Abstract
Melanocytic neoplasms with spitzoid features including spitz nevi, spitz tumors and spitzoid melanomas are commonly encountered in the practice of dermatopathology. Although many cases can be accurately diagnosed on the basis of morphology and histology, a significant number of cases may be difficult to accurately classify. Several studies have now shown that chromosomal copy number aberrations are typical of melanoma while present in only a small percent and to a limited degree in spitz nevi. In this study, we correlated the clinical, histologic and array CGH findings of 10 spiztoid melanocytic neoplasms. Our study shows that the clinical and histologic changes correlate well with the molecular findings by array CGH. Further that array CGH can be used to help classify and predict behavior of spitzoid melanocytic neoplasms. A limited variety of copy number aberrations including gains of 11p and much more rarely 7q may be seen in spitz nevi. Additionally in this report we present the first case of a typical spitz nevus with copy number gains involving both 7q and 11p. Conversely, melanomas with spitzoid features typically have multiple chromsomal copy number aberrations involving a variety of loci. A smaller number of chromosomal aberrations, possibly a single aberrant locus, may be present in spitz tumors, but their presence may predict more aggressive behavior.
Keywords: Spitzoid melanocytic neoplasms, melanoma, dermatopathology, chromsomal copy number aberrations, array, genomic hybridization
Introduction
Comprehensive studies by comparative genomic hybridization (CGH) and array CGH have clearly shown significant genetic differences between histologically benign nevi including Spitz nevi and melanoma [1]. Hence CGH has been adopted as a clinically useful tool for distinguishing benign and malignant melanocytic neoplasms. Specifically isolated gains in 11p have been identified as characteristic of a subset of Spitz nevi where as melanomas may harbor multiple distinct copy number aberrations [2]. Approximately 20% of spitz nevi are said to have an isolated gain in 11p and most of these cases also show mutations in HRAS which is located on the short arm of chromosome 11. Additionally a small number perhaps 5% of spitz nevi without 11p gain may also have this same HRAS mutation [1]. Spitz nevi most likely to harbor 11p gains are those that are large and bulky tumors with an infiltrative pattern with desmoplasia and dispersion to single cells at the base [3]. Additional unique features of Spitz nevi include frequent tetraploidy, which is characteristic of approximately 5% to 10% of spitz nevi [4].
Although rare, isolated gains in 7q have also been reported in Spitz nevi [2, 5]. A specific estimate of the frequency of this finding would require a large study of spitz nevi since studies to date suggest a very low and rare incidence of this finding. Hence while occasional karyotopic abnormalities may be found in spitz nevi, these changes are mostly distinct from melanoma. Based on this difference fluorescence in situ hybridization (FISH) targeting key chromosomal aberrations on 6p, 6q and 11q characteristic of melanoma has also been used as a more targeted method of looking at copy number aberrations in melanocytic neoplasms as a diagnostic tool. In fact in a study of 27 ambiguous melanocytic neoplasms with a differential diagnosis of spitz nevus versus melanoma, the detection of copy number aberrations by FISH involving chromosome 6 or the long arm of chromosome 11 strongly correlated with metastasis. In this study we wished to use array CGH to evaluate 10 melanocytic neoplasms with spitzoid morphology (8 spitz nevi, 1 spitz tumor and 1 spitzoid melanoma) and correlate the array CGH findings to the clinical outcome of these cases.
Material and methods
Specimen material
The cases used in this study were obtained from the files of the Department of Pathology, State University of New York at Buffalo and the Ros-well Park Cancer Institute at Buffalo. Institutional Review Board approval was obtained and resultant guidelines followed throughout the study. Eight spitz nevi, one spitz tumor and one spitzoid melanoma were reviewed by at least two pathologists (TH & RTC) by conventional light microscopy. DNA was isolated from ten-micron sections of nine spitz nevi and one-melanoma using the Gentra Puregene Tissue kit (Qiagen, Inc.), per manufacturer's instructions. Following quantification, 100ngof genomic DNA from each sample was whole genome amplified using the BioScore Screening and Amplification Kit (Enzo Life sciences [7].
Array comparative genomic hybridization
Array CGH was performed on all 10 spitzoid melanocytic neoplasm. Reference genomic DNA and amplified sample DNA (1μg each) was individually fluorescently labeled using the BioArray CGH Labeling System (Enzo Life Sciences). The sample and reference probes were combined, pelleted, resuspended and hybridized to the RPCI 21k BAC array as described [7-9]. The hybridized slides were scanned using a GenePix 4200AL Scanner (Molecular Devices) to generate high-resolution (5 μm) images for both Cy3 (test) and Cy5 (control) channels. Image analysis was performed using the ImaGene (version 8.0.0) software from BioDiscovery, Inc. The log2 test/control ratios were normalized using a subgrid loss. Mapping information was added to the resulting log2 test/control values. The mapping data for each BAC was found by querying the human genome sequence at http://genome.ucsc.edu and BACs in regions of segmental duplication or large-scale variation were flagged.
FISH validation
RPCI-11 BAC clones were chosen from genomic regions where copy number change was identified by the aCGH. DNA from the clones was labeled with Spectrum Orange (Abbott Molecular,Inc) by nick translation using a kit from Abbott Molecular Inc. as per manufacturer's instructions. Labeled DNA was then hybridized to pretreated tissue paraffin sections from the lesion according to standard procedures. A centromere specific probe (Abbott Molecular, Inc.) labeled with Spectrum Green was included as an internal control. Slides were visualized and analyzed on a Nikon Microscope using the Cy-toVision Program (Applied Imaging, Inc.)
Results
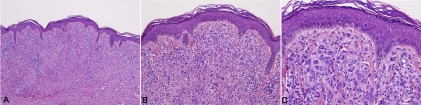
This study included 8 spitz nevi, 1 spitz tumor and 1 spitzoid melanoma. The clinical data is summarized in table 1. The spitz nevi patients ranged in age from 3 to 15 with an average of 8.6 years and included 6 females and 2 males. The patient with the spitz tumor was a 6 year-old male and the patient with the spitzoid melanoma was a 43 year-old male. Histology in the majority of spitz nevi cases showed typical features of spitz nevi, characterized by a symmetric compound proliferation of epithelioid and spindled melanocytes with clefting at the dermal-epidermal junction, and dispersion of single cells with descent in the dermis. Case number 6 showed a primarily dermal and symmetric wedge-shaped proliferation of epithioid melanocytes with prominent collagen fibrosis and dispersion to single cells at the base. The cells had open chromatin and abundant pink cytoplasm and lacked significant nuclear atypia (Composite Figure 1A, B &C).
Figure 1.
Composite figure, case no. 6; (1 A, B, C): Biopsy revealed broad specimen with dermal nests and inflammation. The epidermis lacks acanathosis. Kamino bodies are not evident. Nests of pleomorphic melanocytes with ample amphophillic cytoplasm are noted in the dermis. A patchy lymphohistiocytic infiltrate is present.
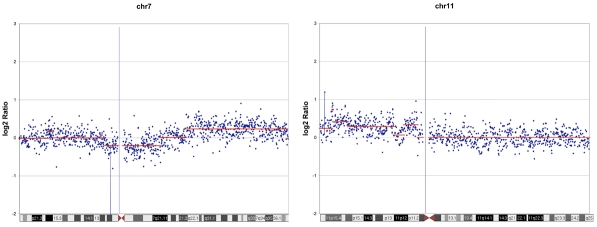
The array CGH data in 7 of these 8 cases was unremarkable with no evidence of any chromosomal aberrations. One case (case 6) which was from an 8 year-old female interestingly showed gains in both 7q and 11p (Figure 2). The 11p gains were validated by FISH analysis as well (Figure 3). From a histologic standpoint this case was a typical Spitz nevus without worrisome histologic changes. The lesion was a 6 mm dome shaped papule. The lesion was symmetric wedge shaped and mostly dermal. The nests in the papillary dermis lacked any worrisome changes, expansile nodular growth or mitotic activity and showed typical spitzoid morphology. There was excellent maturation with descent which included increasing dermal fibroplasia, decrease nest and cell size and dispersion to single cells at the base. The follow up of these 8 cases averaged 3 years and ranged from 2.5 to 3.5 years with all cases having an uneventful course with no evidence of recurrence or subsequent disease following complete excision.
Figure 2.
Case no.6 demonstrating detection of DNA segment copy number changes on chromosome 7 and 11 using 21K BAC aCGH. Average log2 ratios (y-axis) were plotted for all clones based on chromosomal position (x-axis) with the blue bar demarcating the centromere. Horizontal red lines indicate the log2 ratio for each segment as segmented by Circular Binary Segmentation (CBS).
Figure 3.
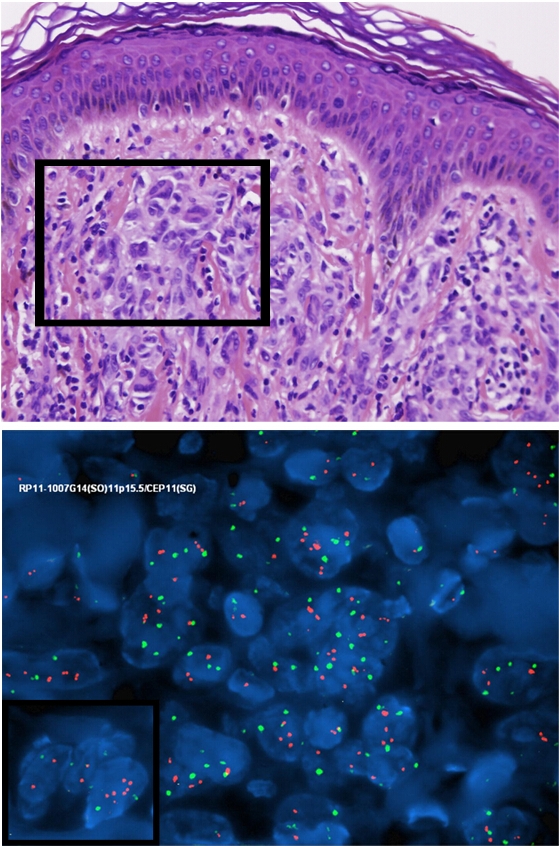
Dual-color FISH with a probe containing chromosome 11p (orange signals) and a reference probe for centromere of chromosome 11 (green signals) showing a central nest of melanocytes with multiple orange signals surrounded by normal cells.
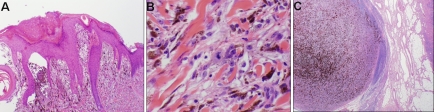
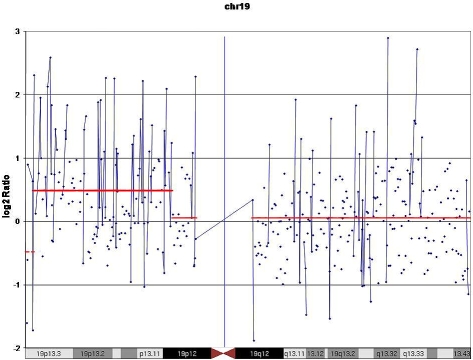
Case 9 was a spitz tumor from a 6 year old male. The lesion was 1 cm nodule with verrucous epidermal hyperplasia on the surface with focal ulceration. There was sheet like proliferation of spitzoid cells into the dermis with a thickness of 1.4mm with focal areas of near confluence. Minimal maturation was noted. A number of mitotic figures approximately 2/mm2 were noted including some near the base of the lesion (Figure 4A and B). The lesion as treated with wide excision and sentinel node biopsy. The sentinel node showed bulky involvement of an inguinal lymph node with spitz tumor (Figure 4C). Subsequent complete lymph node dissection failed to show any further lymph node involvement. Subsequently at 12 months of follow up the child experienced a local recurrence and wide re-excision was again performed with negative margins. The array CGH data in this case showed clear gains in 19p (Figure 5).
Figure 4.
A) Verrucous epidermal hyperplasia on the surface with focal ulceration and proliferation of spitzoid cells into the dermis. B). Rare dermal mitotic figure in the spitz tumor. C). Sentinel lymph node with bulky deposit.
Figure 5.
Case no.9 demonstrating detection of DNA segment copy number changes on chromosome 19 using 21K BAC aCGH. Average log2 ratios (y-axis) were plotted for all clones based on chromosomal position (x-axis) with the blue bar demarcating the centromere. Horizontal red lines indicate the log2 ratio for each segment as segmented by Circular Binary Segmentation (CBS).
Case number 10 was a clear melanoma histologically with spitzoid features. The histology showed confluent melanocytic dermal nests with epitheloid morphology. Numerous abnormal mitotic figures were present. The array CGH data showed amplification of 6p and 17q as well as deletion of 1p, and 15p (Figures not shown). The multiplicity of aberrations with complex karyotype was clearly diagnostic of melanoma as well. Thus far the follow up of this patient includes includes local-regional lymph node and distant metastasis.
Discussion
There has been accumulating evidence that molecular techniques looking at chromosomal copy number aberrations maybe used in conjunction with standard histologic and clinical data as a method of improving diagnosis in ambiguous melanocytic neoplasms [10,11]. In this study we presented the clinical, histologic, and molecular findings in 10 spitzoid neoplasms including 8 typical spitz nevi, 1 spitz tumor and 1 spitzoid melanoma. Interestingly for the most part the molecular changes identified by CGH and FISH correlated well with the clinical and histologic features in these cases.
Seven of the 8 typical spitz nevi in which histology was not atypical and clinical outcome were uneventful; there was no evidence of chromosomal copy number aberrations. However 1 typical spitz nevus showed copy number aberrations which included gains in 11p and 7q. Each of these copy number aberrations has individually been reported in spitz nevi. As mentioned earlier, up to 20% of spitz nevi may harbor isolated 11p gains. Gains in 7q are much more infrequent in spitz nevi, although it has been reported and in general is more typical of melanoma. This case is quite unique in that it is the first example that we are aware of in which a case of a typical spitz nevus without significant atypical features and with a benign outcome has been reported with coexistent gains in 7q and 11p. Hence this case is quite unique and further emphasizes the importance of integrating the clinical, histologic and molecular features in order to make the most optimal diagnosis in melanocytic neoplasms.
Case #9 was histologically a spitz tumor. This case which was from an 6 year-old male who did present with some histologically worrisome changes including dermal confluence of cells with incomplete maturation and multiple mitotic figures with a count of 2/mm2 including some mitoses at the base. Further this case clinically showed more aggressive behavior with lymph node involvement and local recurrence despite removal with wide excision. To date this patient is disease free with very close follow-up for recurrence of skin lesions or lymph node enlargement in two years. The array CGH data in this case revealed gains in 19p. Gains in 19p are finding that can be seen in melanoma and has never been reported in a spitz nevus. This is consistent with the behavior of this case with lymph node involvement and local recurrence. Interestingly this case only had one identifiable copy number aberration by array CGH as opposed to multiple aberrations, which are commonly seen in melanomas. The presence of a single but a definite copy number aberration may be characteristic of some of these intermediate or borderline malignant neoplasms.
The human gene locus c-MEL located on chromosome 19p has been identified by transfection of genomic DNA from the human melanoma cell line NK14. This gene is mapped to a region of short arm of chromosome 19 and is involved in karyotypic abnormalities in a variety of malignancies including melanomas and leukemia [12]. Aberrations on chromosome 19 have been found in multiple types of cancer [13-16]. Similarly involvement of chromosome 19 in the form of t (1; 19) has also been described in advanced malignant melanoma and pre-B cell leukemia [13]. Hence, we would consider the presence of any clonal chromosomal aberration other than the isolated finding of 11p of high concern in a spitz tumor. However, as we have seen in our studies by integrating clinical, morphologic and molecular findings, we may be able to further characterize the spectrum of chromosomal aberrations which may occur in benign spitz nevi which may occasionally include isolated gains in 7q or even the combined presence of gains in 7q with gains in 11p.
Not surprisingly, case # 10 which came from a 43 year-old man and had definitive histologic changes of melanoma with some spitzoid morphology showed multiple karyotypical abnormalities typical of melanoma. Further the outcome of this case showed distant metastasis involvement of distant lymph nodes.
In this study, we have demonstrated a definitive correlation between the clinical, histologic and molecular findings in spitzoid melanocytic neoplasms. Further we show that by integrating clinical, histologic and morphologic features one can make an optimal diagnosis. This was most evident in a unique case of a typical spitz nevus with copy number gains in both 11p and 7q. The appreciation that both of these changes can independently occur in benign spitz nevi combined with the clinical and histologic changes allows one to accurately classify this case as a benign spitz nevus. The classification was further supported by the uneventful follow up in our study. Further this helps us better define the spectrum of karyotypic abnormalities which may be seen in benign spitz nevi. Additionally, we see that in some borderline neoplasms, spitz tumors, which do show intermediate grades of histologic aggressiveness there maybe single or limited karyotypic abnormalities of a smaller degree than seen in typical melanoma. However the presence of even these limited but detectable aberrations can help predict the behavior and classification of such lesions and further may give more of a biologic basis for a category of intermediate grade of malignancy in melanocytic tumors. In summary, we feel that integrating molecular findings with the clinical and histologic data can significantly help in the classification of melanocytic neoplasms and in predicting their behavior.
Acknowledgments
The authors thank Paul Quinn and Mike Henry, for their technical expertise. The authors also want to thank Bill Schiff for his photographic skills.
References
- 1.Bastian BC, LeBoit PE, et al. Mutations and copy number increase of HRAS in Spitz nevi with distinctive histopathological features. Am J Pathol. 2000;157:967–972. doi: 10.1016/S0002-9440(10)64609-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bastian BC, Wesselmann U, Pinkel D, et al. Molecular cytogenetic analysis of Spitz nevi shows clear differences to melanoma. J Invest Dermatol. 1999;113:1065–1069. doi: 10.1046/j.1523-1747.1999.00787.x. [DOI] [PubMed] [Google Scholar]
- 3.Hantschke M, Bastian BC, LeBoit PE. Consumption of the epidermis: a diagnostic criterion for the differential diagnosis of melanoma and Spitz nevus. Am J Surg Pathol. 2004;28(12):1621–5. doi: 10.1097/00000478-200412000-00011. Dec. [DOI] [PubMed] [Google Scholar]
- 4.Isaac AK, Lertsburapa T, Pathria Mundi J, et al. Polyploidy in spitz nevi: a not uncommon karyotypic abnormality identifiable by fluorescence in situ hybridization. Am J Dermatopathol. 2010;32(2):144–8. doi: 10.1097/DAD.0b013e3181b72d6f. Apr. [DOI] [PubMed] [Google Scholar]
- 5.Gerami P, Jewell SS, et al. Fluorescence in situ hybridization (FISH) as an ancillary diagnostic tool in the diagnosis of melanoma. Am J Surg Pathol. 2009;33(8):1146–56. doi: 10.1097/PAS.0b013e3181a1ef36. Aug. [DOI] [PubMed] [Google Scholar]
- 6.Nowak NJ, Snijders A, et al. The BAC Resource: Tools for Array CGH and FISH. Current Protocols in Human Genetics. 2005:1–34. doi: 10.1002/0471142905.hg0413s46. [DOI] [PubMed] [Google Scholar]
- 7.Nowak NJ, Miecznikowski J, et al. Challenges in array comparative genomic hybridization for the analysis of cancer samples. Genet Med. 2007;9(9):585–95. doi: 10.1097/gim.0b013e3181461c4a. Sep. [DOI] [PubMed] [Google Scholar]
- 8.Snijders AM, Nowak N, Segraves R, et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat.Genet. 2001;29:263–264. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- 9.Miliaras D, Conroy J, Pervana S, et al. Karyotypic changes detected by comparative genomic hybridization in a stillborn infant with chorioangioma and liver hemangioma. Birth Defects Res. A Clin.Mol.Teratol. 2007;79:236–241. doi: 10.1002/bdra.20332. [DOI] [PubMed] [Google Scholar]
- 10.Gerami P, Pouryazdanparast P, et al. Molecular analysis of a case of nevus of ota showing progressive evolution to melanoma with intermediate stages resembling cellular blue nevus. Am J Dermatopathol. 2010;32(3):301–5. doi: 10.1097/DAD.0b013e3181b96db7. May. [DOI] [PubMed] [Google Scholar]
- 11.Dalton SR, Gerami P, et al. Use of fluorescence in situ hybridization (FISH) to distinguish intranodal nevus from metastatic melanoma. Am J Surg Pathol. 2010;34(2):231–7. doi: 10.1097/PAS.0b013e3181c805c4. Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nimmo E, Padua RA, Hughes D, et al. Confirmation and refinement of the localisation of the c-MEL locus on chromosome 19 by physical and genetic mapping. Hum Genet. 1989;81(4):382–4. doi: 10.1007/BF00283697. Mar. [DOI] [PubMed] [Google Scholar]
- 13.Parmiter AH, Balaban G, Herlyn M, et al. t(1;19) chromosome translocation in three cases of human malignant melanoma. Cancer Res. 1986;46(3):1526–9. Mar. [PubMed] [Google Scholar]
- 14.Verma RS, Manikal M, Conte RA, Godec CJ. Chromosomal basis of adenocarcinoma of the prostate. Cancer Invest. 1999;17(6):441–7. doi: 10.3109/07357909909021436. Review. [DOI] [PubMed] [Google Scholar]
- 15.Bergman A, Abel F, et al. No germ-line mutations in supposed tumour suppressor genes SAFB1 and SAFB2 in familial breast cancer with linkage to 19p. BMC Med Genet. 2008;13(9):108. doi: 10.1186/1471-2350-9-108. Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larramendy ML, Gentile M, et al. Does comparative genomic hybridization reveal distinct differences in DNA copy number sequence patterns between leiomyosarcoma and malignant fibrous histiocytoma. Hum Pathol. 2007;38(3):400–9. doi: 10.1016/j.cancergencyto.2008.06.005. Mar. [DOI] [PubMed] [Google Scholar]