to V-ATPase dissociation. It should be noted that at present it is not known whether activation of PKA increases assembly or decreases dissociation of the V-ATPase. The mechanism by which protein kinase A affects assembly or blocks dissociation of the V-ATPase in yeast has also not been determined. In insect cells protein kinase A has been shown to stimulate assembly of the V-ATPase and to phosphorylate subunit C of the complex, although interestingly subunit C in the assembled enzyme is not phosphorylated (74). Thus it is unclear what the relationship is between subunit C phosphorylation and V-ATPase assembly in insect cells. In renal epithelial cells phosphatidylinositol 3-kinase (PI-3 kinase) has been shown to mediate glucose-dependent assembly of the V-ATPase (60), although other signaling pathways may also be involved.
One domain of the V-ATPase that has been shown to be important for reversible dissociation of the complex is the “non-homologous” region of the catalytic A subunit. This 90 amino acid region is highly conserved between V-ATPases from different species but is absent from the homologous beta subunit of the F1FO ATP synthase (hence its name). The non-homologous region is present as an insert approximately one third of the way from the N-terminus of subunit A and appears in EM images of the complex as an “ear” sticking out from the A subunit position in the A3B3 hexameric head (24). Mutations in the non-homologous region have been identified which block dissociation of the complex upon glucose depletion without affecting catalytic activity (75). Moreover, when expressed as a separate epitope-tagged construct in the absence of the remainder of the A subunit, binding of the non-homologous region to V0 (without the other V1 subunits) can be detected and occurs in a glucose-dependent manner (76). This suggests the possibility that interaction between the non-homologous region and some portion of the V0 domain (possibly the N-terminal domain of subunit a) may help to control glucose-dependent assembly of the complex.
Reversible dissociation of the V-ATPase in yeast has also been shown to be a sensitive function of the cellular environment in which the complex resides. Thus V-ATPases containing Vph1p and hence localized to the vacuole dissociate upon glucose depletion whereas Stv1p-containing complexes localized to the Golgi do not dissociate. If Stv1p-containing complexes are re-directed to the vacuole through overexpression, dissociation is now observed (38). Moreover, V-ATPase complexes containing either Vph1p or Stv1p which are arrested in intermediate compartments through the use of vps mutants, which disrupt trafficking, show nearly identical behavior with respect to glucose-dependent dissociation (64). Although the environmental factor(s) responsible for this difference in dissociation behavior has not been identified, one property which differs between different compartments and which has been shown to affect dissociation is luminal pH (76). Although membrane environment appears to be more important in controlling reversible dissociation than a subunit isoform in yeast, the relative importance of these factors in mammalian cells is unknown.
Other Regulatory Mechanisms
Proton transport across the apical membrane of a number of polarized cell types is regulated by reversible insertion of fully assembled V-ATPases derived from specialized populations of intracellular vesicles. Increased proton transport across the plasma membrane is achieved by exocytic fusion of these V-ATPase-containing vesicles with the apical membrane whereas a reduction in proton transport results from endocytic retrieval of V-ATPases into this intracellular pool. Examples where this regulatory mechanism plays an important physiological role include control of proton transport across the apical membrane of renal intercalated cells of the late distal tubule and collecting duct (3) and regulation of the pH of the epididymus by epididymal clear cells (77). In the case of renal epithelial cells, a decrease in the cellular pH associated with increased carbon dioxide induces fusion of apical membrane vesicles containing a very high density of V-ATPases with the apical membrane, thus increasing acid secretion into the urine. This process appears to be in part regulated by PI-3 kinase (60). Epididymal clear cells respond to elevated luminal pH by increasing acid secretion across the apical membrane. This is accomplished with the aid of a bicarbonate-sensitive adenylate cyclase (77). As the luminal pH increases cellular levels of bicarbonate increase, activating the bicarbonate-sensitive adenylate cyclase which increases cellular levels of cAMP and hence protein kinase A activity. Activation of protein kinase A leads to increased exocytosis and/or decreased endocytosis of V-ATPases, which shuttle between the plasma membrane and a pool of intracellular vesicles. How protein kinase A regulates this event remains unknown. Recently, it was shown that the metabolic sensor AMP-activated protein kinase (AMPK) as well as PKA directly phosphorylate the V-ATPase A subunit in clear cells. AMPK inhibits both PKA-dependent phosphorylation of the A subunit and cAMP-induced apical membrane accumulation of the V-ATPase. Thus, intracellular trafficking of V-ATPases appears to be regulated by both PKA and AMPK (78). The same co-regulation has been observed in renal intercalated cells (79).
An additional regulatory mechanism that appears to play a role in controlling the differential pH of intracellular compartments is modulation of the coupling efficiency of proton transport and ATP hydrolysis by the V-ATPase (1, 80). Thus comparison of the coupling efficiency of Vph1p-containing complexes with that of Stv1p-containing complexes reveals a 4-5 fold higher efficiency for the former, consistent with the lower pH of the vacuolar compartment (containing Vph1p) relative to the Golgi (which contains Stv1p) (38). Construction of chimeric complexes between Vph1p and Stv1p has demonstrated that coupling efficiency is controlled by the C-terminal hydrophobic domain of subunit a (37). Interestingly, mutations, for example P217V, have been identified in the non-homologous domain of the catalytic A subunit which lead to increased coupling efficiency, even for Vph1p-containing complexes (75). This suggests that the V-ATPase is normally poised between a fully coupled and fully uncoupled state, and proton transport can be altered by either increasing or decreasing the coupling efficiency relative to this starting state. How coupling efficiency may be modulated in vivo remains uncertain.
Another mode of regulation of the V-ATPases is via reversible disulfide bond formation at the catalytic site of subunit A. In yeast, the conserved Cys-261 of the catalytic site and Cys-539 in the same subunit form a disulfide bond which results in reversible inactivation of the V-ATPase by blocking ATP hydrolysis (16). Reversal of inhibition occurs via a disulfide change within subunit A (81-83). Disulfide bond formation between these two conserved cysteine residues would prevent the major conformational change observed in the F1FO ATP synthase β subunit upon transition to the empty state (84). It should also be noted that the redox state of the cytoplasm of the cell can modulate V-ATPase activity by reversible disulfide bond formation in vivo (85).
In addition to V-ATPases, the pH of intracellular compartments is also influenced by other transporters, including passive transport proteins present in intracellular compartments. To dissipate the membrane potential that is generated by the V-ATPase a counterion conductance is required which, in vivo, is frequently provided by intracellular chloride channels (86). In kidney, defects in the chloride channel CLC-5 lead to defective endosomal acidification and cause Dent's disease (87). Failure of another chloride channel protein, CLC-7, causes defective bone resorption by osteoclasts, leading to osteopetrosis (88). The maintenance of the steady-state pH is also determined by passive proton conductance. For example, the luminal pH of the Golgi complex is, in part, controlled by a Zn2+-sensitive proton channel (89). In some cases, members of the Na+/H+ exchanger family may also regulate the pH of intracellular compartments. Thus knock-out of NHX1, the endosomal Na+/H+ exchanger of S. cerevisiae, leads to a decrease in vacuolar pH, suggesting that Nhx1p may be providing a proton leak by exchanging luminal protons for either cytoplasmic Na+ or, more likely, cytoplasmic K+ (90).
Concluding remarks and perspectives
The crucial role that V-ATPases play in normal physiological processes in eukaryotic cells likely accounts for both its structural complexity and the array of regulatory mechanisms employed to control its activity. These mechanisms allow V-ATPases localized to distinct cellular environments or organ systems to be independently regulated. The diversity of subunit isoforms which exist for the V-ATPases in mammalian systems also raises the possibility of selectively targeting particular V-ATPase complexes that are involved in disease processes, such as osteoporosis or cancer, with panels of isoform-specific inhibitors.
Acknowledgments
The authors wish to thank Dr. Ayana Hinton as well as Sarah Bond, Joseph Capecci, Satoko Toei and Rachel Nager for carefully reading the manuscript and their helpful discussions.
This work was supported by National Institutes of Health Grant GM34478 (to M.F.). M.T. was supported by a Postdoctoral Fellowship from the American Heart Association Founder's Affiliate and R.S. was supported through a Postdoctoral Fellowship from the Deutsche Forschungsgemeinschaft (SA 2044/1-1).
Abbreviations
- V-ATPase
vacuolar proton-translocating ATPase
- F-ATPase
F1F0 ATP synthase
- A1A0 ATPase
archaebacterial ATPase
References
- 1.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 2.Kane PM. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol Mol Biol Rev. 2006;70:177–191. doi: 10.1128/MMBR.70.1.177-191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner CA, Finberg KE, Breton S, Marshansky V, Brown D, Geibel JP. Renal vacuolar H+-ATPase. Physiol Rev. 2004;84:1263–1314. doi: 10.1152/physrev.00045.2003. [DOI] [PubMed] [Google Scholar]
- 4.Maxfield FR, McGraw TE. Endocytic recycling. Nat Rev Mol Cell Biol. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- 5.Gu F, Gruenberg J. ARF1 regulates pH-dependent COP functions in the early endocytic pathway. J Biol Chem. 2000;275:8154–8160. doi: 10.1074/jbc.275.11.8154. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh P, Dahms NM, Kornfeld S. Mannose 6-phosphate receptors: new twists in the tale. Nat Rev Mol Cell Biol. 2003;4:202–212. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- 7.Gruenberg J, van der Goot FG. Mechanisms of pathogen entry through the endosomal compartments. Nat Rev Mol Cell Biol. 2006;7:495–504. doi: 10.1038/nrm1959. [DOI] [PubMed] [Google Scholar]
- 8.Smith AN, Skaug J, Choate KA, Nayir A, Bakkaloglu A, Ozen S, Hulton SA, Sanjad SA, Al-Sabban EA, Lifton RP, Scherer SW, Karet FE. Mutations in ATP6N1B, encoding a new kidney vacuolar proton pump 116-kD subunit, cause recessive distal renal tubular acidosis with preserved hearing. Nat Genet. 2000;26:71–75. doi: 10.1038/79208. [DOI] [PubMed] [Google Scholar]
- 9.Toyomura T, Murata Y, Yamamoto A, Oka T, Sun-Wada GH, Wada Y, Futai M. From lysosomes to the plasma membrane: localization of vacuolar-type H+-ATPase with the a3 isoform during osteoclast differentiation. J Biol Chem. 2003;278:22023–22030. doi: 10.1074/jbc.M302436200. [DOI] [PubMed] [Google Scholar]
- 10.Frattini A, Orchard PJ, Sobacchi C, Giliani S, Abinun M, Mattsson JP, Keeling DJ, Andersson AK, Wallbrandt P, Zecca L, Notarangelo LD, Vezzoni P, Villa A. Defects in TCIRG1 subunit of the vacuolar proton pump are responsible for a subset of human autosomal recessive osteopetrosis. Nat Genet. 2000;25:343–346. doi: 10.1038/77131. [DOI] [PubMed] [Google Scholar]
- 11.Pietrement C, Sun-Wada GH, Silva ND, McKee M, Marshansky V, Brown D, Futai M, Breton S. Distinct expression patterns of different subunit isoforms of the V-ATPase in the rat epididymis. Biol Reprod. 2006;74:185–194. doi: 10.1095/biolreprod.105.043752. [DOI] [PubMed] [Google Scholar]
- 12.Sennoune SR, Bakunts K, Martinez GM, Chua-Tuan JL, Kebir Y, Attaya MN, Martinez-Zaguilan R. Vacuolar H+-ATPase in human breast cancer cells with distinct metastatic potential: distribution and functional activity. Am J Physiol Cell Physiol. 2004;286:C1443–1452. doi: 10.1152/ajpcell.00407.2003. [DOI] [PubMed] [Google Scholar]
- 13.Hinton A, Sennoune SR, Bond S, Fang M, Reuveni M, Sahagian GG, Jay D, Martinez-Zaguilan R, Forgac M. Function of a subunit isoforms of the V-ATPase in pH homeostasis and in vitro invasion of MDA-MB231 human breast cancer cells. J Biol Chem. 2009;284:16400–16408. doi: 10.1074/jbc.M901201200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gocheva V, Joyce JA. Cysteine cathepsins and the cutting edge of cancer invasion. Cell Cycle. 2007;6:60–64. doi: 10.4161/cc.6.1.3669. [DOI] [PubMed] [Google Scholar]
- 15.Liu Q, Kane PM, Newman PR, Forgac M. Site-directed mutagenesis of the yeast V-ATPase B subunit (Vma2p) J Biol Chem. 1996;271:2018–2022. doi: 10.1074/jbc.271.4.2018. [DOI] [PubMed] [Google Scholar]
- 16.Liu Q, Leng XH, Newman PR, Vasilyeva E, Kane PM, Forgac M. Site-directed mutagenesis of the yeast V-ATPase A subunit. J Biol Chem. 1997;272:11750–11756. doi: 10.1074/jbc.272.18.11750. [DOI] [PubMed] [Google Scholar]
- 17.MacLeod KJ, Vasilyeva E, Baleja JD, Forgac M. Mutational analysis of the nucleotide binding sites of the yeast vacuolar proton-translocating ATPase. J Biol Chem. 1998;273:150–156. doi: 10.1074/jbc.273.1.150. [DOI] [PubMed] [Google Scholar]
- 18.Vasilyeva E, Liu Q, MacLeod KJ, Baleja JD, Forgac M. Cysteine scanning mutagenesis of the noncatalytic nucleotide binding site of the yeast V-ATPase. J Biol Chem. 2000;275:255–260. doi: 10.1074/jbc.275.1.255. [DOI] [PubMed] [Google Scholar]
- 19.Powell B, Graham LA, Stevens TH. Molecular characterization of the yeast vacuolar H+-ATPase proton pore. J Biol Chem. 2000;275:23654–23660. doi: 10.1074/jbc.M004440200. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Cipriano DJ, Forgac M. Arrangement of subunits in the proteolipid ring of the V-ATPase. J Biol Chem. 2007;282:34058–34065. doi: 10.1074/jbc.M704331200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flannery AR, Graham LA, Stevens TH. Topological characterization of the c, c′, and c″ subunits of the vacuolar ATPase from the yeast Saccharomyces cerevisiae. J Biol Chem. 2004;279:39856–39862. doi: 10.1074/jbc.M406767200. [DOI] [PubMed] [Google Scholar]
- 22.Hirata R, Graham LA, Takatsuki A, Stevens TH, Anraku Y. VMA11 and VMA16 encode second and third proteolipid subunits of the Saccharomyces cerevisiae vacuolar membrane H+-ATPase. J Biol Chem. 1997;272:4795–4803. doi: 10.1074/jbc.272.8.4795. [DOI] [PubMed] [Google Scholar]
- 23.Nishi T, Kawasaki-Nishi S, Forgac M. The first putative transmembrane segment of subunit c″ (Vma16p) of the yeast V-ATPase is not necessary for function. J Biol Chem. 2003;278:5821–5827. doi: 10.1074/jbc.M209875200. [DOI] [PubMed] [Google Scholar]
- 24.Wilkens S, Vasilyeva E, Forgac M. Structure of the vacuolar ATPase by electron microscopy. J Biol Chem. 1999;274:31804–31810. doi: 10.1074/jbc.274.45.31804. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Z, Zheng Y, Mazon H, Milgrom E, Kitagawa N, Kish-Trier E, Heck AJ, Kane PM, Wilkens S. Structure of the yeast vacuolar ATPase. J Biol Chem. 2008;283:35983–35995. doi: 10.1074/jbc.M805345200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arata Y, Baleja JD, Forgac M. Localization of subunits D, E, and G in the yeast V-ATPase complex using cysteine-mediated cross-linking to subunit B. Biochemistry. 2002;41:11301–11307. doi: 10.1021/bi0262449. [DOI] [PubMed] [Google Scholar]
- 27.Iwata M, Imamura H, Stambouli E, Ikeda C, Tamakoshi M, Nagata K, Makyio H, Hankamer B, Barber J, Yoshida M, Yokoyama K, Iwata S. Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole-type ATPase. Proc Natl Acad Sci U S A. 2004;101:59–64. doi: 10.1073/pnas.0305165101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilkens S, Inoue T, Forgac M. Three-dimensional structure of the vacuolar ATPase. Localization of subunit H by difference imaging and chemical cross-linking. J Biol Chem. 2004;279:41942–41949. doi: 10.1074/jbc.M407821200. [DOI] [PubMed] [Google Scholar]
- 29.Féthière J, Venzke D, Diepholz M, Seybert A, Geerlof A, Gentzel M, Wilm M, Böttcher B. Building the stator of the yeast vacuolar-ATPase: specific interaction between subunits E and G. J Biol Chem. 2004;279:40670–40676. doi: 10.1074/jbc.M407086200. [DOI] [PubMed] [Google Scholar]
- 30.Inoue T, Forgac M. Cysteine-mediated cross-linking indicates that subunit C of the V-ATPase is in close proximity to subunits E and G of the V1 domain and subunit a of the V0 domain. J Biol Chem. 2005;280:27896–27903. doi: 10.1074/jbc.M504890200. [DOI] [PubMed] [Google Scholar]
- 31.Imamura H, Nakano M, Noji H, Muneyuki E, Ohkuma S, Yoshida M, Yokoyama K. Evidence for rotation of V1-ATPase. Proc Natl Acad Sci U S A. 2003;100:2312–2315. doi: 10.1073/pnas.0436796100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirata T, Iwamoto-Kihara A, Sun-Wada GH, Okajima T, Wada Y, Futai M. Subunit rotation of vacuolar-type proton pumping ATPase: relative rotation of the G and C subunits. J Biol Chem. 2003;278:23714–23719. doi: 10.1074/jbc.M302756200. [DOI] [PubMed] [Google Scholar]
- 33.Kawasaki-Nishi S, Nishi T, Forgac M. Arg-735 of the 100-kDa subunit a of the yeast V-ATPase is essential for proton translocation. Proc Natl Acad Sci U S A. 2001;98:12397–12402. doi: 10.1073/pnas.221291798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stock D, Gibbons C, Arechaga I, Leslie AG, Walker JE. The rotary mechanism of ATP synthase. Curr Opin Struct Biol. 2000;10:672–679. doi: 10.1016/s0959-440x(00)00147-0. [DOI] [PubMed] [Google Scholar]
- 35.Cross RL, Müller V. The evolution of A-, F-, and V-type ATP synthases and ATPases: reversals in function and changes in the H+/ATP coupling ratio. FEBS Lett. 2004;576:1–4. doi: 10.1016/j.febslet.2004.08.065. [DOI] [PubMed] [Google Scholar]
- 36.Manolson MF, Wu B, Proteau D, Taillon BE, Roberts BT, Hoyt MA, Jones EW. STV1 gene encodes functional homologue of 95-kDa yeast vacuolar H(+)-ATPase subunit Vph1p. J Biol Chem. 1994;269:14064–14074. [PubMed] [Google Scholar]
- 37.Kawasaki-Nishi S, Bowers K, Nishi T, Forgac M, Stevens TH. The amino-terminal domain of the vacuolar proton-translocating ATPase a subunit controls targeting and in vivo dissociation, and the carboxyl-terminal domain affects coupling of proton transport and ATP hydrolysis. J Biol Chem. 2001;276:47411–47420. doi: 10.1074/jbc.M108310200. [DOI] [PubMed] [Google Scholar]
- 38.Kawasaki-Nishi S, Nishi T, Forgac M. Yeast V-ATPase complexes containing different isoforms of the 100-kDa a-subunit differ in coupling efficiency and in vivo dissociation. J Biol Chem. 2001;276:17941–17948. doi: 10.1074/jbc.M010790200. [DOI] [PubMed] [Google Scholar]
- 39.Sun-Wada GH, Toyomura T, Murata Y, Yamamoto A, Futai M, Wada Y. The a3 isoform of V-ATPase regulates insulin secretion from pancreatic beta-cells. J Cell Sci. 2006;119:4531–4540. doi: 10.1242/jcs.03234. [DOI] [PubMed] [Google Scholar]
- 40.Sun-Wada GH, Tabata H, Kawamura N, Futai M, Wada Y. Differential expression of a subunit isoforms of the vacuolar-type proton pump ATPase in mouse endocrine tissues. Cell Tissue Res. 2007;329:239–248. doi: 10.1007/s00441-007-0421-7. [DOI] [PubMed] [Google Scholar]
- 41.Tabata H, Kawamura N, Sun-Wada GH, Wada Y. Vacuolar-type H(+)-ATPase with the a3 isoform is the proton pump on premature melanosomes. Cell Tissue Res. 2008;332:447–460. doi: 10.1007/s00441-008-0597-5. [DOI] [PubMed] [Google Scholar]
- 42.Kawasaki-Nishi S, Yamaguchi A, Forgac M, Nishi T. Tissue specific expression of the splice variants of the mouse vacuolar proton-translocating ATPase a4 subunit. Biochem Biophys Res Commun. 2007;364:1032–1036. doi: 10.1016/j.bbrc.2007.10.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morel N, Dedieu JC, Philippe JM. Specific sorting of the a1 isoform of the V-H+ATPase a subunit to nerve terminals where it associates with both synaptic vesicles and the presynaptic plasma membrane. J Cell Sci. 2003;116:4751–4762. doi: 10.1242/jcs.00791. [DOI] [PubMed] [Google Scholar]
- 44.Hiesinger PR, Fayyazuddin A, Mehta SQ, Rosenmund T, Schulze KL, Zhai RG, Verstreken P, Cao Y, Zhou Y, Kunz J, Bellen HJ. The v-ATPase V0 subunit a1 is required for a late step in synaptic vesicle exocytosis in Drosophila. Cell. 2005;121:607–620. doi: 10.1016/j.cell.2005.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peri F, Nüsslein-Volhard C. Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell. 2008;133:916–927. doi: 10.1016/j.cell.2008.04.037. [DOI] [PubMed] [Google Scholar]
- 46.Hurtado-Lorenzo A, Skinner M, El Annan J, Futai M, Sun-Wada GH, Bourgoin S, Casanova J, Wildeman A, Bechoua S, Ausiello DA, Brown D, Marshansky V. V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat Cell Biol. 2006;8:124–136. doi: 10.1038/ncb1348. [DOI] [PubMed] [Google Scholar]
- 47.Bartkiewicz M, Hernando N, Reddy SV, Roodman GD, Baron R. Characterization of the osteoclast vacuolar H(+)-ATPase B-subunit. Gene. 1995;160:157–164. doi: 10.1016/0378-1119(95)00228-x. [DOI] [PubMed] [Google Scholar]
- 48.Smith AN, Borthwick KJ, Karet FE. Molecular cloning and characterization of novel tissue-specific isoforms of the human vacuolar H(+)-ATPase C, G and d subunits, and their evaluation in autosomal recessive distal renal tubular acidosis. Gene. 2002;297:169–177. doi: 10.1016/s0378-1119(02)00884-3. [DOI] [PubMed] [Google Scholar]
- 49.Imai-Senga Y, Sun-Wada GH, Wada Y, Futai M. A human gene, ATP6E1, encoding a testis-specific isoform of H(+)-ATPase subunit E. Gene. 2002;289:7–12. doi: 10.1016/s0378-1119(02)00542-5. [DOI] [PubMed] [Google Scholar]
- 50.Karet FE, Finberg KE, Nelson RD, Nayir A, Mocan H, Sanjad SA, Rodriguez-Soriano J, Santos F, Cremers CW, Di Pietro A, Hoffbrand BI, Winiarski J, Bakkaloglu A, Ozen S, Dusunsel R, Goodyer P, Hulton SA, Wu DK, Skvorak AB, Morton CC, Cunningham MJ, Jha V, Lifton RP. Mutations in the gene encoding B1 subunit of H+-ATPase cause renal tubular acidosis with sensorineural deafness. Nat Genet. 1999;21:84–90. doi: 10.1038/5022. [DOI] [PubMed] [Google Scholar]
- 51.Paunescu TG, Da Silva N, Marshansky V, McKee M, Breton S, Brown D. Expression of the 56-kDa B2 subunit isoform of the vacuolar H(+)-ATPase in proton-secreting cells of the kidney and epididymis. Am J Physiol Cell Physiol. 2004;287:C149–162. doi: 10.1152/ajpcell.00464.2003. [DOI] [PubMed] [Google Scholar]
- 52.Da Silva N, Shum WW, El-Annan J, Paunescu TG, McKee M, Smith PJ, Brown D, Breton S. Relocalization of the V-ATPase B2 subunit to the apical membrane of epididymal clear cells of mice deficient in the B1 subunit. Am J Physiol Cell Physiol. 2007;293:C199–210. doi: 10.1152/ajpcell.00596.2006. [DOI] [PubMed] [Google Scholar]
- 53.Brown D, Paunescu TG, Breton S, Marshansky V. Regulation of the V-ATPase in kidney epithelial cells: dual role in acid-base homeostasis and vesicle trafficking. J Exp Biol. 2009;212:1762–1772. doi: 10.1242/jeb.028803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paunescu TG, Jones AC, Tyszkowski R, Brown D. V-ATPase expression in the mouse olfactory epithelium. Am J Physiol Cell Physiol. 2008;295:C923–930. doi: 10.1152/ajpcell.00237.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun-Wada GH, Yoshimizu T, Imai-Senga Y, Wada Y, Futai M. Diversity of mouse proton-translocating ATPase: presence of multiple isoforms of the C, d and G subunits. Gene. 2003;302:147–153. doi: 10.1016/s0378-1119(02)01099-5. [DOI] [PubMed] [Google Scholar]
- 56.Smith AN, Jouret F, Bord S, Borthwick KJ, Al-Lamki RS, Wagner CA, Ireland DC, Cormier-Daire V, Frattini A, Villa A, Kornak U, Devuyst O, Karet FE. Vacuolar H+-ATPase d2 subunit: molecular characterization, developmental regulation, and localization to specialized proton pumps in kidney and bone. J Am Soc Nephrol. 2005;16:1245–1256. doi: 10.1681/ASN.2004090761. [DOI] [PubMed] [Google Scholar]
- 57.Lee SH, Rho J, Jeong D, Sul JY, Kim T, Kim N, Kang JS, Miyamoto T, Suda T, Lee SK, Pignolo RJ, Koczon-Jaremko B, Lorenzo J, Choi Y. V-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat Med. 2006;12:1403–1409. doi: 10.1038/nm1514. [DOI] [PubMed] [Google Scholar]
- 58.Smith AN, Francis RW, Sorrell SL, Karet FE. The d subunit plays a central role in human vacuolar H(+)-ATPases. J Bioenerg Biomembr. 2008;40:371–380. doi: 10.1007/s10863-008-9161-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beyenbach KW, Wieczorek H. The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J Exp Biol. 2006;209:577–589. doi: 10.1242/jeb.02014. [DOI] [PubMed] [Google Scholar]
- 60.Sautin YY, Lu M, Gaugler A, Zhang L, Gluck SL. Phosphatidylinositol 3-kinase-mediated effects of glucose on vacuolar H+-ATPase assembly, translocation, and acidification of intracellular compartments in renal epithelial cells. Mol Cell Biol. 2005;25:575–589. doi: 10.1128/MCB.25.2.575-589.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I. Activation of lysosomal function during dendritic cell maturation. Science. 2003;299:1400–1403. doi: 10.1126/science.1080106. [DOI] [PubMed] [Google Scholar]
- 62.Kane PM. Disassembly and reassembly of the yeast vacuolar H(+)-ATPase in vivo. J Biol Chem. 1995;270:17025–17032. [PubMed] [Google Scholar]
- 63.Parra KJ, Kane PM. Reversible association between the V1 and V0 domains of yeast vacuolar H+-ATPase is an unconventional glucose-induced effect. Mol Cell Biol. 1998;18:7064–7074. doi: 10.1128/mcb.18.12.7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qi J, Forgac M. Cellular environment is important in controlling V-ATPase dissociation and its dependence on activity. J Biol Chem. 2007;282:24743–24751. doi: 10.1074/jbc.M700663200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parra KJ, Keenan KL, Kane PM. The H subunit (Vma13p) of the yeast V-ATPase inhibits the ATPase activity of cytosolic V1 complexes. J Biol Chem. 2000;275:21761–21767. doi: 10.1074/jbc.M002305200. [DOI] [PubMed] [Google Scholar]
- 66.Jefferies KC, Forgac M. Subunit H of the vacuolar (H+) ATPase inhibits ATP hydrolysis by the free V1 domain by interaction with the rotary subunit F. J Biol Chem. 2008;283:4512–4519. doi: 10.1074/jbc.M707144200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Diab H, Ohira M, Liu M, Cobb E, Kane PM. Subunit interactions and requirements for inhibition of the yeast V1-ATPase. J Biol Chem. 2009;284:13316–13325. doi: 10.1074/jbc.M900475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu T, Forgac M. Microtubules are involved in glucose-dependent dissociation of the yeast vacuolar [H+]-ATPase in vivo. J Biol Chem. 2001;276:24855–24861. doi: 10.1074/jbc.M100637200. [DOI] [PubMed] [Google Scholar]
- 69.Seol JH, Shevchenko A, Deshaies RJ. Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat Cell Biol. 2001;3:384–391. doi: 10.1038/35070067. [DOI] [PubMed] [Google Scholar]
- 70.Smardon AM, Tarsio M, Kane PM. The RAVE complex is essential for stable assembly of the yeast V-ATPase. J Biol Chem. 2002;277:13831–13839. doi: 10.1074/jbc.M200682200. [DOI] [PubMed] [Google Scholar]
- 71.Smardon AM, Kane PM. RAVE is essential for the efficient assembly of the C subunit with the vacuolar H(+)-ATPase. J Biol Chem. 2007;282:26185–26194. doi: 10.1074/jbc.M703627200. [DOI] [PubMed] [Google Scholar]
- 72.Lu M, Ammar D, Ives H, Albrecht F, Gluck SL. Physical interaction between aldolase and vacuolar H+-ATPase is essential for the assembly and activity of the proton pump. J Biol Chem. 2007;282:24495–24503. doi: 10.1074/jbc.M702598200. [DOI] [PubMed] [Google Scholar]
- 73.Bond S, Forgac M. The Ras/cAMP/protein kinase A pathway regulates glucose-dependent assembly of the vacuolar (H+)-ATPase in yeast. J Biol Chem. 2008;283:36513–36521. doi: 10.1074/jbc.M805232200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Voss M, Vitavska O, Walz B, Wieczorek H, Baumann O. Stimulus-induced phosphorylation of vacuolar H(+)-ATPase by protein kinase A. J Biol Chem. 2007;282:33735–33742. doi: 10.1074/jbc.M703368200. [DOI] [PubMed] [Google Scholar]
- 75.Shao E, Nishi T, Kawasaki-Nishi S, Forgac M. Mutational analysis of the non-homologous region of subunit A of the yeast V-ATPase. J Biol Chem. 2003;278:12985–12991. doi: 10.1074/jbc.M212096200. [DOI] [PubMed] [Google Scholar]
- 76.Shao E, Forgac M. Involvement of the nonhomologous region of subunit A of the yeast V-ATPase in coupling and in vivo dissociation. J Biol Chem. 2004;279:48663–48670. doi: 10.1074/jbc.M408278200. [DOI] [PubMed] [Google Scholar]
- 77.Pastor-Soler N, Beaulieu V, Litvin TN, Da Silva N, Chen Y, Brown D, Buck J, Levin LR, Breton S. Bicarbonate-regulated adenylyl cyclase (sAC) is a sensor that regulates pH-dependent V-ATPase recycling. J Biol Chem. 2003;278:49523–49529. doi: 10.1074/jbc.M309543200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hallows KR, Alzamora R, Li H, Gong F, Smolak C, Neumann D, Pastor-Soler NM. AMP-activated protein kinase inhibits alkaline pH- and PKA-induced apical vacuolar H+-ATPase accumulation in epididymal clear cells. Am J Physiol Cell Physiol. 2009;296:C672–681. doi: 10.1152/ajpcell.00004.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gong F, Alzamora R, Smolak C, Li H, Naveed S, Neumann D, Hallows KR, Pastor-Soler NM. Vacuolar H+-ATPase apical accumulation in kidney intercalated cells is regulated by PKA and AMP-activated protein kinase. Am J Physiol Renal Physiol. 2010 doi: 10.1152/ajprenal.00645.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nelson N. A journey from mammals to yeast with vacuolar H+-ATPase (V-ATPase) J Bioenerg Biomembr. 2003;35:281–289. doi: 10.1023/a:1025768529677. [DOI] [PubMed] [Google Scholar]
- 81.Feng Y, Forgac M. Cysteine 254 of the 73-kDa A subunit is responsible for inhibition of the coated vesicle (H+)-ATPase upon modification by sulfhydryl reagents. J Biol Chem. 1992;267:5817–5822. [PubMed] [Google Scholar]
- 82.Feng Y, Forgac M. A novel mechanism for regulation of vacuolar acidification. J Biol Chem. 1992;267:19769–19772. [PubMed] [Google Scholar]
- 83.Feng Y, Forgac M. Inhibition of vacuolar H(+)-ATPase by disulfide bond formation between cysteine 254 and cysteine 532 in subunit A. J Biol Chem. 1994;269:13224–13230. [PubMed] [Google Scholar]
- 84.Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 85.Oluwatosin YE, Kane PM. Mutations in the CYS4 gene provide evidence for regulation of the yeast vacuolar H+-ATPase by oxidation and reduction in vivo. J Biol Chem. 1997;272:28149–28157. doi: 10.1074/jbc.272.44.28149. [DOI] [PubMed] [Google Scholar]
- 86.Jentsch TJ. Chloride and the endosomal-lysosomal pathway: emerging roles of CLC chloride transporters. J Physiol. 2007;578:633–640. doi: 10.1113/jphysiol.2006.124719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Günther W, Lüchow A, Cluzeaud F, Vandewalle A, Jentsch TJ. ClC-5, the chloride channel mutated in Dent's disease, colocalizes with the proton pump in endocytotically active kidney cells. Proc Natl Acad Sci U S A. 1998;95:8075–8080. doi: 10.1073/pnas.95.14.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kornak U, Kasper D, Bosl MR, Kaiser E, Schweizer M, Schulz A, Friedrich W, Delling G, Jentsch TJ. Loss of the ClC-7 chloride channel leads to osteopetrosis in mice and man. Cell. 2001;104:205–215. doi: 10.1016/s0092-8674(01)00206-9. [DOI] [PubMed] [Google Scholar]
- 89.Schapiro FB, Grinstein S. Determinants of the pH of the Golgi complex. J Biol Chem. 2000;275:21025–21032. doi: 10.1074/jbc.M002386200. [DOI] [PubMed] [Google Scholar]
- 90.Brett CL, Tukaye DN, Mukherjee S, Rao R. The yeast endosomal Na+K+/H+ exchanger Nhx1 regulates cellular pH to control vesicle trafficking. Mol Biol Cell. 2005;16:1396–1405. doi: 10.1091/mbc.E04-11-0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nishi T, Forgac M. The vacuolar (H+)-ATPases-nature's most versatile proton pumps. Nat Rev Mol Cell Biol. 2002;3:94–103. doi: 10.1038/nrm729. [DOI] [PubMed] [Google Scholar]
- 92.Lu M, Sautin YY, Holliday LS, Gluck SL. The glycolytic enzyme aldolase mediates assembly, expression, and activity of vacuolar H+-ATPase. J Biol Chem. 2004;279:8732–8739. doi: 10.1074/jbc.M303871200. [DOI] [PubMed] [Google Scholar]
- 93.Holliday LS, Lu M, Lee BS, Nelson RD, Solivan S, Zhang L, Gluck SL. The amino-terminal domain of the B subunit of vacuolar H+-ATPase contains a filamentous actin binding site. J Biol Chem. 2000;275:32331–32337. doi: 10.1074/jbc.M004795200. [DOI] [PubMed] [Google Scholar]
- 94.Drory O, Frolow F, Nelson N. Crystal structure of yeast V-ATPase subunit C reveals its stator function. EMBO Rep. 2004;5:1148–1152. doi: 10.1038/sj.embor.7400294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vitavska O, Wieczorek H, Merzendorfer H. A novel role for subunit C in mediating binding of the H+-V-ATPase to the actin cytoskeleton. J Biol Chem. 2003;278:18499–18505. doi: 10.1074/jbc.M212844200. [DOI] [PubMed] [Google Scholar]
- 96.Vitavska O, Merzendorfer H, Wieczorek H. The V-ATPase subunit C binds to polymeric F-actin as well as to monomeric G-actin and induces cross-linking of actin filaments. J Biol Chem. 2005;280:1070–1076. doi: 10.1074/jbc.M406797200. [DOI] [PubMed] [Google Scholar]
- 97.Sun-Wada GH, Murata Y, Namba M, Yamamoto A, Wada Y, Futai M. Mouse proton pump ATPase C subunit isoforms (C2-a and C2-b) specifically expressed in kidney and lung. J Biol Chem. 2003;278:44843–44851. doi: 10.1074/jbc.M307197200. [DOI] [PubMed] [Google Scholar]
- 98.Makyio H, Iino R, Ikeda C, Imamura H, Tamakoshi M, Iwata M, Stock D, Bernal RA, Carpenter EP, Yoshida M, Yokoyama K, Iwata S. Structure of a central stalk subunit F of prokaryotic V-type ATPase/synthase from Thermus thermophilus. EMBO J. 2005;24:3974–3983. doi: 10.1038/sj.emboj.7600859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Murata Y, Sun-Wada GH, Yoshimizu T, Yamamoto A, Wada Y, Futai M. Differential localization of the vacuolar H+ pump with G subunit isoforms (G1 and G2) in mouse neurons. J Biol Chem. 2002;277:36296–36303. doi: 10.1074/jbc.M200586200. [DOI] [PubMed] [Google Scholar]
- 100.Sagermann M, Stevens TH, Matthews BW. Crystal structure of the regulatory subunit H of the V-type ATPase of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2001;98:7134–7139. doi: 10.1073/pnas.131192798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Geyer M, Yu H, Mandic R, Linnemann T, Zheng YH, Fackler OT, Peterlin BM. Subunit H of the V-ATPase binds to the medium chain of adaptor protein complex 2 and connects Nef to the endocytic machinery. J Biol Chem. 2002;277:28521–28529. doi: 10.1074/jbc.M200522200. [DOI] [PubMed] [Google Scholar]
- 102.Zhou Z, Peng SB, Crider BP, Slaughter C, Xie XS, Stone DK. Molecular characterization of the 50- and 57-kDa subunits of the bovine vacuolar proton pump. J Biol Chem. 1998;273:5878–5884. doi: 10.1074/jbc.273.10.5878. [DOI] [PubMed] [Google Scholar]
- 103.Zhou Z, Peng SB, Crider BP, Andersen P, Xie XS, Stone DK. Recombinant SFD isoforms activate vacuolar proton pumps. J Biol Chem. 1999;274:15913–15919. doi: 10.1074/jbc.274.22.15913. [DOI] [PubMed] [Google Scholar]
- 104.Liegeois S, Benedetto A, Garnier JM, Schwab Y, Labouesse M. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegans. J Cell Biol. 2006;173:949–961. doi: 10.1083/jcb.200511072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Poea-Guyon S, Amar M, Fossier P, Morel N. Alternative splicing controls neuronal expression of v-ATPase subunit a1 and sorting to nerve terminals. J Biol Chem. 2006;281:17164–17172. doi: 10.1074/jbc.M600927200. [DOI] [PubMed] [Google Scholar]
- 106.Nishi T, Forgac M. Molecular cloning and expression of three isoforms of the 100-kDa a subunit of the mouse vacuolar proton-translocating ATPase. J Biol Chem. 2000;275:6824–6830. doi: 10.1074/jbc.275.10.6824. [DOI] [PubMed] [Google Scholar]
- 107.Nishi T, Kawasaki-Nishi S, Forgac M. Expression and function of the mouse V-ATPase d subunit isoforms. J Biol Chem. 2003;278:46396–46402. doi: 10.1074/jbc.M303924200. [DOI] [PubMed] [Google Scholar]
- 108.Owegi MA, Pappas DL, Finch MW, Jr, Bilbo SA, Resendiz CA, Jacquemin LJ, Warrier A, Trombley JD, McCulloch KM, Margalef KL, Mertz MJ, Storms JM, Damin CA, Parra KJ. Identification of a domain in the V0 subunit d that is critical for coupling of the yeast vacuolar proton-translocating ATPase. J Biol Chem. 2006;281:30001–30014. doi: 10.1074/jbc.M605006200. [DOI] [PubMed] [Google Scholar]
- 109.Sato K, Shikano S, Xia G, Takao J, Chung JS, Cruz PD, Jr, Xie XS, Ariizumi K. Selective expression of vacuolar H+-ATPase subunit d2 by particular subsets of dendritic cells among leukocytes. Mol Immunol. 2006;43:1443–1453. doi: 10.1016/j.molimm.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 110.Sambade M, Kane PM. The yeast vacuolar proton-translocating ATPase contains a subunit homologous to the Manduca sexta and bovine e subunits that is essential for function. J Biol Chem. 2004;279:17361–17365. doi: 10.1074/jbc.M314104200. [DOI] [PubMed] [Google Scholar]
- 111.Malkus P, Graham LA, Stevens TH, Schekman R. Role of Vma21p in assembly and transport of the yeast vacuolar ATPase. Mol Biol Cell. 2004;15:5075–5091. doi: 10.1091/mbc.E04-06-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Supek F, Supekova L, Mandiyan S, Pan YC, Nelson H, Nelson N. A novel accessory subunit for vacuolar H(+)-ATPase from chromaffin granules. J Biol Chem. 1994;269:24102–24106. [PubMed] [Google Scholar]
- 113.Jansen EJ, van Bakel NH, Coenen AJ, van Dooren SH, van Lith HA, Martens GJ. An isoform of the vacuolar (H(+))-ATPase accessory subunit Ac45. Cell Mol Life Sci. 2010;67:629–640. doi: 10.1007/s00018-009-0200-6. [DOI] [PMC free article] [PubMed] [Google Scholar]


