Abstract
By treating the transcript abundance as a quantitative trait, gene expression can be mapped to local or distant genomic regions relative to the gene encoding the transcript. Local expression quantitative trait loci (eQTL) generally act in cis (that is, control the expression of only the contiguous structural gene), whereas distal eQTL act in trans. Distal eQTL are more difficult to identify with certainty due to the fact that significant thresholds are very high since all regions of the genome must be tested, and confounding factors such as batch effects can produce false positives. Here, we compare findings from two large genetic crosses between mouse strains C3H/HeJ and C57BL/6J to evaluate the reliability of distal eQTL detection, including “hotspots” influencing the expression of multiple genes in trans. We found that >63% of local eQTL and >18% of distal eQTL were replicable at a threshold of LOD > 4.3 between crosses and 76% of local and >24% of distal eQTL at a threshold of LOD > 6. Additionally, at LOD > 4.3 four tissues studied (adipose, brain, liver, and muscle) exhibited >50% preservation of local eQTL and >17% preservation of distal eQTL. We observed replicated distal eQTL hotspots between the crosses on chromosomes 9 and 17. Finally, >69% of local eQTL and >10% of distal eQTL were preserved in most tissues between sexes. We conclude that most local eQTL are highly replicable between mouse crosses, tissues, and sex as compared to distal eQTL, which exhibited modest replicability.
GENETIC variations in gene expression contribute to the molecular architecture underlying complex diseases such as atherosclerosis, diabetes, and cancer (Lusis et al. 2008). Detection of gene expression in segregating populations has led to the identification of expression quantitative trait loci (eQTL) (Damerval et al. 1994; de Vienne et al. 1994; Machleder et al. 1997). Global mapping of gene expression provides a useful form of functional variation for positional cloning and can facilitate the modeling of causal interactions and biologic networks (Schadt et al. 2005; Ghazalpour et al. 2006; Rockman and Kruglyak 2006; Cookson et al. 2009; Montgomery and Dermitzakis 2009).
Distinguishing between local (sometimes termed “cis”) and distal (sometimes termed “trans”) eQTL enhances our understanding of the candidate genes underlying the disease phenotype. Due to their close proximity, genetic variants underlying local eQTL most likely affect molecular mechanisms involved in transcription, splicing, or mRNA decay, and most act in cis (Doss et al. 2005). Distal eQTL, on the other hand, generally consist of variants that act in trans. One variety of trans-eQTL, termed “hotspots,” regulates the levels of many different transcripts. For example, one mouse study identified genetic variants influencing the transcription factor RUNX1 affecting the gene expression of a group of distal genes involved in the RUNX1 pathway (Bystrykh et al. 2005). Distal eQTL are of particular interest to complex trait studies since they may perturb entire pathways and mediate complex gene–gene and gene–environment interactions. An important example of a trans-effect relevant to complex disease is sex (van Nas et al. 2009), as QTL studies of males and females have detected striking sex–gene interactions (Wang et al. 2006; Lusis et al. 2008).
In this study, we evaluated the reproducibility of eQTL detection of two large genetic crosses of mouse strains C3H/HeJ and C57BL/6J. We chose these genetic crosses, which had been developed for other studies, because they were quite similar in overall design as well as genetic backgrounds. We evaluated transcript levels in adipose, brain, liver, and muscle of both sexes. The results have both practical and basic importance. A complication with human studies is the difficulty of procuring samples from tissues of interest. Thus an important question is whether accessible tissues such as blood cells act as a proxy for less accessible tissues. A recent human study of several tissues concluded that conservation of eQTL between tissues was low (Dimas et al. 2009). Our results are in agreement for distal eQTL, but they show high conservation of local eQTL. Our results also provide data relevant to basic questions such as conservation of biologic networks between tissue, the global nature of genetic regulation, and the basis of sex differences.
MATERIALS AND METHODS
Mouse inbred strains C57BL/6J and C3H/HeJ F2 reciprocal intercross:
A murine F2 population consisting of ∼300 mice was generated by intercrossing F1 mice of parental strains C57BL/6J (B6) and C3H/HeJ (C3H). Mice were fed a chow diet containing 4% fat until 8 weeks of age and then were placed on a “Western” diet containing 42% fat and 0.15% cholesterol for 12 weeks. After a 12-hr fast, the mice were anesthetized by isoflurane for retroorbital sinus blood collection and subsequently killed at 20 weeks of age for tissue collection of adipose, whole brain, liver, and muscle. The mice were genotyped at ∼1500 single-nucleotide polymorphisms (SNPs) uniformly distributed over the mouse genome at an average density of 1.5 cM (Table 1 and File S1).
TABLE 1.
Comparison of experimental design between the crosses
| Experimental design | Cross I | Cross II | |
|---|---|---|---|
| Number of mice | |||
| Tissue | Sex | ||
| Adipose | Female | 147 | 136 |
| Male | 143 | 133 | |
| Brain | Female | 133 | 140 |
| Male | 118 | 149 | |
| Liver | Female | 149 | 141 |
| Male | 142 | 156 | |
| Muscle | Female | 158 | 123 |
| Male | 139 | 148 | |
| Diets | |||
| Purina Chow containing 4% fat (Ralston Purina, St. Louis) | 8 wk | 8 wk | |
| Western diet 88137 containing 42% fat and 0.15% cholesterol (Harlan, Teklad, Madison, WI) | 16 wk | 12 wk | |
| Duration of fasting | 4 hr | 12 hr | |
| Age of mice when killed | 24 wk | 20 wk | |
| Genotyping platform/density of SNPs | ParAllele 1100 SNP array ± 300 SNPs | ParAllele 2000 SNP array (n = 150); Illumina 1536 SNP array (n = 150) | |
| Number of SNPs in study | 1,326 | 1,326 | |
| Expression array platforms | Agilent Mouse TOE75K Array1 | Agilent Mouse TOE75K Array1 | |
| Number of transcripts | |||
| Adipose | 23,568 | 23,560 | |
| Brain | 23,568 | 23,563 | |
| Liver | 23,568 | 23,558 | |
| Muscle | 23,458 | 23,458 | |
Mouse inbred strains C57BL/6J ApoE−/− and C3H/HeJ ApoE−/− F2 intercross data set:
The genotype and gene expression data from adipose, brain, liver, and muscle of the F2 mouse population used in this study have been previously described (Wang et al. 2006). The C57BL/6J ApoE−/− (B6.ApoE−/−) parental strains were purchased from Jackson Laboratory (Bar Harbor, ME) and the C3H/HeJ ApoE−/− (C3H.ApoE−/−) parental strains were generated by backcrossing B6.ApoE−/− to C3H for 10 generations. The C3H.ApoE−/− parental strains were genotyped using ∼1300 markers throughout the genome and there was no evidence of contaminating regions. Since the B6.ApoE−/− parental strains were purchased from Jackson labs (JAX), no additional genotyping was performed to confirm the purity of the background strains. The region flanking the ApoE gene is derived from strain 129 in both parents. We previously showed that in ApoE−/− from JAX the region extended ∼5–10 Mb proximal and 15–20 Mb distal to the ApoE gene (Lusis et al. 2007). This region did not contain any significant or suggestive (LOD > 2.7) eQTL in the wild-type cross.
The F2 mouse population consisting of 334 mice (169 female, 165 male) was generated by intercrossing F1 mice of parental strains B6.apoE−/− and C3H.apoE−/−. Mice were fed chow diet containing 4% fat until 8 weeks of age and then were placed on a Western diet containing 42% fat and 0.15% cholesterol for 16 weeks. After a 4-hr fast, the mice were anesthetized by isoflurane for retroorbital sinus blood collection and subsequently killed at 24 weeks of age for tissue collection of adipose, whole brain, liver, and muscle. The mice were genotyped at ∼1300 SNPs uniformly distributed over the mouse genome at an average density of 1.5 cM (Table 1 and File S2).
Gene expression analysis:
The RNA and microarray processing, as previously described (He et al. 2003), was performed at Rosetta Inpharmatics (Seattle, WA). The custom ink-jet microarrays (Agilent Technologies, Palo Alto, CA) contain 2186 control probes and 23,574 noncontrol oligonucleotides extracted from mouse Unigene clusters and combined with RefSeq sequences and RIKEN full-length clones. Total RNA was extracted from homogenized mouse tissues with Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Labeled cDNA with either Cy3 or Cy5 was hybridized to at least two microarray slides with fluor reversal and subsequently scanned using a laser confocal scanner. Gene expression changes between two samples were quantified on the basis of spot intensity relative to background, adjusted for experimental variation between arrays using average intensity over multiple channels, and fit to an error model to determine significance (type I error). Gene expression is reported as the ratio of the mean log10 intensity (ml ratio) relative to the pool derived from 150 mice randomly selected from the F2 population. Gene expression data from mice with complete genotype data and at least 95% complete phenotype and array data were used. The microarray data from this study have been deposited in the Gene Expression Omnibus (GEO) under accession nos. GSE2814, GSE3086, GSE3087, GSE3088, GSE11065, GSE11338, GSE12795, and GSE12798.
eQTL replication analysis:
The eQTL within a cross (sex as an interactive covariate) and in female and male mice were calculated utilizing the scanone function in RQTL. One thousand permutations of n = 500 randomly chosen gene transcripts resulted in a significance threshold of LOD > 6. Genes with eQTL of LOD > 2.7 were selected for replication comparison between the crosses, tissues, and sexes. An eQTL detected within 10 Mb from the nearest marker of the gene was identified as a local eQTL, whereas an eQTL outside of 10 Mb from the nearest marker of the gene was defined as a distal eQTL. The eQTL with the highest LOD score per chromosome was chosen for the analysis. For each comparison between crosses, tissues, and sex, the degree of overlap was calculated by ascertaining if the eQTL detected in one group at LOD > 4.3 or LOD > 6 can be detected at LOD > 2.7 in the other group. Specifically, the percentage of overlap was calculated as the number of genes with eQTL of LOD > 4.3 or LOD > 6 of one group that intersect with eQTL at LOD >2.7 of the comparison group over the total number of genes with LOD > 4.3 or LOD > 6 of the first group. The groups that were compared for their eQTL were as follows: I vs. II; adipose vs. brain, adipose vs. liver, adipose vs. muscle, brain vs. liver, brain vs. muscle, and liver vs. muscle; and male vs. female.
The reasons we chose to present our replication results using a less stringent threshold are the following:
Replicating a specific genetic linkage or association requires much more power than finding any significant result in the first place when there are multiple genes that affect trait values. Brian Suarez and colleagues (Suarez et al. 1994) nicely demonstrate this property and others have also demonstrated this same principle. Suarez and colleagues argue that in the initial study, a successful result is the detection of any one (or a few) of a number contributing genes but in the replication a successful result is the much harder problem of finding this (these few) same gene(s) again. A related problem is that the significance of the results in the first cross are actually inflated (the winner's curse problem) and so are more difficult to replicate. Thus we use a lower threshold for the second, replication cross.
The reason for using such a high threshold for the first cross is that we are conducting thousands of tests and need to adjust our significance level accordingly to avoid a severe multiple-testing problem. In the replication, however, we are effectively looking at the limited number of regions that were near significant findings from the first cross; thus the number tests is reduced and we can use a less severe adjustment for multiple testing.
We defined an eQTL hotspot by the highest relative frequency of eQTL at LOD ≥ 5.7. Specifically, we searched for loci with eQTL greater than or equal to three standard deviations above the average frequency of distal eQTL in one tissue. The following thresholds were used to detect hotspot loci: adipose ≥10, brain ≥20, liver ≥13, and muscle ≥8.
RESULTS
To investigate the extent of eQTL preservation, we determined the degree of overlap between two mouse crosses (cross I and cross II) of inbred strains C57BL/6J and C3H/HeJ. Integrating the genotype and gene expression trait data, we utilized the linkage software RQTL cartographer and calculated the eQTL in the adipose, brain, liver, and skeletal muscle tissue of the crosses. We used LOD > 4.3 as a threshold of significance, LOD > 2.7 as a suggestive threshold, and LOD > 6 as a high threshold of significance. We chose significance thresholds at a suggestive, a significant, and a high LOD score to capture the commonly used thresholds to determine if these lead to reasonable replication in eQTL studies. The thresholds LOD > 4.3 and 2.7 were chosen on the basis of a previous study by Lander and Kruglyak, who reported the number of false positives detected in a QTL at various significance thresholds (Lander and Kruglyak 1995). Specifically, a LOD > 4.3 was defined as the baseline of significance, which is approximately equivalent to a P-value <10−5, whereas LOD > 2.7 is equivalent to a P-value <10−3, reflecting a more suggestive threshold. The threshold LOD > 6 was chosen on the basis of the results of permutation (n = 1000) we performed in this analysis on n = 500 randomly chosen transcripts.
For each eQTL at the various LOD score thresholds, we determined if it was detected in both crosses. The eQTL in one cross was considered replicated if it was detected within 10 Mb of an eQTL in the other comparison cross of the same tissue. In this study, local eQTL were defined as loci containing the gene of the transcript abundance that maps within 10 Mb of the nearest QTL. To determine the degree of local and distal eQTL that replicated, we calculated for each LOD score threshold the percentage of eQTL that overlap between the crosses, tissues, and sexes. The eQTL hotspots were determined by the enrichment of gene expression traits that mapped to the same loci.
Replication of local eQTL between crosses:
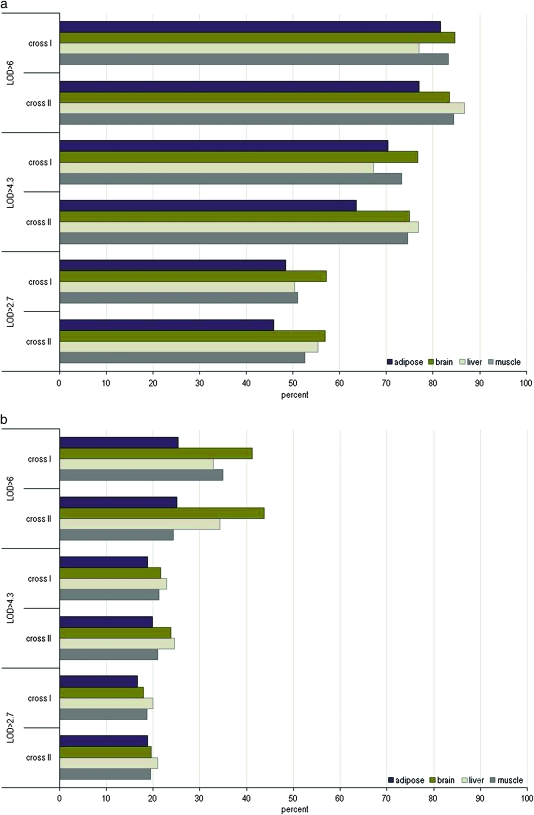
To investigate the degree of local eQTL replication in the mouse crosses, we determined what percent of the local eQTL at LOD > 4.3 or LOD > 6 in one cross could be detected in the other cross at LOD > 2.7. We compared the crosses in this way in both directions, and the results were highly consistent. At LOD > 4.3, our results showed >60% of the local eQTL replicated (63–76%) across all the tissues of the mouse crosses I and II. At LOD > 6, >76% of the local eQTL between crosses I and II replicated at this LOD score threshold, reaching as high as 80% replication (83–86%) in brain, liver, and muscle tissue (Figure 1 and supporting information, Table S1).
Figure 1.—
Replication of local (a) and distal (b) eQTL between crosses. The percentage of eQTL that replicated between crosses at various LOD thresholds is depicted in adipose, brain, liver, and muscle tissue. The eQTL at LOD > 2.7, 4.3, and 6 of the mouse crosses were screened for detection at LOD > 2.7 in their respective comparison crosses. The percentage indicates the degree of replication of eQTL at LOD > 4.3 in cross I that overlap in cross II at LOD > 2.7 over the total number of eQTL at LOD > 4.3 detected in cross I and vice versa. For example, at LOD > 6, >80% of the local eQTL in cross I were preserved in cross II of the adipose tissue.
Replication of distal eQTL between crosses:
To determine the degree of replication of distal eQTL, we ascertained the number of distal eQTL at LOD > 4.3 or LOD > 6 in one cross that could be detected at LOD > 2.7 in the other cross. At LOD > 4.3, we detected >18% replication of distal eQTL over all tissues between crosses I and II, with >21% in brain, liver, and muscle. At LOD > 6 the degree of replication among distal eQTL was greater. Over 25% of the distal eQTL replicated across tissues, reaching >30% in liver and >40% in brain (Figure 1 and Table S1).
Preservation of local eQTL across tissues:
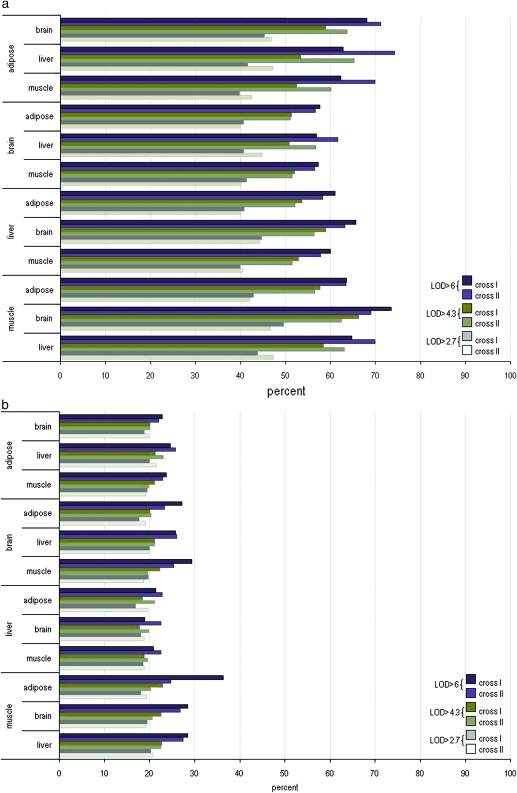
To explore the degree of replication of local eQTL across tissues, we used the same screening strategy as above. At LOD > 4.3, ∼50% of the local eQTL were preserved in tissues. The muscle tissue exhibited the highest degree of preservation (56–66%) compared to the other tissues (adipose, 52–65%; liver, 51–58%) and the brain the lowest degree of preservation (50–56%). At LOD > 6, we observed a similar pattern of preservation, >56% of local eQTL muscle (63–73%), adipose (62–74%), liver (59–65%), and brain tissues (56–61%) (Figure 2 and Table S2).
Figure 2.—
Preservation of local (a) and distal (b) eQTL across tissues. The percentage of eQTL that were preserved across tissues at various LOD thresholds is depicted in adipose, brain, liver, and muscle tissue. The eQTL at LOD > 2.7, 4.3, and 6 of a tissue were screened for detection at LOD > 2.7 in their respective comparison tissues (sex and mouse crosses were combined). The percentage indicates the degree of replication of eQTL at LOD > 4.3 in one tissue that overlap in its comparison tissue at LOD > 2.7 over the total number of eQTL at LOD > 4.3 detected in the first tissue and vice versa. For example, for the local eQTL in cross I, ∼70% of the local eQTL found in adipose at LOD > 6 were detected in brain at LOD > 2.7. The bar graph reflects the eQTL at LOD > 4.3 of the adipose tissue as compared to the eQTL at LOD > 2.7 of the brain, liver, and muscle (brain compared to adipose, liver, and muscle; liver compared to adipose, brain, and muscle; and muscle compared to adipose, brain, and liver).
Preservation of distal eQTL across tissues:
To investigate the degree of distal eQTL replication across tissues, we screened each distal eQTL at LOD > 4.3 and LOD > 6 for detection in the comparison tissue at LOD > 2.7. At LOD > 4.3, ∼18% of the distal eQTL were detected across tissues. Muscle showed the highest preservation (20–22%) compared to the other tissues (adipose, 19–23%; brain, 19–22%; and liver, 18–21%). At LOD > 6 the preservation of distal eQTL was adipose, 23%; brain, 26%; liver, 21%; and muscle, 28% (Figure 2 and Table S2).
Preservation of local eQTL between female and male mice:
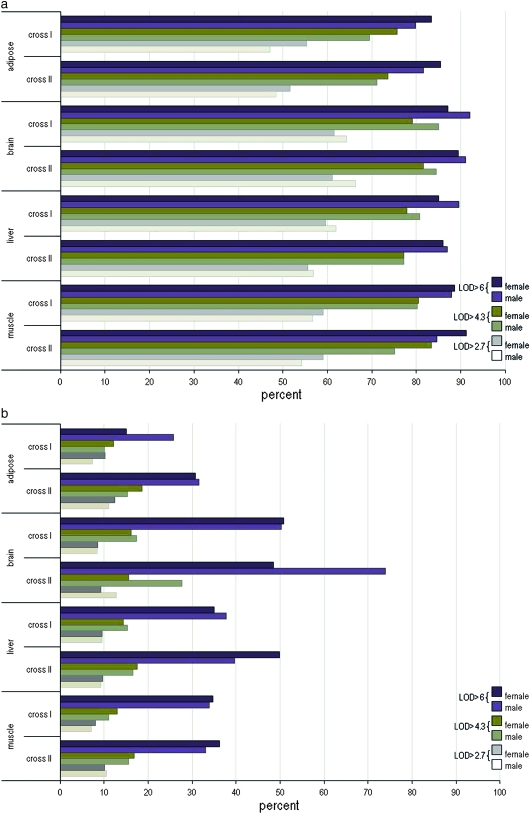
To investigate how sex affects the degree of preservation of eQTL, we determined if eQTL at LOD > 4.3 and LOD > 6 in female mice were detected at LOD > 2.7 in male mice and vice versa. At LOD > 4.3, ∼70% of the local eQTL were preserved between female and male mice: adipose, 69–75%; brain tissue, 79–85%; liver, 77–80%; and muscle, 75–83%. At LOD > 6, >79% of local eQTL (79–92%) were preserved between male and female mice in all tissues: adipose, 79–85%; liver and muscle tissue, 84–91%; and brain, 87–92% (Figure 3 and Table S3).
Figure 3.—
Preservation of local (a) and distal (b) eQTL between females and males. The percentage of eQTL that replicated between sexes at various LOD thresholds is depicted in adipose, brain, liver, and muscle tissue. The eQTL at LOD > 2.7, 4.3, and 6 of one sex were screened for detection at LOD > 2.7 in the opposite sex. The percentage indicates the degree of replication of eQTL at LOD > 4.3 in females that overlap in males at LOD > 2.7 over the total number of eQTL at LOD > 4.3 detected in females and vice versa. For example, at LOD > 6, female adipose in cross I shows >80% of the local eQTL preserved in the male adipose cross I group.
Preservation of distal eQTL between female and male mice:
To investigate the degree of preservation of distal eQTL between females and males, we determined whether each distal eQTL at LOD > 4.3 and LOD > 6 in one sex was detected in the other sex at LOD > 2.7. At LOD > 4.3, >10% of distal eQTL were preserved across tissues between females and males: adipose, 10–18%; brain, 15–27%; liver, 14–17%; and muscle, 10–16%. At LOD > 6, >25% of the distal eQTL were preserved across tissues between females and males, with the exception of female adipose tissue in cross I: adipose, 15–30%; brain, 48–73%; liver, 34–49%; and muscle, 33–36% (Figure 3 and Table S3).
Replication of eQTL hotspots:
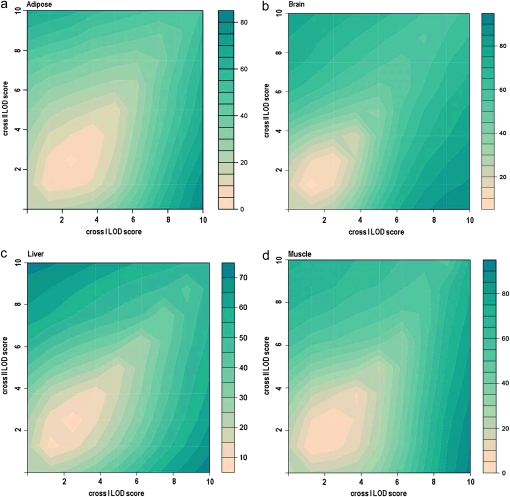
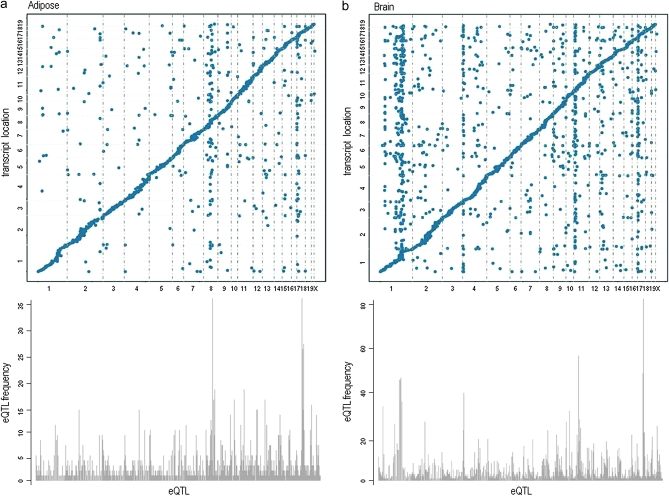
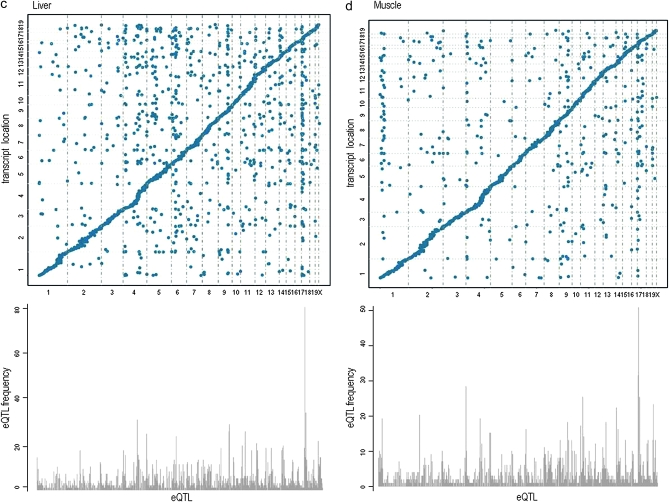
To investigate if the distal eQTL hotspots replicate between the crosses, we determined which chromosomes had the highest relative frequency of distal eQTL at LOD > 6. The brain consisted of the most pronounced replicated distal eQTL hotspots on chromosomes 1, 4, 11, and 17. In the liver tissue replicated hotspots were detected on chromosomes 4, 6, 9, and 17. In muscle, replicated hotspots were found on chromosomes 1, 4, 9, and 17. And in adipose, distal eQTL hotspots replicated on chromosomes 8, 9, and 17. Interestingly, the same hotspot on chromosome 9 (105.18–106.57 Mb) was observed in all tissues, although in varying degrees. We also detected the same hotspot across all tissues on chromosome 17 (24.53 Mb) with the exception of adipose tissue (11.64 Mb). These results show that tissue-specific distal eQTL hotspots as well as those across tissues are replicable in mouse crosses. (Figures 4 and 5).
Figure 4.—
Replication of distal eQTL between crosses at different significance thresholds. The percentage of eQTL that replicated between crosses at various LOD thresholds is depicted in (a) adipose, (b) brain, (c) liver, and (d) muscle tissue. The eQTL at LOD > 2.7–10 of the mouse crosses were screened for detection at LOD > 2.7–10 in their respective comparison crosses. The heat map reflects the degree of replication as the percentage of eQTL at various LOD thresholds in cross I that overlap in cross II at various LOD thresholds and vice versa. For example, in adipose tissue (a), ∼80% of eQTL at LOD > 2.7 in cross II were detected in cross I (bottom right corner).
Figure 5.—
Replication of eQTL hotspots between crosses. The chromosomes were screened for the highest relative frequency of distal eQTL at LOD > 5.7 of the crosses (cross I vs. cross II). The detection of loci with eQTL greater than or equal to three standard deviations above the average frequency of distal eQTL in one tissue alluded to the presence of an eQTL hotspot. (a) Adipose; (b) brain; (c) liver; (d) muscle.
We also determined if the transcripts mapping to these hotspots were enriched for a biological functional category. For hotspots observed across tissues on chromosome 17 (11.64–24.53 Mb), transcripts were modestly enriched for “integral to plasma membrane” (p = 0.021) as well as “non-membrane-bounded organelle” with PDZ domains (common structural domain that mediates protein–protein interactions for signaling complexes) found in diverse signaling proteins (p = 0.025). For hotspots found in only one tissue, transcripts were enriched for tissue-specific functions. For example, at chromosome 11 (36.82 Mb) in brain, two of the transcripts were shown involved in the regulation of “glial cell development” (p = 0.024); in liver at chromosome 6 (94.46 Mb) two genes were found in the pathway for “glucose homeostasis” (p = 0.037) and in adipose at chromosome 8 (84.79–95.42 Mb), seven genes were involved in pathways for “lipid biosynthetic process” (p = 0.0039) and “palmitate” (p = 0.021) (Figure S1 and Table S4).
Additionally, we ascertained if these loci harbored genes responsible for regulating a high frequency of transcripts such as transcription factors. We identified several chromosomes with transcription factors at these hotspots, including chromosome 1 (174.91–178.85 Mb), Usf1, Upstream transcription factor 1 (173.34 Mb) and Atf6, Activating transcription factor 6 (172.62 Mb); chromosome 4 (132.40 Mb), RUNX3, Runt related transcription factor 3 (134.67 Mb); and chromosome 17 (24.53 Mb), E4f1, E4F transcription factor 1 (24.56 Mb) (Figure S1 and Table S4).
Although the majority of these genes were not well annotated, some transcription factors were localized at or near several hotspot loci. In particular, Atf6, one of the primary genes involved in regulating genes in the unfolded protein response system, resides near a hotspot on chromosome 1 (175.34 Mb) that was found to be enriched for transcripts involved in protein synthesis (P ≤ 6.90E-05) (Davenport et al. 2008).
DISCUSSION
Our results have led to several conclusions: First, local eQTL are highly preserved between studies, whereas distal eQTL were only modestly replicable. Second, most local eQTL were not tissue specific, in that an eQTL in one tissue (of the four tissues studied here) was likely to be observed in another tissue. This finding has important implications for human studies where tissue availability is a problem. Third, although thousands of genes differed in expression between males and females, most local eQTL were preserved between sexes whereas distal eQTL were not. Fourth, several hotspots were preserved between crosses, sexes, and multiple tissues. The identity of the underlying genes will be of considerable interest. There were some differences between the two crosses, notably the presence of an ApoE null mutation in cross I, and these undoubtedly decreased the replication of eQTL (Table 1).
Previous eQTL reports have shown similarly high levels of replication of local eQTL between mouse crosses (Peirce et al. 2006). Interestingly, the strongest replicated distal eQTL hotspot we observed in brain on chromosome 1 (174–178 Mb) was previously reported on chromosome 1 (170–174 Mb) in the same tissue (Peirce et al. 2006). However, a recent study detected low replicability between human cell lines from different tissues (Dimas et al. 2009). Since the latter analysis was performed in only 75 individuals, the power was more limited than in our study. Moreover, these authors used a relative high threshold for significance for both identification of the eQTL and replication in a separate tissue.
The importance of eQTL studies has recently been reviewed (Rockman and Kruglyak 2006; Cookson et al. 2009). eQTL analyses allow the modeling of causal interactions that can be used to identify candidate genes influencing biologic pathways (Schadt et al. 2005) and the construction of directed coexpression networks (Chen et al. 2008). The modeling of biologic networks has proved to be particularly informative when combined with genome-wide association studies, making possible the identification of causal genes at a locus, the annotation of genes of unknown function, and the clarification of new candidate genes in biologic pathways.
The high level of replication between crosses of distal as well as local eQTL suggests that systems genetics approaches should be capable of addressing mechanisms governing transcription and RNA processing. For example, it should be possible to examine control of chromatin structure by integrating eQTL data with ChIP-chip or ChIP-seq methodologies (Zhu et al. 2008). Also, massively parallel sequencing of transcripts from genetic crosses (RNA-seq) should result in the mapping of loci controlling RNA splicing as well as transcript levels (Heinzen et al. 2008; Veyrieras et al. 2008). Although similar eQTL approaches are clearly feasible for human studies, genetic heterogeneity and environmental factors result in greatly diminished power (Cookson et al. 2009). It is noteworthy that coexpression networks developed using mouse data exhibit considerable overlap with human networks (Emilsson et al. 2008).
Acknowledgments
We thank Hong-Xiu Qi for assistance in animal husbandry, including animal care, collecting plasma and tissues, and performing the body fat assays; Larry Castellani for assistance in collecting plasma and constructive criticism; Sharda Charugundla for performing the lipid assays; Melenie Rosales, Daria Estrada-Smith, Ping-Zi Wen, Sonal S. Sheth, Sudheer Doss, Xu-Ping Wang, Judy Wu, Xia Yang, and Yi-Shou Shi for their assistance in animal husbandry; Kathleen Ranola, Madlen Ayvasian, and Arpa Ghazarian for assistance in processing the tissues; Eleazar Eskin and Rita Cantor for statistical advice; Calvin Pan for assistance in data management; Sonia Carlson and Stephen Edwards for help in transfer of tissues and organization of data between institutions; and Richard C. Davis for guidance, discussions, and insight. This work was supported by National Institutes of Health (NIH) grants HL28481, HL094322, HL087944, and HL30568 (to A.J.L.) and 5R01DK72206 (to T.D.). A.v.N. was supported by NIH training grant 5T32HD07228. Disclosure statement: The authors have nothing to disclose.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.116087/DC1.
References
- Bystrykh, L., E. Weersing et al., 2005. Uncovering regulatory pathways that affect hematopoietic stem cell function using ‘genetical genomics’. Nat. Genet. 37(3): 225–232. [DOI] [PubMed] [Google Scholar]
- Chen, Y., J. Zhu, P. Y. Lum, X. Yang, S. Pinto et al., 2008. Variations in DNA elucidate molecular networks that cause disease. Nature 452(7186): 429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson, W., L. Liang, G. Abecasis, M. Moffatt and M. Lathrop, 2009. Mapping complex disease traits with global gene expression. Nat. Rev. Genet. 10(3): 184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerval, C., A. Maurice, J. M. Josse and D. de Vienne, 1994. Quantitative trait loci underlying gene product variation: a novel perspective for analyzing regulation of genome expression. Genetics 137 289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport, E. L., G. J. Morgan and F. E. Davies, 2008. Untangling the unfolded protein response. Cell Cycle 7(7): 865–869. [DOI] [PubMed] [Google Scholar]
- de Vienne, D., A. Maurice, J. M. Josse, A. Leonardi and C. Damerval, 1994. Mapping factors controlling genetic expression. Cell. Mol. Biol. 40(1): 29–39. [PubMed] [Google Scholar]
- Dimas, A. S., S. Deutsch, B. E. Stranger, S. B. Montgomery, C. Borel et al., 2009. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 325(5945): 1246–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doss, S., E. E. Schadt, T. A. Drake and A. J. Lusis, 2005. Cis-acting expression quantitative trait loci in mice. Genome Res. 15(5): 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson, V., G. Thorleifsson, B. Zhang, A. S. Leonardson, F. Zink et al., 2008. Genetics of gene expression and its effect on disease. Nature 452(7186): 423–428. [DOI] [PubMed] [Google Scholar]
- Ghazalpour, A., S. Doss, B. Zhang, S. Wang, C. Plaisier et al., 2006. Integrating genetic and network analysis to characterize genes related to mouse weight. PLoS Genet. 2(8): e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y. D., H. Dai, E. E. Schadt, G. Cavet, S. W. Edwards et al., 2003. Microarray standard data set and figures of merit for comparing data processing methods and experiment designs. Bioinformatics 19(8): 956–965. [DOI] [PubMed] [Google Scholar]
- Heinzen, E. L., D. Ge, K. D. Cronin, J. M. Maia, K. V. Shianna et al., 2008. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 6(12): e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E., and L. Kruglyak, 1995. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet. 11(3): 241–247. [DOI] [PubMed] [Google Scholar]
- Lusis, A. J., A. D. Attie and K. Reue, 2008. Metabolic syndrome: from epidemiology to systems biology. Nat. Rev. Genet. 9(11): 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusis, A. J., J. Yu and S. S. Wang, 2007. The problem of passenger genes in transgenic mice. Arterioscler. Thromb. Vasc. Biol. 27(10): 2100–2103. [DOI] [PubMed] [Google Scholar]
- Machleder, D., B. Ivandic, C. Welch, L. Castellani, K. Reue et al., 1997. Complex genetic control of HDL levels in mice in response to an atherogenic diet. Coordinate regulation of HDL levels and bile acid metabolism. J. Clin. Invest. 99(6): 1406–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery, S. B., and E. T. Dermitzakis, 2009. The resolution of the genetics of gene expression. Hum. Mol. Genet. 18(R2): R211–R215. [DOI] [PubMed] [Google Scholar]
- Peirce, J. L., H. Li, J. Wang, K. F. Manly, R. J. Hitzemann et al., 2006. How replicable are mRNA expression QTL? Mamm. Genome 17(6): 643–656. [DOI] [PubMed] [Google Scholar]
- Rockman, M. V., and L. Kruglyak, 2006. Genetics of global gene expression. Nat. Rev. Genet. 7(11): 862–872. [DOI] [PubMed] [Google Scholar]
- Schadt, E. E., J. Lamb, X. Yang, J. Zhu, S. Edwards et al., 2005. An integrative genomics approach to infer causal associations between gene expression and disease. Nat. Genet. 37(7): 710–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez, B. K., C. L. Hampe and P. Van Eerdewegh, 1994. Problems of Replicating Linkage Claims in Psychiatry in Genetic Approaches to Mental Disorders. American Psychiatric Press, Washington, DC.
- van Nas, A., D. Guhathakurta, S. S. Wang, N. Yehya, S. Horvath et al., 2009. Elucidating the role of gonadal hormones in sexually dimorphic gene coexpression networks. Endocrinology 150(3): 1235–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrieras, J. B., S. Kudaravalli et al., 2008. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 4(10): e1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., N. Yehya, E. E. Schadt, H. Wang, T. A. Drake et al., 2006. Genetic and genomic analysis of a fat mass trait with complex inheritance reveals marked sex specificity. PLoS Genet. 2(2): e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., B. Zhang, J. Zhu, B. Zhang, E. N. Smith et al., 2008. Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat. Genet. 40(7): 854–861. [DOI] [PMC free article] [PubMed] [Google Scholar]