Abstract
Recent studies have highlighted the importance of the lysosome in degrading proteins that misfold in neurodegenerative diseases. In this study we explore the role for autophagy in the clearance of an N-terminal caspase-7-generated fragment of ataxin-7, a protein with a pathogenic polyglutamine (polyQ) expansion in the neurodegenerative disease spinocerebellar ataxia 7 (SCA7). Using both cellular and transgenic mouse models of SCA7 we show that the stability of wild-type ataxin-7 is modified by macroautophagy, but not by proteasomal, inhibition, whereas both autophagy and proteasomal degradation have little effect on polyQ-expanded ataxin-7. We also create a post-translational modification-deficient ataxin-7 mutant that has increased protein turnover of both wild-type and polyQ-expanded ataxin-7, mediated through the autophagy pathway. Histological analysis reveals that wild-type ataxin-7 colocalizes with markers of chaperone-mediated autophagy (CMA) and macroautophagy, indicating that both of these mechanisms may play a role in the clearance of ataxin-7. Furthermore, there is an increase in LC3, a marker of autophagy initiation, in the cerebellum of SCA7 transgenic mice. Our findings indicate that the ataxin-7 fragment may be cleared via autophagy and that this process is altered in SCA7. Identification of the different types of autophagy involved in ataxin-7 turnover and the influence of post-translational modifications on these processes will be pursued in future studies.
Keywords: SCA7, caspase-7, autophagy, HDAC, chaperone-mediated autophagy
Autophagy is a lysosomal enzymatic mechanism for degrading long-lived cytosolic proteins, and is constitutively active at low levels in mammalian cells. There are three types of autophagy that the cell employs for degradation: microautophagy, macroautophagy and CMA. In this study we were interested in both macroautophagy, where cytosolic proteins are engulfed and undergo degradation by proteases in the lysosomal lumen, and CMA, which targets specific proteins containing a consensus peptide motif. This motif facilitates interaction with a CMA chaperone protein that transports the substrate to the lysosomal membrane where it interacts with LAMP-2A and various members of a CMA complex, and is translocated into the lysosomal lumen for degradation.
SCA7 is one of nine trinucleotide repeat disorders, with a CAG repeat in the SCA7 gene resulting in an unstable polyQ repeat in the N terminus of the ataxin-7 protein. PolyQ-expansion in ataxin-7 leads to cytotoxicity of distinct neuronal sub-populations (e.g., Purkinje cells in the cerebellum and photoreceptors in the retina). Previous studies in our lab and by others have indicated that it is a caspase-7-cleaved N-terminal fragment of ataxin-7 that accumulates in neuronal inclusions and causes toxicity and neurodegeneration.
A strong link has recently been formed between lysosomal degradation and neurodegeneration, with mice deficient in autophagy resulting in neuronal aggregation of proteins and a neurodegenerative phenotype. Interestingly, mutant huntingtin and other polyQ-expanded proteins can be degraded via macroautophagy in cells, flies and mice, whereas α-Synuclein can be cleared via both macroautophagy and CMA in Parkinson disease models. Although deficiencies in autophagy are linked to neurodegeneration, few studies have looked at lysosomal degradation in the spinocerebellar ataxias. In this study we evaluate the effect of proteasomal and lysosomal clearance mechanisms on the stability of the N-terminal fragment of ataxin-7 and its toxic polyQ-expanded form.
To identify which degradation pathways mediate ataxin-7 protein turnover we treated cells expressing the human ataxin-7 caspase-7 fragment with inhibitors of proteasomal degradation (epoxomycin) and macroautophagy (3-MA). While there were no changes in ataxin-7 fragment turnover with proteasomal inhibition, we saw an increase in ataxin-7 stability with 3-MA treatment. This effect was not seen with the polyQ-expanded ataxin-7 fragment. We also examined the effect of ataxin-7 mutants that are deficient in post-translational modifications, K223R and K257R, that occur close to the caspase-7 cleavage site, as these show enhanced protein turnover compared to the wild-type ataxin-7. Inhibition of macroautophagy, but not proteasomal degradation, increases the stability of the K223R and K257R mutants in both the wild-type and polyQ-expanded proteins. These studies indicate that the N-terminal ataxin-7 fragment is cleared via macroautophagy and that post-translational modifications at K257 and K223 may inhibit this process (see Fig. 1). These findings are consistent with caspase cleavage products being differentially regulated by autophagy, with broader implications in the regulation of cell death pathways. It is possible that caspase cleavage products are not indiscriminately degraded, but in some cases the process is regulated by post-translational modification.
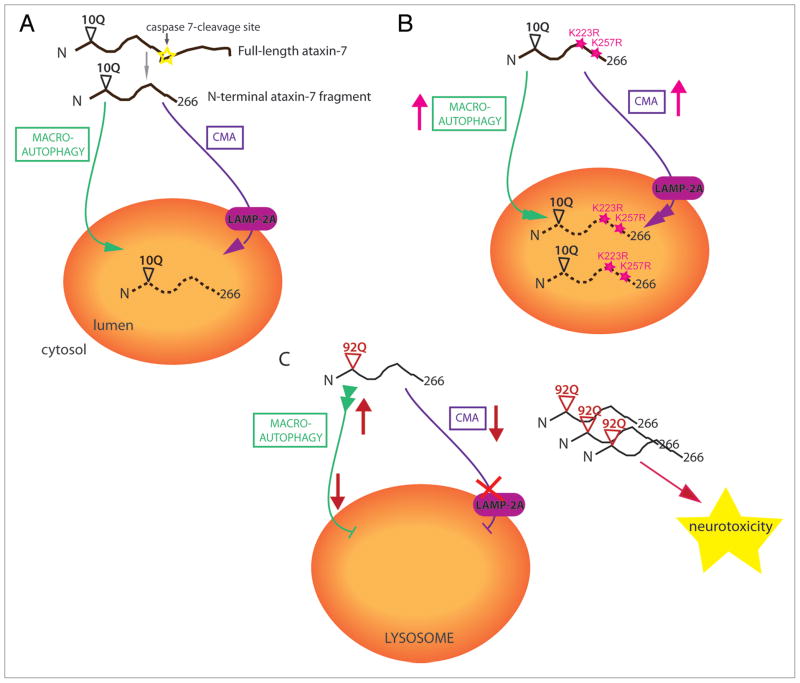
Figure 1.
Proposed model for the clearance of wild-type and polyQ-expanded N-terminal ataxin-7 (1-266) fragment. (A) Following caspase-cleavage, wild-type ataxin-7-10Q (1-266) is cleared by both macroautophagy, where the cytosolic protein is engulfed by the lysosome; and chaperone-mediated autophagy (CMA), which is mediated through the lysosome-associated membrane protein 2A (LAMP-2A). (B) Ataxin-7 post-translational-deficient mutants, K223R and K257R, have increased turnover via both macroautophagy and CMA, indicating that post-translational modification of ataxin-7 normally inhibits lysosomal degradation. (C) We speculate that polyQ-expanded ataxin-7-92Q (1-266) has increased macroautophagy initiation to compensate for an inability to translocate through LAMP-2A into the lysosomal lumen. This enhancement is not sufficient to alter the accumulation of the toxic polyQ-expanded ataxin-7 fragments that also block the CMA pathway for other substrates, resulting in neurodegeneration.
In support of a role for autophagy in ataxin-7 clearance, we find that ataxin-7 colocalizes with LC3, a marker of macroautophagy initiation, in mouse Purkinje cells. In a SCA7 transgenic mouse model we identify a significant increase in LC3 expression per cell, consistent with altered macroautophagy in SCA7. Furthermore, in a cellular model, we detect colocalization between ataxin-7 and LAMP-2A, the lysosome-associated membrane protein that governs the rate-limiting step in CMA. LAMP-2A expression and colocalization with ataxin-7 is dramatically increased in the K257R mutant, indicating that increased turnover of this mutant may occur via both macroautophagy and CMA, processes that these studies indicate are normally inhibited by post-translational modification at this site (see Fig. 1).
Our results demonstrate that ataxin-7 may by degraded by both macroautophagy and CMA and that in SCA7 there is an increase in macroautophagy initiation. We propose that this could be due to a compensatory response to a deficit in CMA, in a mechanism similar to that seen in Parkinson disease models. Researchers show that wild-type α-synuclein can be cleared by both macroautophagy and CMA, whereas the familial Parkinson disease mutant α-synuclein is not cleared by CMA. Not only does this latter form decrease the clearance of mutant α-synuclein, but it has a dominant-negative effect, blocking CMA activity for other substrates and increasing neurotoxicity. Our lab is currently looking at whether ataxin-7 is a substrate for the specific CMA lysosomal degradation pathway, and indeed whether this pathway may be blocked in SCA7 models. This study also shows that acetylation status of ataxin-7 effects its stability. Further studies of post-translational modification effects on autophagy-mediated ataxin-7 turnover are underway, which could prove an important therapeutic target for modifying levels of the polyQ-expanded ataxin-7 fragment in SCA7. Our hypothesis is that impaired turnover of ataxin-7 fragments in SCA7 may be due, in part, to a breakdown of normal lysosomal degradation pathways in this disease.
Acknowledgments
This work was supported by the NIH NS40251 (L.M.E.) and NIH NS40251 (L.M.E.) NIH. C.D. was supported by NIH training grant T32 AG000266.