Abstract
Stomach motility contributes significantly to fullness sensation while eating and cessation of food intake in humans. Genes controlling adrenergic and serotonergic mechanisms (ADRA2A, GNB3 and SLC6A4) affect gastric emptying (GE), volume (GV) and satiation. Fat mass and obesity-associated gene (FTO) is linked with satiety. Our aim was to examine the association of these candidate genes with stomach functions that signal postprandial fullness: GE, GV, and maximum tolerated volume (MTV). These biomarkers constitute a component of the intermediate phenotype of satiation. Sixty-two overweight or obese participants underwent genotyping of the candidate genes, and validated measurements of GE of solids and liquids by scintigraphy, fasting and postprandial change in GV by SPECT, and MTV by nutrient drink test. These markers of satiation were compared for 38 genetic variants in ADRA2A, ADR2C, ADRB3, uncoupling protein [UCP]-2 and -3, GNB3, FTO and SLC6A4 using a recessive model of inheritance. ADRA2A, ADR2C, UCP-3, GNB3, and FTO loci were significantly associated with the intermediate phenotype markers of satiation: ADR2C (Ins-Del322_325) with accelerated GE; GNB3 (rs1047776) with delayed GE; ADRA2A (rs491589 and rs553668) and GNB3 (rs2269355, rs10849527, and rs3759348) with decreased postprandial GV; ADRA2A (rs3750625) and GNB3 (rs4963517 and rs1129649) with increased postprandial GV; UCP-3 (rs1685356) with increased and FTO (rs9939609) decreased MTV. Genetic susceptibility to postprandial satiation can be identified through intermediate phenotype markers. With independent validation, these markers may guide patient selection of weight loss therapies directed at gastric motor functions.
INTRODUCTION
Obesity is a major health problem reaching epidemic proportions in the United States and in most industrialized countries (1) and leads to cardiovascular and metabolic diseases, as well as morbidity, mortality and financial burden.(2) The control of food intake (quantified by meal frequency and size) is a major determinant of an individual’s weight status.(3). Satiation is operationally defined by the symptoms experienced during feeding that results in cessation of eating (4). Satiation is influenced by oropharyngeal tactile, visual, olfactory, gustatory stimuli, by signals from post-ingestive processes and energy stores, (5) and by gastric motor functions. The stomach signals satiation in response to ingested volume and calories (6, 7); the postprandial change in gastric volume is an independent predictor of maximum tolerated volume and therefore satiation (8). In patients with functional dyspepsia and in obese subjects after administration of pharmacological perturbations to mimic dyspepsia, we demonstrated that gastric emptying of solids at 1 and 4 hours postprandially, and fasting gastric volume were associated with postprandial symptom scores such as fullness and bloating, reflecting satiation (9-11).
Intermediate phenotypes, which involve the same biological pathways as diseases, are considered to be less susceptible to confounding environmental influences than the phenotypes used to diagnose disease and therefore more relevant to the gene’s actions (12) In our model (see Figure 1), based on the concept of Huang et al. (12) obesity is the disease or clinical endpoint, satiation is the surrogate endpoint, and gastric motor functions and satiation volume are biomarkers or components of the intermediate phenotype. We are interested in identifying genes that determine decrease satiation, since this impacts the development of obesity. Since gastrointestinal hormones, such as cholecystokinin (CCK), glucagon-like peptide 1 (GLP-1), ghrelin and peptide YY (PYY), are associated with satiation, (13) we plan to expand the intermediate phenotype markers of satiation in future studies.
Figure 1.
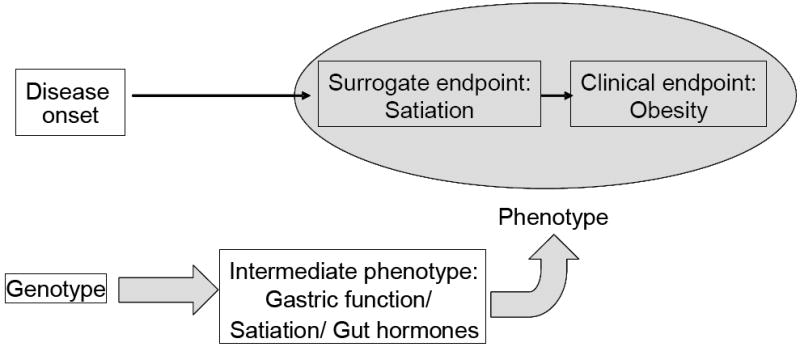
The proposed conceptual pathogenetic model includes the disease of obesity as the clinical endpoint, satiation as the surrogate endpoint, and gastric motor functions and gastrointestinal hormones as biomarkers or endophenotype, subject to modulation by the genotype.
We examined common variants in candidate genes in an attempt to identify variants that alter satiation. The relevance of norepinephrine and serotonin to human gastrointestinal function and appetite is well established in the literature. We have shown there is important α2-adrenergic modulation of gastric function by pharmacological studies (14), genotype-intermediate phenotype association (15) with gastric function, and we have documented the contribution of ADRA2A-1291C<G and GNB3 C825T or 5-HTTLPR with pharmacological response to sibutramine (16, 17). These SNPs may be simple biomarkers that are in linkage disequilibrium with the causative genetic variant that imparts differential function to the respective genes. Therefore, we evaluated further genotype-phenotype interactions in adrenergic mechanisms (including G protein) and serotonergic reuptake (SLC6A4) that may determine stomach function and satiation. Other genes associated with obesity (e.g. fat mass and obesity-associated gene, FTO [18]) are also linked with satiety, but their association with gastric functions is unknown.
The aim of the current study is to explore the hypothesis that genetic variations in the control of ADRA2A, ADR2C, ADRB3, GNB3, 5-HTTLPR, UCP-2 and UCP-3 and FTO are associated with changes in gastric function and satiation. This aim was pursued by comparing the associations with gastric functions for the three genotypes using the recessive model.
MATERIALS AND METHODS
Selection of participants
We used a database of 234 normal weight (NW), overweight (OW) or obese (Ob) subjects who participated in previous studies in our laboratory in order to estimate the minor allele frequency in a representative Caucasian population from the same geographic region of the upper mid-West of the United States. We obtained approval to use stored DNA by the Mayo Clinic Institutional Review Board. This approval was in accordance with written informed consent from the participants to use their stored DNA. The origination of this database through multiple studies has been described in detail elsewhere.(15) As 10 participants in the DNA collection subsequently withdrew consent to use their medical records for research, the final sample was 224 people.
For the specific aim of this study, 62 Caucasian, normal weight (NW), overweight (BMI: 25–29.9 kg/m2) and obese subjects (BMI >30 kg/m2) were included; they had previously participated in our studies of the pharmacogenetics of sibutramine (16) and our studies of gastric sensorimotor functions in obesity (8, 15). Men and women between the ages of 18 and 65 years residing in Olmsted County, Minnesota who were not currently on treatment for cardiac, pulmonary, gastrointestinal, hepatic, renal, hematologic, neurologic, endocrine (other than hyperglycemia not requiring medical therapy), or unstable psychiatric disease were eligible. Participants were excluded if their weight exceeded 137 kg (because of a weight limit of the SPECT camera) and if they had a positive history of previous abdominal surgery other than appendectomy, cholecystectomy, Caesarian section, or tubal ligation. They also were excluded if they had a positive history of any systemic disease that could affect gastrointestinal motility; or used medications that may alter gastrointestinal motility, appetite, or absorption (e.g. orlistat). Permissible medications were multivitamins, birth control pills, estrogen, and thyroxine replacement, all at stable doses for at least 30 days prior to the physiological studies. Women of childbearing potential (not postmenopausal/not surgically sterilized) had a negative pregnancy test within 48 hours before the studies that included radioisotopes. The study was approved by the Mayo Clinic Institutional Review Board.
Gastric emptying, accommodation and satiation data were available from the previous studies in 50 participants (46 with solid GE, 45 with liquid GE, 49 with gastric volumes, and 50 with satiation test data). These studies were completed in the remaining 12 participants.
Experimental protocol
All participants gave written informed consent and on different days, they presented to the Mayo Clinic Clinical Research Unit at 7 am after an 8 hour fasting period and underwent a gastric emptying scintigraphic study, a nutrient drink satiation test and a gastric accommodation SPECT study in this order. Gastric emptying and SPECT studies were performed at least 72 hours apart in order to avoid interference by 111In from the meal ingested during the gastric emptying study with the measurement of gastric volume by SPECT.
Gastric emptying study with scintigraphy
1.0 mCi 99mTc-sulfur colloid was added to two raw eggs during the scrambling, cooking process. The eggs were served on one slice of buttered bread with 240 ml of 1% milk (total calories: 296 kcal, 32% protein, 35% fat, 33% carbohydrate) labeled with 0.1 mCi 111In-DTPA. Anterior and posterior gamma camera images were obtained immediately after radiolabeled meal ingestion, every 15 minutes for the first 2 hours, then every 30 minutes for the next 2 hours (total 4 hours after the radiolabeled meal).(8) Data were analyzed as in previous studies.(19) Geometric mean of decay-corrected counts in anterior and posterior gastric regions of interest were used to estimate the proportion of 99mTc or 111In emptied at each time point. Gastric emptying endpoints of analysis were t1/2 for solids and liquids.
Gastric volume and accommodation assessment with 99mTc-SPECT
We used a noninvasive method to measure gastric volume during fasting and at 32 minutes after 300 mL of Ensure® (316 Kcal) using single photon emission computed tomography. The method has been validated in detail elsewhere.(20) Intravenous injection of 99mTc sodium pertechnetate, which is taken up by the parietal and non-parietal cells of the gastric mucosa, allows visualization of the stomach wall. Tomographic images of the gastric wall are obtained throughout the long axis of the stomach using a dual-head gamma camera (SMV SPECT System*, Twinsburg, Ohio, U.S.A.) that rotates around the body. This allows assessment of the radiolabeled circumference of the gastric wall rather than the intra-gastric content. Using the AVW 3.0 (Biomedical Imaging, Mayo Foundation) image processing libraries, a three-dimensional rendering of the stomach is obtained and its volume (mL) calculated. There is high intra-observer reproducibility to measure gastric volume with this technique.(21) Intra- and inter-individual coefficients of variation (at average 9 months) are ~12%.(22)
Nutrient drink satiation test
We used a standardized and validated nutrient drink test (23) to measure satiation and postprandial symptoms when drinking a liquid nutrient at constant rate (30 mL/min Ensure®: 1 Kcal/mL, 11% fat, 73% carbohydrate and 16% protein). Every 5 minutes, subjects scored level of fullness or satiation using a horizontal scale combining verbal descriptors and numbers (0 = no symptoms; 5 = maximum or unbearable fullness/satiation). Nutrient intake was stopped when subjects reached the score of 5, and caloric intake to achieve maximum satiation was recorded. Postprandial fullness, nausea, bloating and pain, were measured 30 minutes after the meal using 100mm horizontal visual analog scales (VAS), with the words ‘none’ and ‘worst ever’ anchored at the left and right ends of the lines for each symptom.
Rationale for selection of candidate genes
At least 17 loci have been reported to be associated with variation of BMI.(24) We have previously studied the association of ADRA2A-1291C<G and GNB3 C825T or SLC6A4 with the intermediate phenotype of postprandial satiation and with pharmacological response to sibutramine (16, 17). In Table 1, we include these and other genetic variations in ADRA2A, GNB3 and SLC6A4, and summarize a literature review of the main polymorphisms associated with BMI and obesity phenotypes in different populations.
Table 1.
Summary of main polymorphisms associated with BMI and obesity phenotypes in different populations
| Gene/Name | MAF | Variation location role | Associations with BMI | BMI association, p= | N | Age | Sex | Ethnic Group | Study Design (reference) |
|---|---|---|---|---|---|---|---|---|---|
|
ADRA2A A1780G |
0.16 | 3’-UTR | AA: 24.5, AG:35, GG:37 (BMI kg/m2 in African Americans) | 0.05* | 235 | 18-49 | both | White and African Americans | cross-sectional single center (36) |
|
ADRB3 Trp64Arg |
0.09 | non-synonymous | +0.24 (BMI kg/m2 Δ between carrier and WT) | 0.0002* | 44833 | NA | both | Overall | meta-analysis of 97 studies (26) |
| ADRB3 Trp64Arg |
0.09 | non-synonymous | +0.08 (BMI kg/m2 Δ between carrier and WT) | 0.363 | 23198 | NA | both | Caucasian | meta-analysis of 53 studies (26) |
| ADRB3 Trp64Arg |
0.09 | non-synonymous | +0.31 (BMI kg/m2 Δ between carrier and WT) | 0.001* | 16061 | NA | both | East Asian | meta-analysis of 90 studies (26) |
| ADRB3 Trp64Arg |
0.09 | non-synonymous | +0.27 (BMI kg/m2 Δ between carrier and WT) | 0.258 | 5574 | NA | both | Other | meta-analysis of 29 studies (26) |
|
UCP-3 Tyr210Tyr |
0.42 | Exon 5 | +0.29/+1.23 (BMI kg/m2 Δ of heterozygotes/homozygotes from WT) | HT 0.457 H 0.019* |
382 | 40-80 | Male | Dutch Caucasian | cross-sectional single center (32) |
| UCP-3 | 0.45 | Exon 7 | +0.38/+1.11 (BMI kg/m2 Δ of heterozygotes/ homozygotes from WT) | HT: 0.363 H: 0.033 |
382 | 40-80 | Male | Dutch Caucasian | cross-sectional single center (32) |
| UCP-2 +3474 45bp-insdel | 0.24 | 3’-UTR, exon 8 | +2/+3 (BMI kg/m2 Δ of hetero/ homozygotes from WT) | HT: NS H: 0.005 |
603 | Adult | Both (289F) | German Caucasian | cross-sectional single center (30) |
| UCP-2 - 866G>A | 0.372 | Intergenic | +0.4/+0.5 (BMI kg/m2 Δ of heterozygotes/ homozygotes from WT) | 0.036 | 737 | 7-8 | Both (347F) | Korean | cross-sectional single center (31) |
| UCP-2 -866G>A | 0.372 | Intergenic | +0.4 (BMI kg/m2 Δ from WT) | 0.22 | 732 | 20-74 | both (295F) | Korean | cross-sectional single center (31) |
| FTO | 0.44 | Intron | +0.7/+1.3 (BMI kg/m2 Δ of heterozygotes/ homozygotes from WT) | 0.013 | 947 | Adult | both | American (Utah pedigrees) | longitudinal single center study (37) |
| FTO | 0.44 | Intron | +0.08/+0.49 (BMI kg/m2 Δ of hetero-homozygotes vs. WT) | 0.003 | 2422 | 4-10 | both | Scottish Caucasian | cross-sectional single center (38) |
|
GNB3 C825T |
0.303 | synonymous | higher percentage of homozygotes with BMI>25 or >27 kg/m2 | BMI>25: 0.03 BMI>27: 0.01 |
277 | 23.9±3.4 | male | German Caucasian | multicenter study (39) |
| GNB3 C825T |
0.303 | synonymous | different genotype distributions by weight | <0.001 | 960 | 24.6±4.4 | male | Chinese | multicenter study (39) |
| GNB3 C825T |
0.303 | synonymous | different genotype distributions by weight | <0.05 | 713 | 22 mean | male | Black Africans | multicenter study (39) |
|
SLC6A4 5-HTTLPR |
0.41 | 44bp del/ins in Promoter | 1.47±1.09 vs. 0.51±1.4, (BMI SS+SL vs. LL). OR for overweight: 1.85 | SS+SL<0.002 LL <0.02 |
172 | 16±2 | both | Argentina, rural area, ? ethnicity | population-based, cross- sectional high school students (40) |
| SLC6A4 5-HTTLPR |
0.41 | 44bp deletion/insertion in promoter | different genotype distribution in different BMI groups, OR for obesity=1.36, SS vs. LL | Distribution: <0.0002* OR: 0.026* |
1329 | 34.6±0.3 | both | Argentina, factory workers, ? ethnicity | population-based crossover study (41) |
| SLC6A4 5-HTTLPR |
0.41 | 44bp deletion/insertion in promoter | Males overall: OR for overweight & obese=1.69 (SS vs. LL): White males: OR=2.01 SS vs. SL; Hispanic males: OR 1.37 SS vs. LL; NS for females | OR M: 0.04 overall M 0.01 White 0.0007 Hispanic 0.047 |
1584 | Adolescent | both | USA, Caucasian and Hispanic origin, unrelated | subset of the National Longitudinal Study of Adolescent Health (42) |
BMI=body mass index; del-ins=deletion/insertion; Δ=difference; HT=heterozygote; H=homozygous variant; M=males;OR=odds ratio; WT=“wild” type, more frequent polymorphism
Indirect epidemiological and experimental data also support the hypothesis that other candidate mechanisms may be involved in control of gastrointestinal functions that lead to satiation. These mechanisms include:
β3-adrenoreceptor (β3-AR), and the rationale for inclusion is based on tissue expression in gastrointestinal organs (25) and the association of Trp64Arg ADRB3 genetic variant with BMI in a meta-analysis 44,000 people.(26)
Fat mass and obesity associated gene (FTO), SNP rs1421085, which is significantly associated with childhood and adult onset obesity;(27) SNP rs9939609 is also associated with satiety.(18, 28)
Uncoupling proteins (UCP) comprise a family of mitochondrial transporters that control the process of adaptive thermogenesis. Two proteins of relevance are UCP-2, which is widely expressed in human tissues including the stomach,(29) and UCP-3 which is predominantly expressed in skeletal muscle, is involved in adaptive thermogenesis in humans, and interacts with norepinephrine. Variants in the genes controlling UCP-2 and -3 are epidemiologically linked to BMI or satiation.(30-32) Furthermore, the central hypothalamic effects of ghrelin appear to be mediated by a UCP-2-dependent pathway; since ghrelin affects gastric motility (33) and appetite in the short term, it is relevant to assess whether genetic variations in UCP-2 alter gastric function and satiation.
Since previous studies identified ADRA2A- 1291C<G and GNB3 C825T in association with gastric motor functions (14-17), we used the Genome Variation Server sponsored by Seattle SNPS (http://gvs.gs.washington.edu/GVS/) to identify TagSNPs in the ADRA2A, GNB3, UCP2 and UCP3 genes expanding the amount of variation studied in those genes. TagSNPs for the genes of interest were selected to capture SNPs with a minor allele frequency of 10% with an R2 of 0.9. Analysis of linkage disequilibrium among the SNPs of the candidate genes was analyzed using HaploView 4.1. (34).
Genetic testing
Genomic DNA was isolated from peripheral blood leukocytes in previous studies (using the Gentra Purgene Blood Kit (Qiagen, Valencia, CA). We used assays for 38 variations in the genes of interest.
Genotyping of α2C adrenoreceptor (Del322-325) polymorphism [ADRA2C (Del332-325)], 5-HTTLPR polymorphism was performed using previously published method (15). For UCP-2 +3474 45bp-insdel, the A 45bp insertion/deletion was identified by PCR-based fragment length polymorphisms and confirmed by random direct sequencing. We used oligonucleotide primers flanking the polymorphic region forward (5’-CAG TGA GGG AAG TGG GAG G-3’) and reverse (5’ - GGG GCA GGA CGA AGA TTC – 3’) as used by Walder et al. (35).
Genotyping of the remaining 35 SNPs was performed by TaqMan® SNP Genotyping Assays (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions, using 10-20ng DNA. Following PCR amplification, end reactions were read on the ABI 7500 Fast Real-Time PCR system using Sequence Detection Software version 1.3.1 (Applied Biosystems).
Statistical methods
We assessed the associations of allelic subtype for 38 variations in the genes of ADRA2A, ADR2C, ADRB3, UCP-2, UCP-3, GNB3, FTO and 5-SLC6A4 with intermediate phenotype using an analysis of covariance (ANCOVA, adjusting for age and gender) separately for each gene. Since some genes exhibited a low minor allele genotype (<5%), the analysis combined the homozygous minor genotype with the respective heterozygous allele type yielding the recessive gene model in the ANCOVA analyses. The α level for testing genotype association was set at 0.05 in light of the exploratory hypothesis-generating nature of the analyses. Thus, no correction was made for multiple genes examined.
Statistical Power
The univariate association of genotype with intermediate phenotype response can be considered via comparison of response values among the genotypes for a given gene (i.e. a comparison of group mean values). Table 2 summarizes data for primary response measures and uses the relative variation (CV%) to estimate the effect size detectable with 80% power based on a two sample t-test at a two-sided α level of 0.05. The effect size is the difference in group means as a percentage of the overall mean for each response. The total number of subjects with specific physiological (intermediate phenotype) measurements varied between 49 and 62 depending on the measurement and the effect sizes detectable for any specific gene depend on the number in each subtype (minor homozygous plus heterozygous vs. major homozygous). In general, assuming roughly 10, or separately 25, in the minor allele/heterozygous subtype implies the remainder (52 or 37) are in the other subtype. The ANCOVA models provided similar power for somewhat smaller differences by incorporating relevant covariates (e.g. gender in the case of gastric emptying, and maximum tolerated volume). The data for satiety testing in healthy volunteers, for scintigraphic gastric emptying and for SPECT have been published elsewhere (19,20,23). The observed variation (CV%) among subjects in this study was very similar to those listed in Table 2.
Table 2.
Primary response measures
| Response Type | Mean | CV % | Effect Size (%) Detectable with 80% Power (α= 0.05) | |
|---|---|---|---|---|
| N=25† vs. 37 | N=10† vs. 52 | |||
| Solid (egg meal) gastric emptying t1/2 | 112 | 32 | 24 | 32 |
| Satiety volume (ml) | 1350 | 25 | 19 | 25 |
| Fasting gastric volume (ml) by SPECT | 215 | 31 | 23 | 31 |
| Post-meal gastric volume (ml) by SPECT | 673 | 16 | 12 | 16 |
| Δ (PP-Fasting) gastric volume by SPECT | 716 | 17 | 13 | 17 |
Number in the minor allele subgroup
RESULTS
Demographics of participants
The demographic data and flow of participants, all of whom were European Americans, are summarized in Figure 2.
Figure 2.
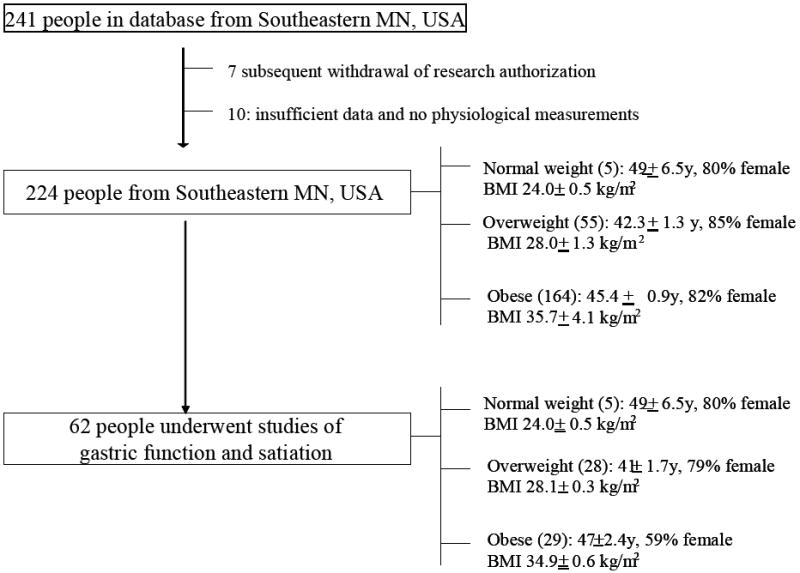
Study flow chart. Note that the age, gender and BMI distributions of the 62 people who underwent intermediate phenotyping were similar to those of the entire cohort of people from southeastern Minnesota, USA.
Allelic frequency of candidate SNPs in people in southeastern Minnesota
Minor allele frequencies of 38 SNPs pertaining to the genes of ADRA2A, ADRA2C, ADRB3, GNB3, SLC6A4, FTO, and UCP 2 and 3 have been estimated in 234 adults (N=5 normal weight, N=55 overweight, and N=164 obese). Data are summarized in Table 3. The allelic frequencies were in Hardy-Weinberg equilibrium (Table 3).
Table 3.
Minor allele frequencies (MAF) of 38 gene polymorphisms for 234 people of southeastern Minnesota
| Gene | Name | Rs | HW p value | MAF | Function |
|---|---|---|---|---|---|
| ADRA2C | ADRA2C-Del322-325 | 1 | 0.05 | Del-ins | |
| ADRB3 | Trp64Arg | rs4994 | 0.41 | 0.09 | Non-synonymous |
| ADRA2A | rs17128356 | 0.55 | 0.058 | intergenic | |
| ADRA2A | rs7096359 | 1 | 0.083 | intergenic | |
| ADRA2A | rs491589 | 0.62 | 0.156 | intergenic | |
| ADRA2A | -1291(C-G) | rs1800544 | 0.88 | 0.295 | near gene |
| ADRA2A | rs1800545 | 0.76 | 0.124 | 5’ UTR | |
| ADRA2A | rs11195419 | 0.57 | 0.132 | 3’ UTR | |
| ADRA2A | rs553668 | 0.81 | 0.16 | 3’ UTR | |
| ADRA2A | rs3750625 | 0.49 | 0.053 | 3’ UTR | |
| ADRA2A | rs602618 | 0.76 | 0.299 | intergenic | |
| ADRA2A | rs34665740 | 0.23 | 0.034 | intergenic | |
| UCP-2 | +3474 45bp-insdel | 0.62 | 0.24 | Del-ins | |
| UCP-2 | Ala55Val | rs660339 | 1 | 0.423 | Non-synonymous |
| UCP-2 | rs659366 | 0.89 | 0.372 | intergenic | |
| UCP-3 | rs590336 | 0.15 | 0.447 | ||
| UCP-3 | rs15763 | 1 | 0.25 | 3’ UTR | |
| UCP-3 | rs647126 | 0.15 | 0.447 | 3’ UTR | |
| UCP-3 | rs1685356 | 0.43 | 0.429 | near gene | |
| UCP-3 | rs2632723 | 0.42 | 0.201 | near gene | |
| UCP-3 | rs11235971 | 0.2 | 0.286 | near gene | |
| UCP-3 | rs1626521 | 0.41 | 0.267 | near gene | |
| UCP-3 | Tyr210 | rs2075577 | 0.11 | 0.404 | synonymous |
| UCP-3 | rs2734827 | 0.064 | 0.357 | inton | |
| UCP-3 | Tyr99 | rs1800006 | 0.3 | 0.256 | synonymous |
| GNB3 | rs2269355 | 1 | 0.432 | intron | |
| GNB3 | rs10849527 | 0.63 | 0.288 | intron | |
| GNB3 | rs3759348 | 0.75 | 0.286 | near gene | |
| GNB3 | rs4963517 | 0.6 | 0.429 | near gene | |
| GNB3 | rs4963516 | 1 | 0.135 | near gene | |
| GNB3 | Ile685Thr | rs1129649 | 0.57 | 0.346 | nonsense/missense |
| GNB3 | rs1047776 | 0.62 | 0.073 | 3’ UTR | |
| GNB3 | rs5439 | 0.45 | 0.096 | 3’ UTR | |
| GNB3 | rs5440 | 0.52 | 0.487 | 3’ UTR | |
| GNB3 | G814A (Gly272Ser) | rs5442 | 0.61 | 0.068 | Non-synonymous |
| GNB3 | C825T (Ser275) | rs5443 | 0.76 | 0.303 | synonymous |
| FTO | rs9939609 | 0.51 | 0.44 | intron | |
| 5-HTTLPR | LS/SS (-SLC6A4) | 44 bp ins | 0.69 | 0.41 | Del-ins |
Association of candidate genes with indices of satiation and gastric motor function
In 62 of these people (5 NW, 28 OW, and 29 Ob), ADRA2A, ADRB3, UCP-2, UCP-3, GNB3, FTO and SLC6A4A loci were significantly associated with altered gastric functions or satiation. Data for associations (adjusted for age and gender) for each SNP tested are summarized in Table 4.
Table 4.
Summary of significant genotype-intermediate phenotype associations
| Entire cohort available | Cohort with intermediate phenotype | Δ Vol | MTV | Solid GE (t1/2) | p-value† | |
|---|---|---|---|---|---|---|
| ADRA2C_Del322_325 | ||||||
| ins/ins | 200 | 52 | 487.9±13.7 | 1247.4 ± 40.8 | 123.5 ± 5.4 | .0271 ↑(Solid GE) |
| ins/del; del/del | 24; 0 | 10; 0 | 486.3±24.8 | 1060.5±113.7 | 101.8±7.5 | .1136 ↓ (MTV) |
| ADRA2A-rs491589 CC | 158 | 46 | 503.1±13.3 | 1207.6±48.2 | 117.7±5.0 | .0395 ↓(ΔVol) |
| CT_TT | 62; 4 | 14;2 | 449.0±23.8 | 1245.0±65.5 | 125.6±12.0 | |
| ADRA2A-rs553668 GG | 157 | 45 | 503.1±13.3 | 1210.8±49.2 | 117.6±5.1 | .0395 ↓ (ΔVol) |
| AG_AA_ | 62; 5 | 15;2 | 449.0±23.8 | 1234.4±62.4 | 125.6±11.2 | |
| ADRA2A-rs3750625 CC | 200 | 55 | 477.6±12.5 | 1216.8±42.0 | 122.7±5.1 | .0275 ↑ (ΔVol) |
| AC_AA | 23; 1 | 7;0 | 559.8±27.5 | 1220.9±121.5 | 94.8±10.1 | |
| ADRA2A-rs34665740 AA | 209 | 57 | 480.9±12.6 | 1223.9±41.8 | 120.5±5.1 | .0900 ↑ (ΔVol) |
| AC_CC | 14; 1 | 5;0 | 547.2±33.6 | 1141.0±113.0 | 109.9±12.8 | |
| UCP_2 +347445bp-insdel | ||||||
| ins/ins | 111 | 31 | 498.5±18.1 | 1217.9±48.5 | 112.1±5.7 | All p>0.15 |
| ins/del; del/del | 98;15 | 28;3 | 475.3±15.7 | 1216.6±62.9 | 126.9±7.5 | |
| UCP_3-rs590336 TT | 62 | 16 | 462.4±17.5 | 1055.3±62.3 | 130.1±11.1 | .0750 ↑ (MTV) |
| CT_ CC | 124;38 | 38;8 | 496.7±15.0 | 1273.6±45.9 | 115.8±5.1 | |
| UCP_3-rs1685356 CC | 69 | 19 | 480.7±17.6 | 1044.3±53.6 | 127.0±10.1 | .0261 ↑ (MTV) |
| CT_ TT | 118;37 | 35; 8 | 491.0±15.9 | 1293.7±47.4 | 116.5±5.3 | |
| UCP_3-rs11235971 TT | 110 | 29 | 513.2±15.9 | 1244.3±55.6 | 115.1±6.5 | .0676 ↓ (ΔVol) |
| CT_ CC | 101;13 | 30;3 | 466.8±16.9 | 1193.1±56.0 | 124.1±7.0 | |
| UCP_3-rs2075577 TT | 73 | 18 | 456.6±16.4 | 1024.4±59.3 | 128.4±10.4 | .0805 ↑ (ΔVol) |
| CT_ TT | 122;29 | 37;7 | 501.3±15.4 | 1296.1±45.1 | 115.9±5.1 | .0058 ↑ (MTV) |
| GNB3-rs2269355 CC | 74 | 26 | 517.2±17.0 | 1196.9±68.2 | 126.7±8.3 | .0212 ↓ (ΔVol) |
| CG_GG | 112; 38 | 23;13 | 463.5±15.8 | 1232.0±47.3 | 114.9±5.7 | |
| GNB3-rs10849527 CC | 116 | 35 | 512.9±15.0 | 1213.9±55.7 | 124.4±6.8 | .0029 ↓ (ΔVol) |
| CT_ TT | 90; 18 | 22;5 | 456.6±17.8 | 1221.6±55.5 | 114.1±6.6 | |
| GNB3-rs3759348 CC | 116 | 32 | 508.3±16.9 | 1231.8±60.4 | 119.3±7.3 | .0156↓ (ΔVol) |
| CT_TT | 91; 17 | 25;5 | 454.2±16.2 | 1201.7±50.5 | 120.3±6.4 | |
| GNB3-rs4963517 CC | 67 | 20 | 449.7±22.6 | 1188.4±54.8 | 109.6±7.3 | .0288 ↑ (ΔVol) |
| CT_ TT | 116; 41 | 30; 12 | 504.4±13.5 | 1231.0±52.2 | 124.7±6.1 | |
| GNB3-rs1129649 TT | 99 | 24 | 521.5±18.8 | 1238.8±73.1 | 123.0±9.2 | .0021 ↑ (ΔVol) |
| CC_CT | 99;26 | 30;8 | 462.2±14.2 | 1203.7±45.4 | 117.9±5.5 | |
| GNB3-rs1047776 GG | 190 | 54 | 480.5±13.0 | 1205.8±42.6 | 116.1±4.3 | .0042 ↓ (Solid GE) |
| AG_AA | 34;0 | 8; 0 | 524.3±30.3 | 1294.6±105.0 | 151.7±26.0 | |
| GNB3-rs5440 GG | 51 | 19 | 452.8±26.5 | 1196.7±53.5 | 118.6±8.6 | All p>0.15 |
| AA_AG | 63;110 | 17; 26 | 500.2±13.0 | 1226.3±51.9 | 120.3±5.8 | |
| GNB3-rs5443 (C825T) CC | 108 | 29 | 500.4±17.6 | 1307.0±66.0 | 122.6±8.7 | All p>0.15 |
| CT_TT | 100;16 | 30;3 | 472.0±15.7 | 1138.4±42.3 | 117.7±5.4 | |
| FTO-rs9939609 AA | 48 | 17 | 508.0±20.1 | 1372.8±80.6 | 108.8±8.0 | .0075 ↑ (MTV) |
| TT_ AT | 73;10 | 16;29 | 480.3±14.7 | 1158.5±42.2 | 124.0±5.8 | |
| 5_HTTLPR LL | 70 | 23 | 478.9±19.5 | 1118.7±62.9 | 124.3±7.1 | .1059 ↑ (MTV) |
| LS_SS | 109; 45 | 29; 10 | 491.9±15.3 | 1275.4±48.6 | 117.2±6.4 | |
Δ Vol=difference between postprandial and fasting gastric volume,
MTV=maximum tolerated volume during nutrient drink test,
GE=gastric emptying
↑ = acceleration of gastric emptying rate or increased volume (MTV or Δ Vol),
↓ = delay in gastric emptying rate, or decreased volume
=for test of association of response with genotype categories from the ANCOVA (adjusting for age and gender); unless otherwise stated, p>0.15
Using the recessive model, associations were identified between intermediate phenotypes and the following candidate genes:
ADR2C (Ins-Del 322_325) had accelerated GE of solids; and GNB3 (rs1047776) had delayed GE;
ADRA2A (rs491589 and rs553668) and GNB3 (rs2269355, rs10849527, and rs3759348) had decreased postprandial GV;
ADRA2A (rs3750625) and GNB3 (rs4963517 and rs1129649) had increased postprandial GV;
UCP-3 (rs1685356) had increased MTV;
FTO (rs9939609) had decreased MTV
Linkage disequilibrium
Haploview was used to assess whether there was linkage disequilibrium of SNPs within the different candidate genes. These are illustrated in Figure 3. Two haploblocks were identified within GNB3, three within the UCP2/UCP3 locus and one within ADRA2A. Note that there is expected linkage among TagSNPs selected for study within a gene (e.g. GNB3, ADRA2A). Of the SNPs listed in Table 4 which are associated (p<0.10) with gastric motor function or satiation, the following also occur within a haplotype block: GNB3 rs10849527, and rs3759348 in block 1; UCP3 rs590336 in block 4; UCP3 rs1685356, rs11235971, rs2075577 in block 5; and ADRA2A rs491589, rs553668, rs3750625 in block 6.
Figure 3.
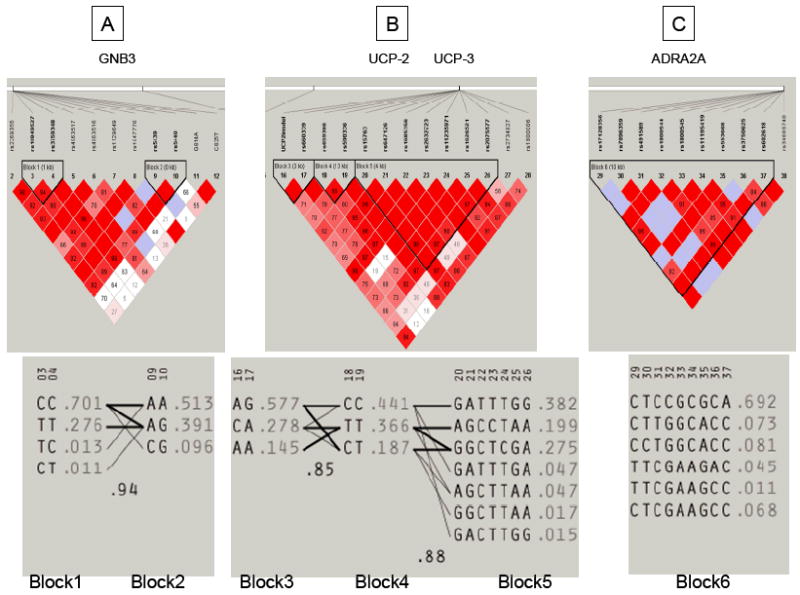
Haploview analysis of haploblocks within the candidate genes studied. Two haploblocks were identified within GNB3 (A), three within the UCP2/UCP3 locus (B) and one within ADRA2A (C). Population frequencies are shown next to each haplotype and lines show the most common crossings from one block to the next, with thicker lines showing more common crossings than thinner lines. Shown beneath the crossing lines is multi-locus D’.
Whereas, GNB3 rs5439 and rs5440 were in linkage disequilibrium in block 2, and UCP2 +3474 45bp-insdel and rs660339 were in linkage disequilibrium in block 3, none of these SNPs were associated with gastric motor function or satiation at p<0.10.
DISCUSSION
In this preliminary study, we explored the association between genetic variations of 38 candidate genes associated with obesity, and gastric motor functions and satiation in overweight and obese European Americans. We detected (Table 4) significant associations of certain genetic variations with the intermediate phenotypes of satiation and measures of gastric motor functions. These findings, which are not corrected for multiple comparisons, are preliminary and serve to generate hypotheses for future studies.
The number of genotype and genome-wide association studies with obesity has steadily increased during recent years; however, data on genotype-intermediate phenotype associations in obesity are lacking. This study provides new evidence on the potential relevance of genetic modulation of gastric motor functions that may determine satiation in obesity. These include slower gastric emptying, reduced fasting gastric volume and postprandial (accommodation) gastric volume and lower maximum tolerated volume. We had previously demonstrated these functions were associated with postprandial fullness and bloating in obesity and dyspepsia (9-11). These symptoms would be expected to lead to feeding cessation and hence impact on body weight through their effect on calorie ingestion.
Although the results of the present study are promising, they should be regarded as hypothesis-generating and replication of the results in larger studies is warranted. It is also relevant to note that the magnitude of differences in the physiological endpoints (e.g. ins/del ADRA2C Del 322-325 was associated with faster t1/2 gastric emptying of solids by an average of 21 minutes) may not be sufficient to induce clinical symptoms, such as symptoms of dumping syndrome. However, this genetic variation may be relevant in the context of calorie intake, since Delgado-Aros et al. (10) showed that maximum tolerated volume changed by 78 ± 36 mL for each 10% change in the rate of gastric emptying. Being able to tolerate 150 kcal per fully satiating meal in patients with heterozygous variant of ADRA2C Del 322-325 may conceivably impact calorie intake and body weight.
The other genetic variants were associated with a single intermediate phenotype marker. Therefore, the precise physiological mechanism underpinning the lower MTV with genetic variations related to UCP-3 and GNB3 genes cannot be determined in the current studies. For example, it is conceivable that the association of UCP-3 and satiation is unrelated to gastric functions and is more closely or directly related to the effects of norepinephrine on adipocytes, which change appetite control and calories ingested. Despite these limitations, the current data provide the basis for future larger studies to address the genotype- intermediate phenotype of satiation. Such studies have the potential to identify susceptibility genes to select patients and identify targets for therapy in obesity and metabolic syndrome. Future studies will also include studies of association of the candidate genes with gastrointestinal peptides and hormones that mediate satiation or satiety, such as CCK, GLP-1 and PYY.
Limitations and strengths
In view of the small sample size, it is possible that there is a type II error, and underestimation of the association strength of some gene polymorphisms cannot be excluded. Moreover, the positive findings may be potentially spurious because of the multiple endpoints and the relatively large number of genetic variations evaluated. Rather than evaluating all the genetic loci linked to obesity in the literature, we limited our study to a comprehensive assessment of all the genetic variations of candidate genes that are associated with satiation or satiety based on epidemiological or experimental evidence. Use of validated physiological endpoints rather than clinical endpoints, like BMI or weight change, allows for much smaller sample sizes, and the current pilot study has provided important information to plan the size of future studies. Thus, we have estimated that for each candidate gene, we require 25 major genotype and 25 minor genotype to detect (with 80% power) gene associated effect sizes of 18% in solid gastric emptying t1/2, 14% in satiation volume, 18% in fasting gastric volume and 9 % in post-meal gastric volume by SPECT. The sample sizes are therefore practical in the setting of a single-center study employing comprehensive, specialized physiological testing to study genotype-intermediate phenotype associations.
In fact, the effect sizes demonstrable with the samples studied appear large when compared with the reported effect of disease-associated variants on disease predisposition. However, this study has used intermediate phenotypes with known clinical significance. For example, the 20% effect size demonstrable for the t1/2 gastric emptying of solids is in the range that differentiates health from gastroparesis.
One of the strengths of this study is the extensive use of validated, reproducible methods with known coefficients of variation in the assessment of gastric motor functions and satiation, supporting comparisons in relatively small samples of people of candidate genetic differences with previously demonstrated biological functions. The single-center design with the advantage of a uniform, European American group provides a reliable estimation of MAF for the genes of interest in southeastern Minnesota. These MAFs were also in Hardy-Weinberg equilibrium and consistent with allele frequencies in reports in the literature or publicly available databases. More importantly, this is the first study that addresses possible associations of ADRA2A, ADR2C, UCP- 3, and FTO polymorphisms with gastric intermediate phenotype in obesity. The availability of original MAF data for the southeastern Minnesota population is also valuable for sample size estimation in these studies. Thus, with a relatively large MAF and associations with BMI from epidemiological studies, the intermediate phenotype approach allows study of selected polymorphisms to identify those that predispose to significant and clinically meaningful alteration of gastric motor function or satiation.
In conclusion, inherited variation in genes of ADRA2A, ADR2C, UCP-3, GNB3, and FTO are associated with changes in gastric motor functions and satiation. These data support the hypothesis that intermediate phenotype markers can be used to explore genetic predisposition to decreased satiation, and if confirmed, would identify susceptibility genes to select patients for peripherally targeted weight loss therapies. These may include devices or medications that reduce gastric reservoir volume, or medications that alter gastric emptying.
Acknowledgments
Dr. Camilleri is funded in part by grants RO1-DK-67071 and K24-DK-02638 from National Institutes of Health. These studies were facilitated by the patient care and Endoscopy and Imaging Laboratory of the Mayo Clinic CTSA grant RR0024150 from National Institutes of Health. Dr. Papathanasopoulos was funded by an international grant of the Hellenic Society of Gastroenterology.
References
- 1.Ogden CL, Carroll MD, Curtin LR, McDowell MA, Tabak CJ, Flegal KM. Prevalence of overweight and obesity in the United States, 1999-2004. JAMA. 2006;295:1549–1555. doi: 10.1001/jama.295.13.1549. [DOI] [PubMed] [Google Scholar]
- 2.National Task Force on the Prevention and Treatment of Obesity. Long-term pharmacotherapy in the management of obesity. JAMA. 1996;276:1907–1915. doi: 10.1001/jama.1996.03540230057036. [DOI] [PubMed] [Google Scholar]
- 3.Blundell JE, Gillett A. Control of food intake in the obese. Obes Res. 2001;9(Suppl 4):263S–270S. doi: 10.1038/oby.2001.129. [DOI] [PubMed] [Google Scholar]
- 4.Hill AJ, Magson LD, Blundell JE. Hunger and palatability: tracking ratings of subjective experience before, during and after the consumption of preferred and less preferred food. Appetite. 1984;5:361–371. doi: 10.1016/s0195-6663(84)80008-2. [DOI] [PubMed] [Google Scholar]
- 5.Woods SC. Signals that influence food intake and body weight. Physiol Behav. 2005;86:709–716. doi: 10.1016/j.physbeh.2005.08.060. [DOI] [PubMed] [Google Scholar]
- 6.Deutsch JA. The role of the stomach in eating. Am J Clin Nutr. 1985;42:1040–1043. doi: 10.1093/ajcn/42.5.1040. [DOI] [PubMed] [Google Scholar]
- 7.Deutsch JA, Young WG, Kalogeris TJ. The stomach signals satiety. Science. 1978;201:165–167. doi: 10.1126/science.663647. [DOI] [PubMed] [Google Scholar]
- 8.Vazquez Roque MI, Camilleri M, Stephens DA, et al. Gastric sensorimotor functions and hormone profile in normal weight, overweight, and obese people. Gastroenterology. 2006;131:1717–1724. doi: 10.1053/j.gastro.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 9.Delgado-Aros S, Camilleri M, Castillo EJ, et al. Effect of gastric volume or emptying on meal-related symptoms after liquid nutrients in obesity: a pharmacologic study. Clin Gastroenterol Hepatol. 2005;3:997–1006. doi: 10.1016/s1542-3565(05)00285-5. [DOI] [PubMed] [Google Scholar]
- 10.Delgado-Aros S, Camilleri M, Cremonini F, Ferber I, Stephens D, Burton DD. Contributions of gastric volumes and gastric emptying to meal size and postmeal symptoms in functional dyspepsia. Gastroenterology. 2004;127:1685–1694. doi: 10.1053/j.gastro.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Delgado-Aros S, Cremonini F, Castillo JE, et al. Independent influences of body mass and gastric volumes on satiation in humans. Gastroenterology. 2004;126:432–440. doi: 10.1053/j.gastro.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Huang GH, Hsieh CC, Chen CH, Chen WJ. Statistical validation of endophenotypes using a surrogate endpoint analytic analogue. Genet Epidemiol. 2009 doi: 10.1002/gepi.20407. in press. [DOI] [PubMed] [Google Scholar]
- 13.Cummings DE, Overduin J. Gastrointestinal regulation of food intake. J Clin Invest. 2007;117:13–23. doi: 10.1172/JCI30227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thumshirn M, Camilleri M, Choi MG, Zinsmeister AR. Modulation of gastric sensory and motor functions by nitrergic and alpha2-adrenergic agents in humans. Gastroenterology. 1999;116:573–585. doi: 10.1016/s0016-5085(99)70179-4. [DOI] [PubMed] [Google Scholar]
- 15.Grudell AB, Camilleri M, Carlson P, et al. An exploratory study of the association of adrenergic and serotonergic genotype and gastrointestinal motor functions. Neurogastroenterol Motil. 2008;20:213–219. doi: 10.1111/j.1365-2982.2007.01026.x. [DOI] [PubMed] [Google Scholar]
- 16.Grudell AB, Sweetser S, Camilleri M, et al. A controlled pharmacogenetic trial of sibutramine on weight loss and body composition in obese or overweight adults. Gastroenterology. 2008;135:1142–1154. doi: 10.1053/j.gastro.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vazquez Roque MI, Camilleri M, Clark MM, et al. Alteration of gastric functions and candidate genes associated with weight reduction in response to sibutramine. Clin Gastroenterol Hepatol. 2007;5:829–837. doi: 10.1016/j.cgh.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 18.Wardle J, Carnell S, Haworth CM, Farooqi IS, O’Rahilly S, Plomin R. Obesity associated genetic variation in FTO is associated with diminished satiety. J Clin Endocrinol Metab. 2008;93:3640–3643. doi: 10.1210/jc.2008-0472. [DOI] [PubMed] [Google Scholar]
- 19.Cremonini F, Mullan BP, Camilleri M, Burton DD, Rank MR. Performance characteristics of scintigraphic transit measurements for studies of experimental therapies. Aliment Pharmacol Ther. 2002;16:1781–1790. doi: 10.1046/j.1365-2036.2002.01344.x. [DOI] [PubMed] [Google Scholar]
- 20.Bouras EP, Delgado-Aros S, Camilleri M, et al. SPECT imaging of the stomach: comparison with barostat, and effects of sex, age, body mass index, and fundoplication. Single photon emission computed tomography. Gut. 2002;51:781–786. doi: 10.1136/gut.51.6.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Schepper H, Camilleri M, Cremonini F, Foxx-Orenstein A, Burton D. Comparison of gastric volumes in response to isocaloric liquid and mixed meals in humans. Neurogastroenterol Motil. 2004;16:567–573. doi: 10.1111/j.1365-2982.2004.00533.x. [DOI] [PubMed] [Google Scholar]
- 22.Delgado-Aros SBD, Brinkmann BH, Camilleri M. Reliability of a semi automated analysis to measure gastric accommodation using SPECT in humans. Gastroenterology. 2001;120:A287. abstract. [Google Scholar]
- 23.Chial HJ, Camilleri C, Delgado-Aros S, et al. A nutrient drink test to assess maximum tolerated volume and postprandial symptoms: effects of gender, body mass index and age in health. Neurogastroenterol Motil. 2002;14:249–253. doi: 10.1046/j.1365-2982.2002.00326.x. [DOI] [PubMed] [Google Scholar]
- 24.Hofker M, Wijmenga C. A supersized list of obesity genes. Nat Genet. 2009;41:139–140. doi: 10.1038/ng0209-139. [DOI] [PubMed] [Google Scholar]
- 25.Evans BA, Papaioannou M, Bonazzi VR, Summers RJ. Expression of beta 3-adrenoceptor mRNA in rat tissues. Br J Pharmacol. 1996;117:210–216. doi: 10.1111/j.1476-5381.1996.tb15176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurokawa N, Young EH, Oka Y, et al. The ADRB3 Trp64Arg variant and BMI: a meta-analysis of 44 833 individuals. Int J Obes (Lond) 2008;32:1240–1249. doi: 10.1038/ijo.2008.90. [DOI] [PubMed] [Google Scholar]
- 27.Meyre D, Delplanque J, Chevre JC, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 28.Wardle J, Llewellyn C, Sanderson S, Plomin R. The FTO gene and measured food intake in children. Int J Obes (Lond) 2009;33:42–45. doi: 10.1038/ijo.2008.174. [DOI] [PubMed] [Google Scholar]
- 29.Fleury C, Neverova M, Collins S, et al. Uncoupling protein-2: a novel gene linked to obesity and hyperinsulinemia. Nature Genet. 1997;15:269–272. doi: 10.1038/ng0397-269. [DOI] [PubMed] [Google Scholar]
- 30.Evans D, Minouchehr S, Hagemann G, et al. Frequency of and interaction between polymorphisms in the beta3-adrenergic receptor and in uncoupling proteins 1 and 2 and obesity in Germans. Int J Obes Relat Metab Disord. 2000;24:1239–1245. doi: 10.1038/sj.ijo.0801402. [DOI] [PubMed] [Google Scholar]
- 31.Jun HS, Kim IK, Lee HJ, et al. Effects of UCP2 and UCP3 variants on the manifestation of overweight in Korean children. Obesity. 2009;17:355–362. doi: 10.1038/oby.2008.531. [DOI] [PubMed] [Google Scholar]
- 32.van Abeelen AF, de Krom M, Hendriks J, Grobbee DE, Adan RA, van der Schouw YT. Variations in the uncoupling protein-3 gene are associated with specific obesity phenotypes. Eur J Endocrinol. 2008;158:669–676. doi: 10.1530/EJE-07-0834. [DOI] [PubMed] [Google Scholar]
- 33.Tack J, Depoortere I, Bisschops R, et al. Influence of ghrelin on interdigestive gastrointestinal motility in humans. Gut. 2006;55:327–333. doi: 10.1136/gut.2004.060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 35.Walder K, Norman RA, Hanson RL, et al. Association between uncoupling protein polymorphisms (UCP2-UCP3) and energy metabolism/obesity in Pima indians. Hum Mol Genet. 1998;7:1431–1435. doi: 10.1093/hmg/7.9.1431. [DOI] [PubMed] [Google Scholar]
- 36.Lima JJ, Feng H, Duckworth L, et al. Association analyses of adrenergic receptor polymorphisms with obesity and metabolic alterations. Metabolism. 2007;56:757–765. doi: 10.1016/j.metabol.2007.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunt SC, Stone S, Xin Y, et al. Association of the FTO gene with BMI. Obesity. 2008;16:902–904. doi: 10.1038/oby.2007.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cecil JE, Tavendale R, Watt P, Hetherington MM, Palmer CN. An obesity-associated FTO gene variant and increased energy intake in children. N Engl J Med. 2008;359:2558–2566. doi: 10.1056/NEJMoa0803839. [DOI] [PubMed] [Google Scholar]
- 39.Siffert W, Forster P, Jockel KH, et al. Worldwide ethnic distribution of the G protein beta3 subunit 825T allele and its association with obesity in Caucasian, Chinese, and Black African individuals. J Am Soc Nephrol. 1999;10:1921–1930. doi: 10.1681/ASN.V1091921. [DOI] [PubMed] [Google Scholar]
- 40.Sookoian S, Gemma C, Garcia SI, et al. Short allele of serotonin transporter gene promoter is a risk factor for obesity in adolescents. Obesity. 2007;15:271–276. doi: 10.1038/oby.2007.519. [DOI] [PubMed] [Google Scholar]
- 41.Sookoian S, Gianotti TF, Gemma C, Burgueno A, Pirola CJ. Contribution of the functional 5-HTTLPR variant of the SLC6A4 gene to obesity risk in male adults. Obesity (Silver Spring) 2008;16:488–91. doi: 10.1038/oby.2007.64. [DOI] [PubMed] [Google Scholar]
- 42.Fuemmeler BF, Agurs-Collins TD, McClernon FJ, et al. Genes implicated in serotonergic and dopaminergic functioning predict BMI categories. Obesity (Silver Spring) 2008;16:348–55. doi: 10.1038/oby.2007.65. [DOI] [PMC free article] [PubMed] [Google Scholar]


