Abstract
The possibility of independently manipulating the affinity and efficacy of pore-blocking ligands of sodium channels is of interest for the development of new drugs for the treatment of pain. The analgesic µ-conotoxin KIIIA, a 16-residue peptide with three disulfide bridges, is a pore-blocker of voltage-gated sodium channels, including the neuronal subtype NaV1.2 (Kd of 5 nM). At saturating concentrations, µ-KIIIA incompletely blocks the sodium current of NaV1.2, leaving a 5% residual current (rINa). Lys7 is an important residue: the mutation K7A decreases both the efficacy (i.e., increases rINa to 23%) and the affinity of the peptide (Kd, 115 nM). In this report, various replacements of residue 7 were examined to determine whether affinity and efficacy were inexorably linked. Because of their facile chemical synthesis, KIIIA analogs were used that had as a core structure the disulfide-depleted KIIIA[C1A,C2U,C9A,C5U] (where U is selenocysteine) or ddKIIIA. The analogs ddKIIIA and ddKIIIA[K7X], where X represents one of nine different amino acids, were tested on voltage-clamped Xenopus oocytes expressing rat NaV1.2 or NaV1.4. Their affinities ranged from 0.01 to 36 µM and rINa's from 2 to 42%, and these two variables appeared uncorrelated. Instead, rINa varied inversely with side chain size, and remarkably charge and hydrophobicity appeared inconsequential. The ability to manipulate a µ-conopeptide's affinity and efficacy, as well as its capacity to interfere with subsequent tetrodotoxin-binding, greatly expands its scope as a reagent to probe sodium channel structure and function, and may also lead to the development of µ-conotoxins as safe analgesics.
Voltage-gated sodium channels (VGSCs) are responsible for the upstroke of action potentials, which mediate rapid communication of electrical signals in neuron and muscle (1). Mammals have nine isoforms of the pore-forming α-subunit of VGSCs (NaV1.1 through 1.9) and four isoforms of associated β-subunits (β1 through β4) (2–4). The importance of VGSCs is reflected by the fact that they are targets of many neurotoxins that have convergently evolved in a variety of organisms (5–8). These neurotoxins are of considerable interest not only as probes to investigate the functioning of these channels but also as potential therapeutics for neurological disorders including neuropathic pain and epilepsy (9, 10).
Four families of VGSC-targeting conopeptides, each with a distinct mechanism of action, are known: µ-, µO-, δ- and ι-conotoxins. The first two are antagonists, and the latter two are agonist (8, 11). These conopeptides have multiple disulfide bridges, with members of each family having a characteristic disulfide framework. Of these families, µ-conotoxins were the first to be discovered and like the guanidinium alkaloids, tetrodotoxin (TTX) and saxitoxin (STX), are pore blockers that act at neurotoxin receptor site 1 (8, 12–15). Among the more recently discovered members of this family is µ-conotoxin KIIIA (16). KIIIA has several distinctive features when compared with µ-conotoxin GIIIA, the first- and best-studied member of the family. 1. GIIIA has 22 residues, whereas KIIIA has 16 and is the smallest µ-conopeptide described so far (see Fig. 1). 2. Unlike GIIIA, which has highest affinity for NaV1.4 (the skeletal muscle-specific VGSC), KIIIA has highest affinity for NaV1.2 (a neuronal VGSC) (17). Furthermore, KIIIA also exhibits analgesic activity (17). 3. When GIIIA binds to NaV1.4, it blocks the sodium current (INa) essentially completely; in contrast, when KIIIA binds NaV1.2, a small residual sodium current (rINa, 5% of control) persists (18). 4. When the R13Q mutant of GIIIA, GIIIA[R13Q], binds to NaV1.4, a finite rINa of 28% is observed in single-channel recordings in planar lipid bilayers (19). A loss-of-positive-charge mutation at the presumed homologous location in KIIIA, K7A (see Fig. 1), likewise increases rINa (18). However, unlike the rINa of GIIIA[R13Q], which could not be blocked by decarbamoly-STX (19), the rINa of both KIIIA and KIIIA[K7A] can be blocked by TTX (18) and decarbamoly-STX (Zhang et al., in preparation); thus, guanidinium alkaloid, KIIIA (or KIIIA[K7A]), and NaV1.2 can form a ternary complex.
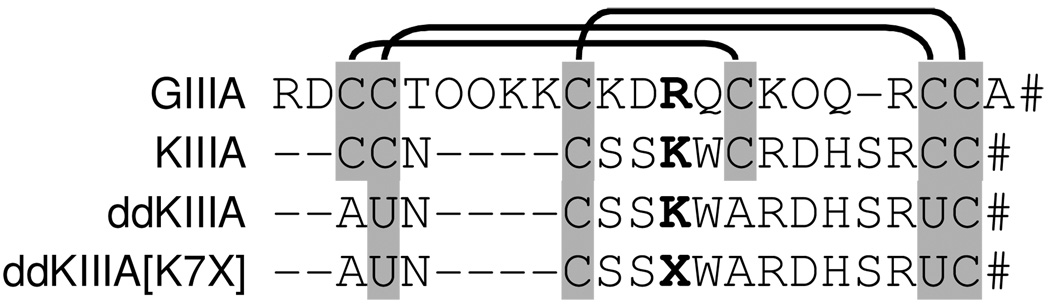
Figure 1.
Aligned sequences of µ-conotoxin GIIIA, KIIIA, ddKIIIA, and ddKIIIA[K7X]. GIIIA and KIIIA have the three canonical disulfide-bridges of µ-conotoxins: between 1st and 4th, 2nd and 5th, and 3rd and 6th cysteine residues (as indicated by lines above the sequences). ddKIIIA and ddKIIIA[K7X] have the same bridge framework except the first disulfide bridge was deleted and the second was replaced with a diselenide bridge. The long moniker of ddKIIIA[K7X] is KIIIA[C1A,C2U,K7X,C9A,C5U], where in the present report, residue X at position 7 was either Ala, Asp, Gly, Leu, Lys (i.e., ddKIIIA), Phe, Ser, Thr, Val, or diaminoproprionate (dap). O, hydroxyproline; #, amide; U, selenocysteine; shading, disulfide- or diselenide-bridged residues.
The K7A mutation in KIIIA not only increased rINa but also increased the peptide's Kd for NaV1.2 (18); thus, the mutation decreased both efficacy and affinity, raising the possibility that affinity and efficacy might be immutably linked. In this report, we investigated this further by positional scanning. Exploration of the structure-activity relationships of multiply disulfide-bonded peptides, such as µ-conotoxins, requires correctly folding each synthetic peptide, a time consuming and laborious process. Therefore, we employed a newly developed method for the facile synthesis of disulfide-bonded peptides that exploits two recent observations: 1, the first disulfide bridge, that formed by the 1st and 4th cysteines of KIIIA, can be removed with only modest attenuation of functional activity (20); and 2, a diselenide bridge can be substituted for one of the disulfide bridges, which results in a construct that is significantly easier to fold (21). Regarding the latter, it is noteworthy that the selenopeptide derivatives of α-, ω- and µ-conopeptides have recently been shown to retain bioactivities of their native conotoxin counterparts (22–24). Incorporation of both modifications into KIIIA results in the disulfide-depleted peptide KIIIA[C1A,C2U,C9A,C5U] (where U represents selenocysteine, see Fig. 1) or ddKIIIA, which retains functional activity (25). Thus, ddKIIIA served as a more readily synthesized surrogate of KIIIA and was used as a core structure for positional scanning; specifically, the residue at position 7 was mutated to form ddKIIIA[K7X], where X was one of nine amino acids differing in size, charge, or hydrophobicity. This investigation showcases the usefulness disulfide-depleted, selenocysteine-substituted, peptides and at the same time reveals that derivatives of KIIIA with divergent affinities versus efficacies could be synthesized. These properties are particularly interesting, since the KIIIA is an analgesic (17) with possibilities as a lead compound in the development potential therapeutics for the treatment of pain.
MATERIALS AND METHODS
Oocyte Electrophysiology
Oocytes expressing NaV1.2 or 1.4 were prepared and voltage-clamped essentially as previously described (11, 17). Briefly, a given oocyte was injected with 30 nl cRNA of rat NaV1.2 or rat NaV1.4 (1.5 or 0.6 ng, respectively) and incubated 1 to 4 days at 16 °C in ND96 composed (in mM) of: NaCl (96), KCl (2), CaCl2 (1.8) MgCl2 (1), and HEPES (5), pH 7.5. The incubation medium also contained the antibiotics penicillin (100 units/ml), streptomycin (0.1 mg/ml), amikacin (0.1 mg/ml), and Septra (0.2 mg/ml). Oocytes in ND96 were two-electrode voltage-clamped using microelectrodes containing 3M KCl (<0.5 MΩ) and held a membrane potential of −80 mV. Sodium channels were activated by stepping the potential to −10 mV (unless otherwise specified) for 50 ms every 20 sec. Current signals were filtered at 2 KHz, digitized at a sampling frequency of 10 KHz, and leak-subtracted by a P/8 protocol using in-house software written in LabVIEW (National Instruments, Austin, TX).
The oocyte-recording chamber consisted of a well (4 mm diameter, 30 µl volume) in Sylgard (Dow Corning, Midland MI), a silicone elastomer. Oocytes were exposed to toxin by applying 3 µl of toxin at 10-times its final concentration with a pipettor and manually stirring the bath for a few seconds by gently aspirating and expelling a few µl of the bath fluid several times with the pipettor. Toxin exposures were in a static bath to conserve material. Toxin-containing solution was washed out of the well by perfusion with ND 96, initially at a speed of 1.5 ml/min for ~20 s, then at 0.5 ml/min thereafter. Recordings were done at room temperature.
Toxins
All peptides were synthesized on solid support using the (N-(9-flourenyl)methoxycarbonyl) (Fmoc) chemistry as described elsewhere (20). In particular, ddKIIIA and its derivatives were synthesized as recently described (25). Briefly, selenocysteine residues were protected with p-methoxybenzyl groups (Chem-Impex International, Wood Dale, IL, USA). Analogs were removed from the resin by a 3 h treatment with a modified reagent K consisting of trifluoroacetic acid (TFA)/thioanisole/phenol/water (90:2.5:7.5:2.5, v/v/v/v) and 1.3 equivalents DTNP (2,2′-dithiobis(5-nitropyridine). The selenoconopeptide analogs were treated for 1 h with DTT (dithiothreitol, 100 mM), Tris (tris(hydroxymethyl)aminomethane, 0.1M), EDTA (ethylenediaminetetraacetic acid, 1 mM), pH 7.5 at room temperature. The peptides were purified by reversed-phase HPLC. Oxidative folding was performed with a mixture of oxidized and reduced glutathione (1 mM GSSG and 1 mM GSH) in Tris-HCl (0.1 M, pH 7.5), EDTA (1 mM) at room temperature. The oxidized peptide analogs were purified by C18 semi-preparative HPLC. Chemical identity of each analog was confirmed by mass spectrometry. TTX was purchased from Alomone Labs (Jerusalem, Israel). Lyophilized peptides were dissolved in ND96, and lyophilized TTX was dissolved in water, and both were stored frozen until used.
Data analysis
Unless otherwise specified, values of kon were obtained from slopes of kobs versus [peptide] plots, and koff values were obtained from single-exponential fits of recovery from block following peptide-washout, as previously described (26). Unless otherwise indicated, averaged data in graphs and tables are mean ± S.D. (N ≥ 3 oocytes).
RESULTS
Affinities and Efficacies of ddKIIIA and nine ddKIIIA[K7X] Analogs in Blocking NaV1.2
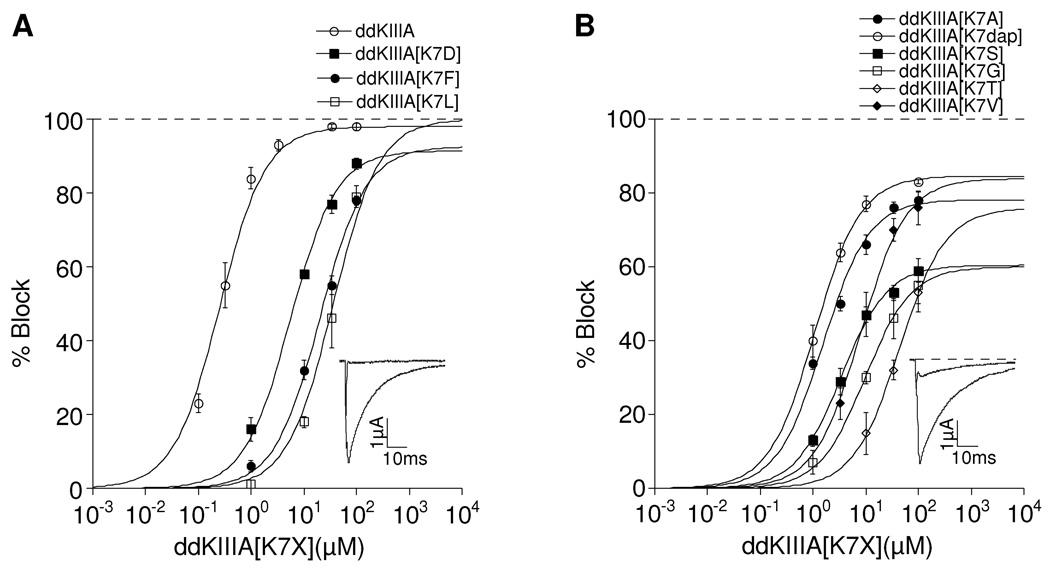
Synthetic peptides were synthesized and tested for their ability to inhibit voltage-gated sodium currents (INa) of voltage-clamped Xenopus oocytes expressing NaV1.2 as described in Materials and Methods. When tested at various concentrations, the resulting dose-response curves were well-fit by the Langmuir adsorption isotherm (Fig. 2), consistent with a bimolecular reaction between peptide and channel; furthermore, the steady-state IC50 values were similar to the kinetically derived Kd values (Table 1).
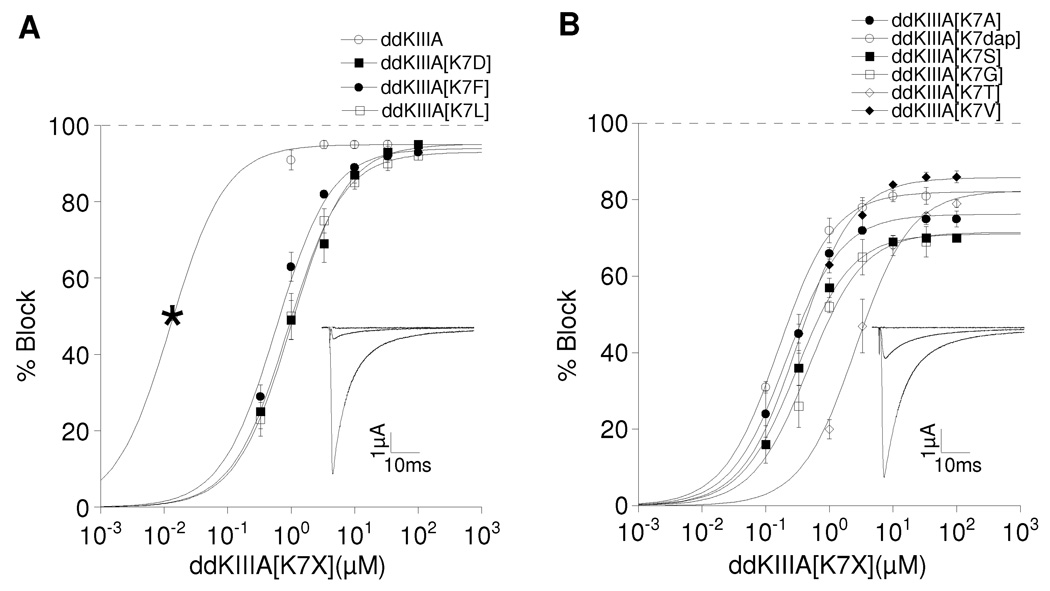
Figure 2.
Saturating concentrations of ddKIIIA or ddKIIIA[K7X] block NaV1.2 incompletely. Oocytes were voltage-clamped as described in Materials and Methods and exposed to peptide until steady-state block of sodium currents was achieved. Superimposed plots of analogs with: A, small (5 – 7%) rINa, where X = K, D, F, or L; and B, large (14 – 30%) rINa, where X = A, S, G, T, V or dap (diaminoproprionate). Asterisk in A (left-most data point for ddKIIIA) represents Kd, calculated from koff/kon (Table 1), since slow rates of block by ddKIIIA in that range of concentrations precluded accurate measurement of steady-state levels of block. Data points represent mean ± S.D. (N ≥ 4). Solid curves represent fit of data (including asterisk) to the equation for the Langmuir adsorption isotherm, Y = 100 • plateau/[1+ (IC50/[peptide])]. IC50 and plateau (i.e., rINa) values are listed in Table 1. The insets show representative traces in response to a depolarizing step to −10 mV from a holding potential of −80 mV before (control, large trace), in the presence of peptide (attenuated trace) either 10 µM ddKIIIA (A) or 30µM ddKIIIA[K7A] (B), and finally in the presence of both peptide and TTX (flat trace), where residual current that persisted in the presence of peptide was obliterated by TTX (100 and 10 µM in A and B, respectively) (see also Fig. 4C & D). Data in A and B are plotted separately for visual clarity.
Table 1.
Kinetics, affinities and efficacies of ddKIIIA and analogs in blocking NaV1.2a
| Peptide | kon | koff | Kd | IC50 (95% C.I.) | rINad |
|---|---|---|---|---|---|
| µM−1•min−1 | min−1 | µM | µM | % | |
| ddKIIIA | 0.75 ± 0.015 | 0.01 ± 0.0015 | 0.013 ± 0.002 | N.A.b | 5 ± 1.1 |
| ddKIIIA[K7A] | 0.69 ± 0.11 | 0.13 ± 0.03 | 0.19 ± 0.05 | 0.21 (0.17 – 0.25) | 24 ± 2.1 |
| ddKIIIA[K7D] | 0.08 ± 0.005 | 0.083 ± 0.02 | 1.04 ± 0.26 | 1 (0.85 – 1.2) | 5 ± 2.4 |
| ddKIIIA[K7S] | 0.77 ± 0.07 | 0.11 ± 0.02 | 0.14 ± 0.03 | 0.31 (0.26 – 0.37) | 29 ± 1.7 |
| ddKIIIA[K7F] | 0.55 ± 0.05 | 0.3 ± 0.04 | 0.55 ± 0.09 | 0.6 (0.52 – 0.7) | 6 ± 2.0 |
| ddKIIIA[K7G] | 1.2 ± 0.1 | 0.67 ± 0.11 | 0.56 ± 0.1 | 0.47 (0.36 – 0.6) | 29 ± 2.6 |
| ddKIIIA[K7L] | 0.39 ± 0.15 | 0.2 ± 0.009 | 0.51 ± 0.2 | 0.87 (0.76 – 1.0) | 7 ± 1.7 |
| ddKIIIA[K7V] | 0.33 ± 0.018 | 0.091 ± 0.014 | 0.28 ± 0.045 | 0.34 (0.3 – 0.38) | 14 ± 1.4 |
| ddKIIIA[K7T] | 0.1 ± 0.015 | 0.09 ± 0.006 | 0.9 ± 0.15 | 2.3 (1.8 – 3.0) | 19 ± 3.3 |
| ddKIIIA[K7dap] | 1.2 ± 0.06 | 0.06 ± 0.008 | 0.05 ± 0.007 | 0.13 (0.09 – 0.19) | 18 ± 1.3 |
| KIIIAb | 0.3 ± 0.03 | 0.0016 ± 0.0016 | 0.0053 ± 0.005 | N.A.c | 5 ± 3 |
| KIIIA[K7A]b | 0.13 ± 0.006 | 0.015 ± 0.005 | 0.115 ± 0.038 | N.A.c | 23 ± 1 |
| KIIIA[K7D] | 0.02 ± 0.002 | 0.008 ± 0.001 | 0.4 ± 0.06 | N.A.c | 6 ± 2.0 |
Rate constants were determined as described under Materials and Methods. Steady-state IC50 and rINa values were obtained with data and equation presented in Fig. 2. Standard deviation and 95% Confidence Intervals (95% C.I.) were calculated from at least three independent experiments using Prism software. For comparison, also shown are data for KIIIA and KIIIA[K7A] from ref. (18).
Data from ref. (18).
IC50 value was not obtained, because the rates of block using peptide concentrations in the range of the expected IC50 were too slow to provide accurate steady-state levels of block within the time frame of the experiments.
rINa for ddKIIIA, KIIIA, and KIIIA[K7A] were directly measured using a saturating peptide concentration, and those for the remaining peptides represent the plateaus of the best-fit curves as described in Fig. 2.
At saturating concentrations, the maximum block produced by all of the peptides plateaued at a level below 100%; i.e., a residual current (rINa) persisted when peptide was bound to the channel. In this regard, the peptides could be placed into two categories, those that blocked >90% (rINa <10%), and those that were unable to block more than 86% (rINa ≥ 14%) (Table 1). For clarity, the dose-response curves of the peptides in each category are plotted separately in Fig. 2.
The residual current resembled the control current in all respects except amplitude. Specifically, the time course as well as the I-V curve of the rINa with ddKIIIA or ddKIIIA[K7A] were essentially the same as those of sodium currents under control conditions (Fig. 3). These results show that the kinetics and voltage sensitivities of the rINa are essentially indistinguishable from those of the control INa and are consistent with the peptides acting purely as pore blockers.
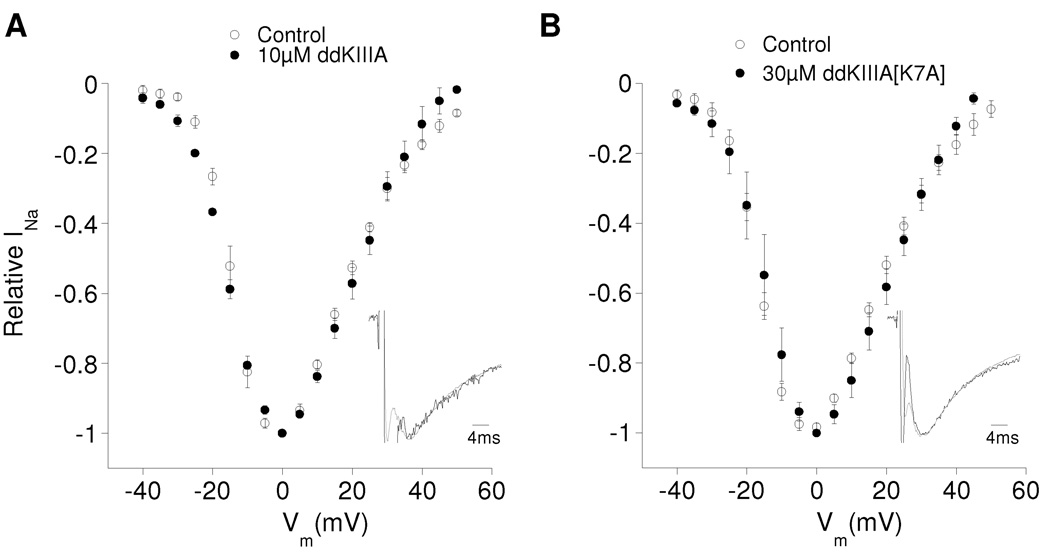
Figure 3.
Kinetics and voltage-dependence of rINa of NaV1.2 with ddKIIIA or ddKIIIA[K7A] are essentially the same as those of control INa. Oocytes expressing NaV1.2 were voltage-clamped and exposed to toxin essentially as in Fig. 2. I-V curves of control INa (closed circles) and rINa (open circles) with ddKIIIA (A) and ddKIIIA[K7A] (B) have essentially the same profile and indicate that the gating properties of the channel were minimally, if at all, affected by the presence of the peptides. To insure that control INa and rINa were obtained with comparable voltage-clamp fidelity, control currents were obtained from oocytes that provided small INa with amplitudes comparable to those of rINa (about 200 and 500 nA for ddKIIIA and ddKIIIA[K7A], respectively). Insets, normalized averaged traces of control INa (gray) and rINa (black) with ddKIIIA (A) and ddKIIIA[K7A] (B) illustrating that rINa and control INa have essentially the same time course (first few ms of the traces represent capacitative transients to be ignored). Data are mean ± S.D. from ≥ 3 oocytes.
TTX blocks the rINa with all peptides examined
The residual current, which persisted in saturating concentrations of all peptides tested, could be blocked by TTX, albeit more slowly than the rate at which TTX blocked control currents (Fig. 4, Table 2).
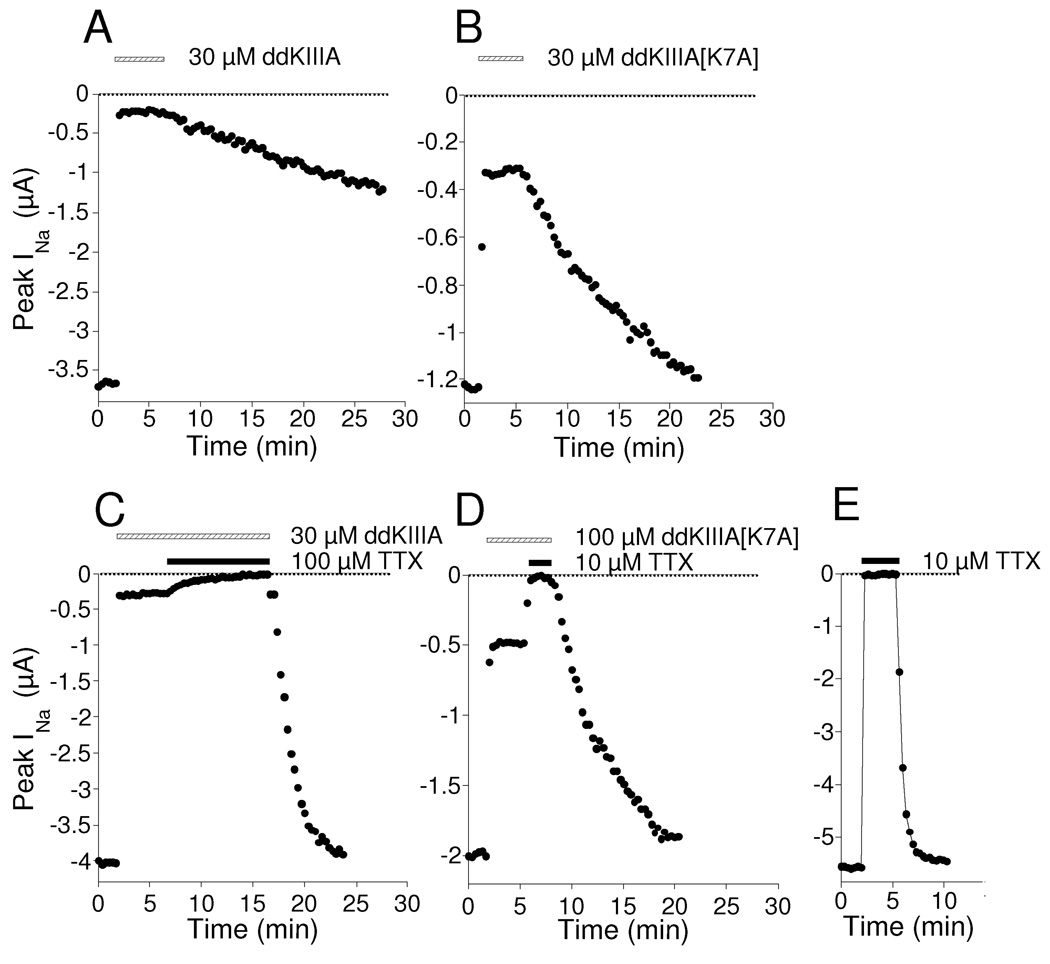
Figure 4.
Kinetics of block of INa of NaV1.2 by ddKIIIA or ddKIIIA[K7A], and kinetics of block of respective rINa's by TTX. Oocytes expressing NaV1.2 were voltage-clamped and current recordings were obtained every 20 s as described in Materials and Methods. Example plot of peak sodium currents (INa) versus time during exposure to and washout of a saturating concentration (30 µM) of ddKIIIA (A) or ddKIIIA[K7A] (B). Block by ddKIIIA and ddKIIIA[K7A] leveled off at <100% (leaving rINa of ~5 and 25%, respectively), and washout of ddKIIIA[K7A] was faster than that of ddKIIIA. The preceding was repeated, except that after steady-state block by each peptide had been obtained, the bath was supplemented with TTX, either 100 or 10 µM with ddKIIIA (C) and ddKIIIA[K7A] (D), respectively. In each case, the rINa was completely blocked by TTX, albeit more slowly than when exposure was to 10 µM TTX alone (E). Following washout of both peptide and TTX, INa fully recovered with a time course faster than that following washout of peptide alone (compare A with C and B with D) but more slowly than that following TTX alone (compare C and D with E). For all panels, presence of TTX is indicated by an closed bar and that of peptide by a open bar. A different oocyte was used to acquire the data for each panel. Rate constants of block of rINa by TTX (kobs) and recovery (koff) for ddKIIIA and all ddKIIIA[K7X] derivatives are summarized in Table 2.
Table 2.
Rate constants of block of rINa by TTX and recovery from block following simultaneous washout of TTX and peptide
| Peptide | [TTX] kobs of block of rINa by TTX |
koff of peptide•TTX•NaVa |
koff of peptide•NaVb |
Estimated konc |
Estimated kond |
|
|---|---|---|---|---|---|---|
| µM | min−1 | min−1 | min−1 | µM−1•min−1 | µM−1•min−1 | |
| ddKIIIA | 100 | 0.44 ± 0.13 | 0.58 ± 0.03 | 0.01 ± 0.0015 | −0.0014 ± 0.0013e |
0.0044 ± 0.0013 |
| ddKIIIA[K7A] | 10 | 4.4 ± 1.3 | 0.27 ± 0.035 | 0.13 ± 0.03 | 0.41 ± 0.13 | 0.44 ± 0.13 |
| ddKIIIA[K7D] | 10 | 1.7 ± 0.4 | 0.081 ± 0.013 | 0.083 ± 0.02 | 0.16 ± 0.04 | 0.17 ± 0.04 |
| ddKIIIA[K7S] | 10 | 4.6 ± 1.6 | 0.13 ± 0.015 | 0.11 ± 0.02 | 0.45 ± 0.16 | 0.46 ± 0.16 |
| ddKIIIA[K7F] | 10 | 2.2 ± 0.5 | 0.27 ± 0.01 | 0.3 ± 0.04 | 0.19 ± 0.05 | 0.22 ± 0.05 |
| ddKIIIA[K7G] | 3.3f | 6.5 ± 1.0 | 0.48 ± 0.085 | 0.67 ± 0.11 | 1.82 ± 0.3 | 1.97 ± 0.3 |
| ddKIIIA[K7L] | 10 | 0.73 ± 0.06 | 0.17 ± 0.01 | 0.2 ± 0.009 | 0.056 ± 0.006 | 0.073 ± 0.006 |
| ddKIIIA[K7V] | 10 | 1.2 ± 0.05 | 0.082 ± 0.01 | 0.091 ± 0.014 | 0.11 ± 0.005 | 0.12 ± 0.005 |
| ddKIIIA[K7T] | 10 | 1.3 ± 0.2 | 0.1 ±0.009 | 0.09 ± 0.006 | 0.12 ± 0.02 | 0.13 ± 0.02 |
| ddKIIIA[K7dap] | 3.3f | 1.8 ± 0.5 | 0.55 ± 0.02 | 0.06 ± 0.008 | 0.38 ± 0.15 | 0.54 ± 0.15 |
| KIIIAg | NA | NA | 0.017 ± 0.002 | 0.0016 ± 0.0016 | 0.3 ± 0.03h | NA |
| KIIIA[K7A]g | NA | NA | 0.024 ± 0.004 | 0.015 ± 0.005 | 0.13 ± 0.006h | NA |
| KIIIA[K7D] | 10 | 2.3 ± 0.6 | 0.01 ± 0.003 | 0.008 ± 0.001 | 0.23 ± 0.06 | 0.23 ± 0.06 |
Rate constant of recovery from block of ternary complex, peptide•TTX•NaV1.2, when both peptide and TTX were simultaneously washed out.
Binary complex values from Table 1, reproduced here for ready comparison with ternary complex values.
Calculated assuming bimolecular reaction kinetics (kon = (kobs − koff)/[TTX]) and TTX dissociated from the ternary peptide•TTX•NaV complex with a rate constant no larger than the koff of peptide•TTX•NaV (i.e., 5th column in table). Rationale for this assumption is that if during the washout of both toxins TTX dissociated from the complex before peptide did, then a rapid recovery of rINa would be expected, followed by the slower recovery from block by peptide alone; instead, the recovery from block followed essentially a single exponential time course (see Figs. 4C & D). For the most part, the koff value for a given ternary peptide•TTX•NaV complex was either larger or approximately equal to the koff value for the corresponding binary peptide•NaV complex (compare columns 4 and 5).
Upper limit of kon obtained by assuming TTX could not dissociate from the ternary complex, in which case estimated kon = kobs/[TTX]. Note, both kon estimates gave similar values.
Negative value for estimated kon suggests koff of recovery from block of the ternary complex is a gross overestimate of koff of TTX in this instance.
Lower concentration than elsewhere was used because of larger relative kobs.
Data from ref. (18).
Obtained from slope of kobs versus [ligand] plot.
Comparison of koff for recovery from block following toxin-washout of the binary peptide•NaV complex with that of the ternary peptide•TTX•NaV complex indicates that presence of TTX accelerated the recovery from block when the peptide was ddKIIIA (58-fold increase in koff), ddKIIIA[K7dap] (9-fold increase in koff), or ddKIIIA[K7A] (2-fold increase in koff). However, the presence of TTX did not significantly affect the koff values for the remaining dd-peptides (Table 2).
Affinities and Efficacies of KIIIA and Its Analogs in Blocking NaV1.4
To examine the block by KIIIA and its analogs of a sodium channel isoform other than NaV1.2, the effects of KIIIA, KIIIA[K7A], ddKIIIA and all nine ddKIIIA[K7X] peptides were also tested on NaV1.4. The behavior of these peptides on NaV1.4 qualitatively resembled those on NaV1.2. Thus, the peptides could be placed in either of two categories with respect to rINa size (Fig. 5, Table 3); furthermore, the peptides belonging to each rINa-size category were the same for NaV1.2 and 1.4.
Figure 5.
Saturating concentrations of ddKIIIA and ddKIIIA[K7X] block NaV1.4 incompletely. Superimposed plots of analogs with small (< 9%) rINa, where X = K, D, F, or L (A); and large (16 – 42%) rINa, where X = A, S, G, V, T, or dap (B). Data were acquired and plotted as in Fig. 2. Except for ddKIIIA, data points of peptides do not extend to saturating concentrations because of supply limitations; thus, rINa values were obtained from plateaus extrapolated by curve-fitting. Resulting IC50 and rINa values are listed in Table 3. The insets show representative traces in response to a depolarizing step to −10 mV from a holding potential of −80 mV before (control, large trace), in the presence of peptide (attenuated trace) either 10 µM ddKIIIA (A) or 30 µM ddKIIIA[K7A] (B); the zero-current level is shown by the dashed line in B but not shown in A because the peptide-attenuated current trace would obscure it.
Table 3.
Kinetics, affinities and efficacies of ddKIIIA analogs in blocking NaV1.4a
| Peptide | kon | koff | IC50 (95% C.I.) | rINa |
|---|---|---|---|---|
| µM−1•min−1 | min−1 | µM | %b | |
| ddKIIIA | 1.4 ± 0.15 | 0.36 ± 0.06 | 0.25 (0.22 – 0.29) | 2 ± 1.0 |
| ddKIIIA[K7A] | 0.17 ± 0.03 | 1.2 ± 0.3 | 1.6 (1.3 – 1.9) | 22 ± 2.3 |
| ddKIIIA[K7D] | 0.18 ± 0.02 | 1.5 ± 0.3 | 5.6 (4.7 – 6.6) | 9 ± 2.6 |
| ddKIIIA[K7S] | N.A. | 2.4 ± 0.7 | 3.4 (2.8 – 4.1) | 40 ± 2.3 |
| ddKIIIA[K7F] | N.A. | 4.6 ± 1.0 | 20 (17 – 25) | 7 ± 4.7 |
| ddKIIIA[K7G] | N.A. | 6.9 ± 1.6 | 9.8 (6.0 – 16) | 40 ± 6.0 |
| ddKIIIA[K7L] | N.A. | 9.5 ± 3.8 | 36 (30 – 44) | --c |
| ddKIIIA[K7V] | N.A. | 2.6 ± 0.8 | 8.1 (6.0 – 11) | 16 ± 5.2 |
| ddKIIIA[K7T] | N.A. | 2.8 ± 0.7 | 45.3 (26 – 78) | 23 ± 13 |
| ddKIIIA[K7dap] | N.A. | 3.1 ± 0.8 | 1.1 (0.92 – 1.3) | 16 ± 2.2 |
| KIIIA | 0.97 ± 0.09 | 0.047 ± 0.015 | 0.082 (0.067 – 0.1) | 1 ± 0.6 |
| KIIIA[K7A] | 0.36 ± 0.08 | 0.37 ± 0.08 | 1.1 (0.95 – 1.3) | 30 ± 2.1 |
| KIIIA[K7D] | 0.11 ± 0.015 | 0.15 ± 0.02 | 1.5 (1.1 – 2.0) | 6 ± 3.5 |
Kinetic and steady-state values were obtained as in Table 1, and data are partially illustrated in Fig. 5. For comparison, also shown are data for KIIIA, KIIIA[K7A], and KIIIA[K7A] from experiments performed for this report.
Values of rINa were directly obtained at saturating [peptide] for ddKIIIA, KIIIA and KIIIA[K7A], and for remaining peptides curve-fit plateau values were used to provide estimates of rINa, because low affinities experienced with NaV1.4 required concentrations of peptide >100 µM to fully saturate NaV1.4, and such concentrations called for quantities of peptides unavailable to us.
Curve-fit plateau value was > 100%; therefore, no estimate of rINa is given. N.A. indicates on-rate was too fast to be accurately measured at the high peptide concentrations necessary to observe significant block of sodium current.
The rINa with ddKIIIA, like that with native KIIIA, was smaller with NaV1.4 than NaV1.2. However, for the rest of the peptides, the rINa with NaV1.4 was about the same or greater than that with NaV1.2. A detailed examination of the rINa with NaV1.4 was not performed, as was done with NaV1.2, for mainly two reasons. First, the ddKIIIA analogs had much lower affinities for NaV1.4, therefore accurate measurements of rINa required very high concentrations (>100 µM) of the peptides, and such amounts were unavailable for cost reasons. Moreover, the peptides' off rates with NaV1.4 were larger than those with NaV1.2 by order of magnitude or more, and this made the determination of the formation of peptide•TTX•NaV1.4 ternary complexes problematical.
Properties of KIIIA[K7D]
We previously compared the affinities and efficacies of KIIIA and KIIIA[K7A] (18). In the present report, we also synthesized and examined KIIIA[K7D] to document the consequences of having a negatively charged residue at position 7 in the native peptide. The kinetic constants, affinities, and efficacies of KIIIA[K7D] in blocking NaV1.2 or 1.4 are listed, along with those of KIIIA and KIIIA[K7A], in Tables 1 through 4.
Table 4.
Comparison of kinetics of TTX- or peptide-block of NaV1.2 and of TTX-block of binary peptide•NaV1.2 complexes
| Ligand | Target | kon | koff | Kd | Reference |
|---|---|---|---|---|---|
| µM−1•min−1 | min−1 | nM | |||
| TTX | NaV1.2 | 58 ± 5.3b | 2.2 ± 0.3 | 38 ± 6 | (18) |
| KIIIA | NaV1.2 | 0.3 ± 0.03b | 0.0016 ± 0.0016 | 5.3 ± 5 | (18) |
| KIIIA[K7A] | NaV1.2 | 0.13 ± 0.006b | 0.015 ± 0.005 | 115 ± 38 | (18) |
| KIIIA[K7D] | NaV1.2 | 0.02 ± 0.002 | 0.008 ± 0.001 | 0.4 ± 0.06 | this report |
| ddKIIIA[2–5C]a | NaV1.2 | 1.8 ± 0.39c | 0.014 ± 0.002 | 7.8 ± 2 | (27) |
| ddKIIIA | NaV1.2 | 0.75 ± 0.015b | 0.01 ± 0.0015 | 13 ± 2 | this report |
| ddKIIIA[K7A] | NaV1.2 | 0.69 ± 0.11b | 0.13 ± 0.03 | 190 ± 50 | this report |
| ddKIIIA[K7D] | NaV1.2 | 0.08 ± 0.005 | 0.083 ± 0.02 | 1.04 ± 0.26 | this report |
| TTX | KIIIA•NaV1.2 | 0.003 ± 0.0004b | N.A. | N.A. | (18) |
| TTX | KIIIA[K7A]•NaV1.2 | 0.56 ± 0.08b | N.A. | N.A. | (18) |
| TTX | KIIIA[K7D]•NaV1.2 | 0.23 ± 0.06c | N.A. | N.A. | this report |
| TTX | ddKIIIA•NaV1.2 | 0.0044 ± 0.0013c | N.A. | N.A. | this report |
| TTX | ddKIIIA[K7A]•NaV1.2 | 0.44 ± 0.13c | N.A. | N.A. | this report |
| TTX | ddKIIIA[K7D]•NaV1.2 | 0.17 ± 0.04c | N.A. | N.A. | this report |
ddKIIIA[2–5C] is ddKIIIA with disulfide, instead of diselenide, bridge (i.e., KIIIA[C1A,C9A]).
Obtained from slope of kobs versus [ligand] plot.
Obtained from kobs at single [ligand] (see Table 2). N.A. indicates value not available, because it could not be directly determined experimentally.
DISCUSSION
Comparison of disulfide-depleted KIIIA with KIIIA
The use of ddKIIIA as a core structure for these experiments was motivated by the observation that the structures and functional activities of KIIIA and its disulfide-depleted counterpart, KIIIA[C1A,C9A], are much the same (20, 27); that is: A, the solution structures of both peptides are quite similar, indicating that the Cys1-Cys9 disulfide bond could be removed without significant distortion of the alpha-helix bearing the known key residues; B, the rank order of potency of KIIIA[C1A,C9A] in blocking the sodium currents of all six NaV isoforms tested were essentially the same as that of KIIIA. Thus, we concluded that the key residues for VGSC binding resided mostly on an alpha-helix in the C-terminal half of the peptide and that the first disulfide bond can be removed without significantly affecting the structure of this helix (27).
More recently, the replacement of one of the two remaining disulfide bridges of the disulfide-depleted KIIIA[C1A,C9A] with a diselenide bridge to yield the "ddKIIIA" of the present report simplified the synthesis of the core structure even further (25). The activities of KIIIA, KIIIA[K7A], KIIIA[K7D] and their disulfide-depleted counterparts on NaV1.2 are summarized in Table 4. ddKIIIA (i.e., ddKIIIA[2–5U]) and ddKIIIA[2–5C] had similar Kd's (13 and 7.8 nM, respectively), with the former having a slower kon and koff than those of the latter. Since the substitution of the disulfide bridge with a diselenide bridge had only a minimal effect, we felt justified in using the peptide with the diselenide bridge as the core structure for positional scanning studies, particularly in view of the vastly accelerated synthesis of its analogs. This view is further supported by the observation that the functional differences between ddKIIIA[K7A] and ddKIIIA found in this study paralleled those between KIIIA[K7A] and KIIIA (18).
To further investigate the role of electrical charge at position 7 of KIIIA, KIIIA[K7D] was tested for comparison with KIIIA and KIIIA[K7A], as well as with ddKIIIA[K7A]. In each case, whether KIIIA[K7X] or ddKIIIA[K7X], the major effect of replacing Lys 7 with Ala or Asp was about a ten-fold increase in koff (Table 1). The rINa levels with KIIIA, KIIIA[K7A] and KIIIA[K7D] mirrored those of their respective disulfide-deficient counterparts, as discussed further below. Finally, TTX blocked the rINa of NaV1.2 with either KIIIA or ddKIIIA at about the same rate, which was about 300-fold slower than that with either KIIIA[K7D] or ddKIIIA[K7D] and about 100-fold slower than that with either KIIIA[K7A] or ddKIIIA[K7A]; these in turn were 100- to 300-fold slower than the rate at which TTX blocked the control INa (compare kon data in first row and last six rows in Table 4). In summary, for each pair tested on NaV1.2, when the ddKIIIA[K7X] is compared to its KIIIA[K7X] counterpart, for X = K, A, and D: 1) kon is larger by factor ranging from 2.5 to 5.3, 2) koff is larger by a factor ranging from 6.2 to 10, 3) Kd is larger by factor ranging from 1.6 to 2.6, 4) efficacy (or rINa) changes by ≤ 17%, and 5) the rate of TTX-block of rINa differs by only 1.3- to 1.5-fold. Thus, the alteration of functional parameters upon converting "native" into "dd-peptides" occurred in a parallel fashion for all three pairs of peptides.
Kinetics and affinities in the formation of binary ddKIIIA[K7X]•NaV1.2 complexes
Positional scanning of Lys7 of ddKIIIA with the nine amino acids tested revealed that affinity could be attenuated to varying degrees (Figs. 2 & 5), in all cases koff was increased (Tables 1 & 3), consistent with the notion that the loss of the positive charge of Lys destabilized the bound complex (18); dap also has a positive charge, but on a much shorter side chain than Lys, and the dap-containing analog had the lowest koff after ddKIIIA when tested against NaV1.2 (but not NaV1.4). Note however, the analog with reversed charge, ddKIIIA[K7D], did not have the fastest koff of the peptides tested against NaV1.2, although it had the slowest kon (Table 1).
Compared to KIIIA, KIIIA[K7A] and KIIIA[K7D], most ddKIIIA analogs had a larger kon. A possible explanation for this is that the disulfide-deficiency endowed the peptides with greater structural flexibility and therefore they more readily assumed a conformation that favored a faster on-rate, as we previously speculated (27).
Like KIIIA, ddKIIIA blocked with an rINa that was small: 5% on NaV1.2 (Table 1) and ~2% on NaV1.4 (Table 3). KIIIA[K7D] had a small 6% rINa on NaV1.2 and 1.4 (Tables 1 and 4, respectively), and ddKIIIA[K7D] also blocked NaV1.2 and 1.4 with a small rINa (5 and 9%, respectively, Tables 1 and 4). In contrast, the K7A analogs of each of these produced an rINa that was large: 23% with KIIIA[K7A] and 24% with ddKIIIA[K7A] on NaV1.2 (Table 1), and 30% with KIIIA[K7A] and 22% with ddKIIIA[K7A] on NaV1.4 (Table 3). Thus, the disulfide-deficiency affected the efficacies of these peptides only marginally.
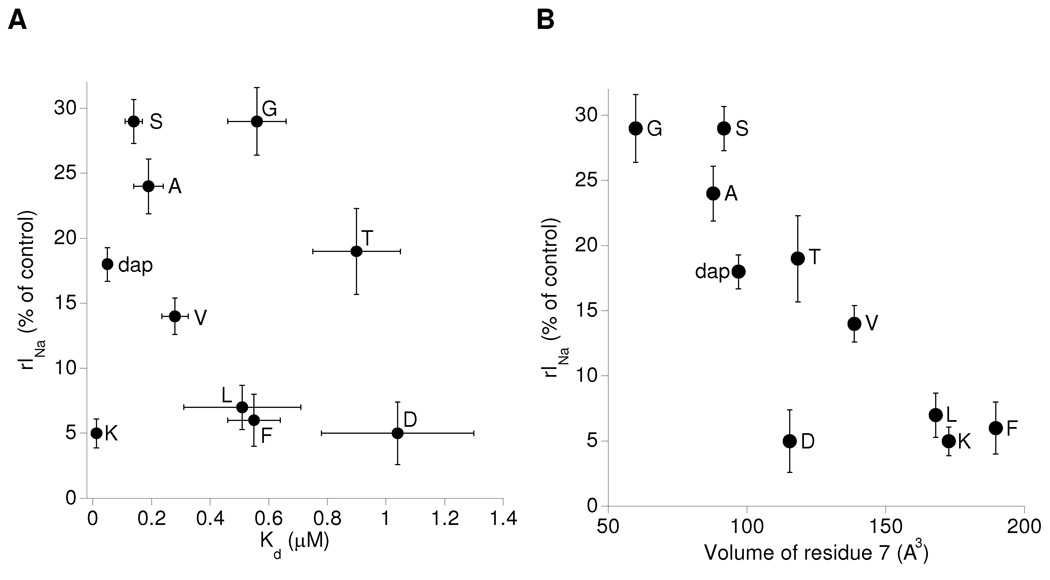
Residual currents correlate inversely with the size of the residue at position 7
The rINa values of ddKIIIA[K7X] did not seem to correlate with either their affinities (Fig. 6A), kon, or koff values (Tables 1 & 3); instead, rINa correlated inversely with the size of the side chain of residue 7, with both NaV1.2 (Fig 6B) and NaV1.4 (not graphically illustrated, but see Table 3). Thus, the impediment to passage of sodium ions increased as the size of residue 7 of the bound peptide increased.
Figure 6.
Relationships between NaV1.2 rINa versus Kd and volume of ddKIIIA[K7X]'s residue 7. Plots of data in Table 1. Each data point is labeled with amino acid corresponding to residue 7 of ddKIIIA[K7X]. A, rINa versus Kd shows that efficacy and affinity are apparently uncorrelated. B, rINa versus volume of residue 7 shows that rINa varies inversely with size of the residue; remarkably, rINa seems insensitive to side-chain charge or hydrophobicity. AA volumes are from Table 1 of Perkins (38); dap volume was estimated by subtracting the volume of three methylene groups from volume of Lys, where volume of a methylene group was estimated from the average difference in volume between Glu and Asp, and Gln and Asn.
The correlation of blocking efficacy with side chain size is reminiscent of the actions of µ-conotoxin GIIIA[R13X] mutants on NaV1.4, where X was one of seven residues with different side chain lengths and charges; for a given charge, efficacy progressively increased with length, and for a given length, efficacy progressively increased as charge was varied from −1, 0, to +1 (28). This last behavior differs from that of the ddKIIIA[K7X] analogs, where charge was largely immaterial and suggests that functionally Lys7 of KIIIA is not strictly homologous to Arg13 of GIIIA.
Block of rINa by TTX
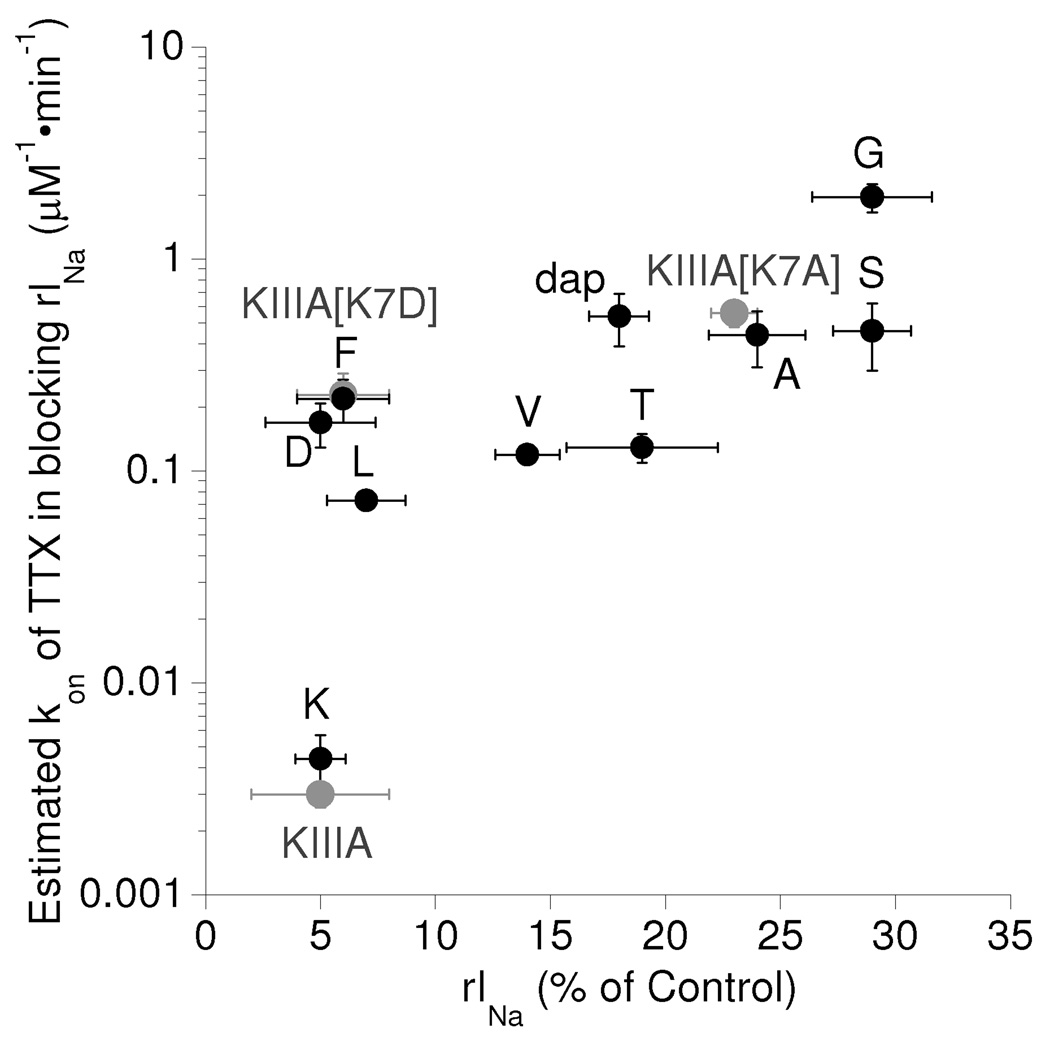
In all cases, the presence of bound peptide not only increased the impediment to passage of Na+, but apparently also that of TTX. That is, TTX blocked the rINa with ddKIIIA and all ddKIIIA[K7X] derivatives at a rate significantly slower than control INa (Fig. 4, Table 2). Thus, TTX appears to be able to 'sneak by' the bound peptides only with difficulty and block the residual current, as was originally suggested to occur with KIIIA and KIIIA[K7A] (18).
When the rates of TTX-block of the rINa's are compared with the magnitudes of the rINa's, a positive correlation is evident (Fig. 7). Among the peptides with a small rINa (≤ 7% of control), TTX's kon appears to depend on charge, suggesting that an electrostatic attraction between the positively charged guanidinium of TTX and the negativity of Asp and the pi-electrons of Phe may account for the elevated values of kon of the mutants with those residues. In this regard, it might be noted that a cation-pi electron interaction is observed between Tyr401 of NaV1.4 and bound TTX (29); furthermore, the NaV1.4[Y401C] mutation results in a very large decrease in TTX affinity that is mostly due to a decrease in the kon of TTX (30). Conversely, the very low kon value of TTX with bound KIIIA or ddKIIIA may be a reflection of the positive charge of Lys7, which would repel TTX. Note however, dap, although positively charged, did not retard TTX strongly, possibly because the ammonium group of dap is on a very short tether compared to that of Lys; i.e., one versus four methylene groups link the α-carbon with the side-chain ammonium group.
Figure 7.
Relationship between susceptibility of rINa to be blocked by TTX and size of residue 7 of ddKIIIA[K7X]. Value of kon for block of rINa with NaV1.2 by TTX was estimated from observed rate constant of block (kobs) of rINa at a single concentration of TTX and assuming koff to be zero (see last column in Table 2). Data points are labeled as in Fig. 6. The plot suggests a general tendency that the smaller the rINa, the more TTX is impeded. Gray circles represent data for KIIIA and KIIIA[K7A] from (ref. 18), and KIIIA[K7D] (Tables 1 and 4); note, the data point for each peptide lies close to that of its disulfide-depleted counterpart.
Stability of the ternary peptide•TTX•NaV1.2 complex
Presence of TTX accelerated the unbinding of KIIIA from NaV1.2 by a factor of ten (18), and such an effect of TTX was also only evident with ddKIIIA and ddKIIIA[K7X] when X was dap (compare columns 4 and 5 of Table 2). The destabilizing effect of TTX is consistent with an electrostatic repulsion between the positive charge of TTX and that at residue 7 in the peptide. The minimal effect the charge on dap had in retarding TTX-block, as discussed above, suggests that the impediment to passage of TTX is sensitive to the position of the positive charge at residue 7, whereas the destabilization of the ternary complex is relatively insensitive to the position of the charge. It might be noted that the koff of ddKIIIA[K7D] (where Asp confers a negative charge at residue 7) was the same whether or not TTX was present, but this could be a reflection of the relatively slow koff of ddKIIIA[K7D] from NaV1.2 to begin with (Table 2).
In the experiments thus far, only position 7 of KIIIA was scrutinized, and further experiments are necessary to obtain a clearer picture of the structural elements of the peptide responsible for its affinity for the channel and its efficacy in interfering with TTX-block versus sodium-permeation. Positional scanning at other locations with positively charged residues, such as R10 and R14, may be enlightening in this regard. Furthermore, determination of the NaV subtype selectivity profiles of the various derivatives of KIIIA would be informative, particularly since the channel(s) responsible for KIIIA's analgesic activity (17) remain unidentified.
Exploiting µ-conopeptides that have limited efficacies
The possibility of independently manipulating the affinity and efficacy of a pore-blocking ligand of VGSCs is of interest for the development of possible drugs for therapeutic intervention. Consider a µ-conopeptide that has been engineered to have: 1, high affinity for VGSCs; 2, highly-attenuated efficacy in blocking INa; and 3, high efficacy in retarding the block by TTX. Such a peptide could serve as a contratoxin, or antidote, against tetrodotoxin poisoning (18). The analogs with lowest efficacy against NaV1.4 were ddKIIIA[K7G] and ddKIIIA[K7S], with rINa near 40% (Table 3). It would be interesting to see if these are able to shield the channel against TTX and therefore prevent TTX from blocking action potentials in adult skeletal muscle, which normally have NaV1.4 exclusively (31), a situation that might be expected in muscle fibers with a safety factor of greater than two for action potential propagation.
The possibility of using TTX and STX congeners as local analgesic agents has recently been explored (32–34). However, for more general therapeutic use of VGSC blockers, a major quandary is that all but two of the mammalian NaV1 isoforms appear to be critical for survival. The exceptions are NaV1.8 and NaV1.9, which apparently serve largely in the signaling of nociceptive information and for which null mouse mutations are non-lethal (35, 36). Thus, to be therapeutically useful, a VGSC-blocker should not only have appropriate subtype-selectivity, but its dosage must also be carefully regulated. The latter constraint may be circumvented by developing a selective µ-conopeptide that has been engineered to have high affinity but reduced efficacy. Such a drug would be expected to have a large therapeutic index and may be of use to treat the signs of gain-of-function channelopathies such as those implicated in some epilepsies and pain disorders (37).
ACKNOWLEGEMENT
We thank Prof. Alan Goldin for providing the NaV clones and Dr. Layla Azam for producing cRNA from them. We also thank Drs. Robert Schackmann and Scott Endicott from the DNA/Peptide Synthesis Core Facility at the University of Utah for the synthesis of peptides in general and Corey Jensen for the synthesis of ddKIIIA[K7D].
†This work was supported by National Institutes of Health grants PO1 GM48677 (G.B., B.M.O. and D.Y.) and R21 NS055845 (G.B. and D.Y.).
Abbreviations
- dap
diaminoproprionate
- ddKIIIA
disulfide-depleted µ-conotoxin KIIIA; i.e., KIIIA[C1A,C2U,C9A,C5U], where U is selenocysteine
- ddKIIIA[K7X]
ddKIIIA with residue X in position 7
- ddKIIIA•NaV
binary complex of ddKIIIA and NaV
- ddKIIIA[K7X]•NaV
binary complex of ddKIIIA[K7X] and NaV
- ddKIIIA[K7X]•TTX•NaV
ternary complex of ddKIIIA[K7X]
- TTX and NaV
GIIIA, µ-conotoxin GIIIA
- INa
sodium current
- KIIIA
µ-conotoxin KIIIA
- KIIIA[K7A]
µ-conotoxin KIIIA[K7A]
- NaV
α-subunit of voltage-gated sodium channel
- rINa
residual sodium current
- TTX
tetrodotoxin
- TTX•NaV
the binary complex of TTX and NaV
- VGSC
voltage-gated sodium channel
Footnotes
Conflict of Interest Disclosures
BMO is a cofounder of Cognetix, Inc. GB is a cofounder of NeuroAdjuvants, Inc.
REFERENCES
- 1.Hille B. Ion Channels of Excitable Membranes. Third ed. Sunderland, MA U.S.A.: Sinauer Associates; 2001. [Google Scholar]
- 2.Catterall WA, Goldin AL, Waxman SG. International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol Rev. 2005;57:397–409. doi: 10.1124/pr.57.4.4. [DOI] [PubMed] [Google Scholar]
- 3.Brackenbury WJ, Isom LL. Voltage-gated Na+ channels: potential for beta subunits as therapeutic targets. Expert opinion on therapeutic targets. 2008;12:1191–1203. doi: 10.1517/14728222.12.9.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu FH, Catterall WA. Overview of the voltage-gated sodium channel family. Genome Biol. 2003;4:207. doi: 10.1186/gb-2003-4-3-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Billen B, Bosmans F, Tytgat J. Animal peptides targeting voltage-activated sodium channels. Current pharmaceutical design. 2008;14:2492–2502. doi: 10.2174/138161208785777423. [DOI] [PubMed] [Google Scholar]
- 6.Daly JW. Marine toxins and nonmarine toxins: convergence or symbiotic organisms? J Nat Prod. 2004;67:1211–1215. doi: 10.1021/np040016t. [DOI] [PubMed] [Google Scholar]
- 7.Fry BG, Roelants K, Champagne DE, Scheib H, Tyndall JD, King GF, Nevalainen TJ, Norman JA, Lewis RJ, Norton RS, Renjifo C, de la Vega RC. The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annual review of genomics and human genetics. 2009;10:483–511. doi: 10.1146/annurev.genom.9.081307.164356. [DOI] [PubMed] [Google Scholar]
- 8.Terlau H, Olivera BM. Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol Rev. 2004;84:41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- 9.French RJ, Terlau H. Sodium channel toxins--receptor targeting and therapeutic potential. Curr Med Chem. 2004;11:3053–3064. doi: 10.2174/0929867043363866. [DOI] [PubMed] [Google Scholar]
- 10.Wood JN, Boorman JP, Okuse K, Baker MD. Voltage-gated sodium channels and pain pathways. Journal of neurobiology. 2004;61:55–71. doi: 10.1002/neu.20094. [DOI] [PubMed] [Google Scholar]
- 11.Fiedler B, Zhang MM, Buczek O, Azam L, Bulaj G, Norton RS, Olivera BM, Yoshikami D. Specificity, affinity and efficacy of iota-conotoxin RXIA, an agonist of voltage-gated sodium channels Na(V)1.2, 1.6 and 1.7. Biochemical pharmacology. 2008;75:2334–2344. doi: 10.1016/j.bcp.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cestele S, Catterall WA. Molecular mechanisms of neurotoxin action on voltage-gated sodium channels. Biochimie. 2000;82:883–892. doi: 10.1016/s0300-9084(00)01174-3. [DOI] [PubMed] [Google Scholar]
- 13.Cruz LJ, Gray WR, Olivera BM, Zeikus RD, Kerr L, Yoshikami D, Moczydlowski E. Conus geographus toxins that discriminate between neuronal and muscle sodium channels. The Journal of biological chemistry. 1985;260:9280–9288. [PubMed] [Google Scholar]
- 14.Moczydlowski E, Olivera BM, Gray WR, Strichartz GR. Discrimination of muscle and neuronal Na-channel subtypes by binding competition between [3H]saxitoxin and mu-conotoxins. Proc Natl Acad Sci U S A. 1986;83:5321–5325. doi: 10.1073/pnas.83.14.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato K, Ishida Y, Wakamatsu K, Kato R, Honda H, Ohizumi Y, Nakamura H, Ohya M, Lancelin JM, Kohda D, et al. Active site of mu-conotoxin GIIIA, a peptide blocker of muscle sodium channels. The Journal of biological chemistry. 1991;266:16989–16991. [PubMed] [Google Scholar]
- 16.Bulaj G, West PJ, Garrett JE, Watkins M, Zhang MM, Norton RS, Smith BJ, Yoshikami D, Olivera BM. Novel conotoxins from Conus striatus and Conus kinoshitai selectively block TTX-resistant sodium channels. Biochemistry. 2005;44:7259–7265. doi: 10.1021/bi0473408. [DOI] [PubMed] [Google Scholar]
- 17.Zhang MM, Green BR, Catlin P, Fiedler B, Azam L, Chadwick A, Terlau H, McArthur JR, French RJ, Gulyas J, Rivier JE, Smith BJ, Norton RS, Olivera BM, Yoshikami D, Bulaj G. Structure/function characterization of micro-conotoxin KIIIA, an analgesic, nearly irreversible blocker of mammalian neuronal sodium channels. The Journal of biological chemistry. 2007;282:30699–30706. doi: 10.1074/jbc.M704616200. [DOI] [PubMed] [Google Scholar]
- 18.Zhang MM, McArthur JR, Azam L, Bulaj G, Olivera BM, French RJ, Yoshikami D. Synergistic and antagonistic interactions between tetrodotoxin and mu-conotoxin in blocking voltage-gated sodium channels. Channels (Austin) 2009;3:32–38. doi: 10.4161/chan.3.1.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.French RJ, Prusak-Sochaczewski E, Zamponi GW, Becker S, Kularatna AS, Horn R. Interactions between a pore-blocking peptide and the voltage sensor of the sodium channel: an electrostatic approach to channel geometry. Neuron. 1996;16:407–413. doi: 10.1016/s0896-6273(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 20.Han TS, Zhang MM, Walewska A, Gruszczynski P, Robertson CR, Cheatham TE, 3rd, Yoshikami D, Olivera BM, Bulaj G. Structurally minimized mu-conotoxin analogues as sodium channel blockers: implications for designing conopeptide-based therapeutics. ChemMedChem. 2009;4:406–414. doi: 10.1002/cmdc.200800292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walewska A, Zhang MM, Skalicky JJ, Yoshikami D, Olivera BM, Bulaj G. Integrated oxidative folding of cysteine/selenocysteine containing peptides: improving chemical synthesis of conotoxins. Angewandte Chemie (International ed. 2009;48:2221–2224. doi: 10.1002/anie.200806085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armishaw CJ, Daly NL, Nevin ST, Adams DJ, Craik DJ, Alewood PF. Alpha-selenoconotoxins, a new class of potent alpha7 neuronal nicotinic receptor antagonists. The Journal of biological chemistry. 2006;281:14136–14143. doi: 10.1074/jbc.M512419200. [DOI] [PubMed] [Google Scholar]
- 23.Gowd KH, Yarotskyy V, Elmslie KS, Skalicky JJ, Olivera BM, Bulaj G. Site-specific effects of diselenide bridges on the oxidative folding of a cystine knot peptide, omega-selenoconotoxin GVIA. Biochemistry. 2010;49:2741–2752. doi: 10.1021/bi902137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muttenthaler M, Nevin ST, Grishin AA, Ngo ST, Choy PT, Daly NL, Hu SH, Armishaw CJ, Wang CI, Lewis RJ, Martin JL, Noakes PG, Craik DJ, Adams DJ, Alewood PF. Solving the alpha-conotoxin folding problem: efficient selenium-directed on-resin generation of more potent and stable nicotinic acetylcholine receptor antagonists. Journal of the American Chemical Society. 2010;132:3514–3522. doi: 10.1021/ja910602h. [DOI] [PubMed] [Google Scholar]
- 25.Han TS, Zhang MM, Gowd KH, Walewska A, Yoshikami D, Olivera BM, Bulaj G. Disulfide-Depleted Selenoconopeptides: Simplified Oxidative Folding of Cysteine-Rich Peptides. ACS Med Chem Lett. doi: 10.1021/ml900017q. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.West PJ, Bulaj G, Garrett JE, Olivera BM, Yoshikami D. Mu-conotoxin SmIIIA, a potent inhibitor of tetrodotoxin-resistant sodium channels in amphibian sympathetic and sensory neurons. Biochemistry. 2002;41:15388–15393. doi: 10.1021/bi0265628. [DOI] [PubMed] [Google Scholar]
- 27.Khoo KK, Feng ZP, Smith BJ, Zhang MM, Yoshikami D, Olivera BM, Bulaj G, Norton RS. Structure of the analgesic mu-conotoxin KIIIA and effects on the structure and function of disulfide deletion. Biochemistry. 2009;48:1210–1219. doi: 10.1021/bi801998a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hui K, Lipkind G, Fozzard HA, French RJ. Electrostatic and steric contributions to block of the skeletal muscle sodium channel by mu-conotoxin. The Journal of general physiology. 2002;119:45–54. doi: 10.1085/jgp.119.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santarelli VP, Eastwood AL, Dougherty DA, Horn R, Ahern CA. A cation-pi interaction discriminates among sodium channels that are either sensitive or resistant to tetrodotoxin block. The Journal of biological chemistry. 2007;282:8044–8051. doi: 10.1074/jbc.M611334200. [DOI] [PubMed] [Google Scholar]
- 30.Penzotti JL, Fozzard HA, Lipkind GM, Dudley SC., Jr Differences in saxitoxin and tetrodotoxin binding revealed by mutagenesis of the Na+ channel outer vestibule. Biophysical journal. 1998;75:2647–2657. doi: 10.1016/S0006-3495(98)77710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trimmer JS, Cooperman SS, Agnew WS, Mandel G. Regulation of muscle sodium channel transcripts during development and in response to denervation. Developmental biology. 1990;142:360–367. doi: 10.1016/0012-1606(90)90356-n. [DOI] [PubMed] [Google Scholar]
- 32.Lattes K, Venegas P, Lagos N, Lagos M, Pedraza L, Rodriguez-Navarro AJ, Garcia C. Local infiltration of gonyautoxin is safe and effective in treatment of chronic tension-type headache. Neurological research. 2009;31:228–233. doi: 10.1179/174313209X380829. [DOI] [PubMed] [Google Scholar]
- 33.Marcil J, Walczak JS, Guindon J, Ngoc AH, Lu S, Beaulieu P. Antinociceptive effects of tetrodotoxin (TTX) in rodents. British journal of anaesthesia. 2006;96:761–768. doi: 10.1093/bja/ael096. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez-Navarro AJ, Lagos N, Lagos M, Braghetto I, Csendes A, Hamilton J, Figueroa C, Truan D, Garcia C, Rojas A, Iglesias V, Brunet L, Alvarez F. Neosaxitoxin as a local anesthetic: preliminary observations from a first human trial. Anesthesiology. 2007;106:339–345. doi: 10.1097/00000542-200702000-00023. [DOI] [PubMed] [Google Scholar]
- 35.Akopian AN, Souslova V, England S, Okuse K, Ogata N, Ure J, Smith A, Kerr BJ, McMahon SB, Boyce S, Hill R, Stanfa LC, Dickenson AH, Wood JN. The tetrodotoxin-resistant sodium channel SNS has a specialized function in pain pathways. Nature neuroscience. 1999;2:541–548. doi: 10.1038/9195. [DOI] [PubMed] [Google Scholar]
- 36.Amaya F, Wang H, Costigan M, Allchorne AJ, Hatcher JP, Egerton J, Stean T, Morisset V, Grose D, Gunthorpe MJ, Chessell IP, Tate S, Green PJ, Woolf CJ. The voltage-gated sodium channel Na(v)1.9 is an effector of peripheral inflammatory pain hypersensitivity. J Neurosci. 2006;26:12852–12860. doi: 10.1523/JNEUROSCI.4015-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Catterall WA, Dib-Hajj S, Meisler MH, Pietrobon D. Inherited neuronal ion channelopathies: new windows on complex neurological diseases. J Neurosci. 2008;28:11768–11777. doi: 10.1523/JNEUROSCI.3901-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perkins SJ. Protein volumes and hydration effects. The calculations of partial specific volumes, neutron scattering matchpoints and 280-nm absorption coefficients for proteins and glycoproteins from amino acid sequences. European journal of biochemistry / FEBS. 1986;157:169–180. doi: 10.1111/j.1432-1033.1986.tb09653.x. [DOI] [PubMed] [Google Scholar]