Abstract
Any established or aspiring model organism must justify itself using two criteria: does the model organism offer experimental advantages not offered by competing systems? And will any discoveries made using the model be of wider relevance? This review addresses these issues for the social amoeba Dictyostelium and highlights some of the organisms more recent applications. These cover a remarkably wide gamut, ranging from sociobiological to medical research with much else in between.
DICTYOSTELIUM came into the world of research later than many other model organisms. The type species, Dictyostelium discoideum, was discovered as recently as 1935, and much of the early interest centered around its multicellular development. This remains a major area of research, but there has been a distinct gravitation to earlier developmental stages and also to the growing cell. Unusually, multicellularity is achieved by cellular aggregation, which occurs by chemotaxis of cells toward pulses of cAMP released from the cells in the center of each aggregation territory. Much of the emphasis in the field has shifted into studying the molecular basis of chemotaxis. This has been extremely informative for understanding the process in animal cells but has been reviewed extensively elsewhere (Franca-Koh et al. 2006; Van Haastert and Veltman 2007; Kay et al. 2008; Kolsch et al. 2008).
Fueled in part by the chemotaxis bonanza, realization has grown that Dictyostelium offers distinct advantages for studying those cell biological processes that the two commonly studied yeasts do not undertake and for studying those genes that they do not possess. The genome sequence has been a major stimulant in the latter respect, because a number of important gene families are now known to fall into this class. Another four Dictyostelid genome sequences are close to completion or complete. These will aid in analyzing gene expression and function, and they can be used to understand how the diverse developmental forms displayed by the Dictyostelids evolved. Another, more general evolutionary issue that is being vigorously investigated is that of cellular altruism: What are the forces that maintain the 20% stalk-to-spore ratio over evolutionary time? At the other end of the spectrum, there are some very practical uses for the organism. Dictyostelium, for example, is being used to determine how cells interact with bacterial agents of infectious diseases. What are the virtues of the organism that have fueled these diverse applications?
THE DICTYOSTELIUM CELL
Dictyostelium is a microbe that can be rapidly grown to high cell densities in an inexpensive medium. Therefore, analyses on a biochemical scale are normally very straightforward. It is also an amoebozoan with a cell biology that is, in several important respects, closer to that of animal cells than that of yeast cells. It has a flexible plasma membrane rather than a rigid cell wall. This permits Dictyostelium cells to be highly motile and very active in phagocytosis and pinocytosis. The cells are only a little smaller than the average animal cell, and they are excellent subjects for all forms of light microscopy. Fueled by the GFP revolution, there has been a flowering of informative and memorable images of Dictyostelium cells engaged in various cellular processes (http://dictybase.org/). Dictyostelium has two further important attributes for any model system: a large knowledge base with many characterized genes and ready means of isolating, manipulating, and analyzing them.
DICTYOSTELIUM GENOMICS AND MOLECULAR GENETICS
The Dictyostelium genome is divided among six haploid chromosomes and has a total length of 34 Mb. The genome has many complex repeats, which complicated the genome sequencing, and one class of clustered repeat is suggested to constitute the centromeric sequence (Eichinger et al. 2005). Interestingly, telomeres appear to compose partial copies of the extrachromosomal ribosomal DNA element. The original estimate from the genome sequence suggested 13,500 genes, separated by an average spacing of 2.5 kb. A later estimate, discounting a number of very small genes and potential pseudogenes, suggests the lower number of 10,300 (Olsen 2005). Even assuming the lower estimate to be more correct, this is still a much higher number of genes than the ∼6000 genes encoded by Saccharomyces cerevisiae and is much nearer to the 13,000 genes encoded by Drosophila melanogaster (Kumar et al. 2002; Hahn et al. 2007). The coding regions are AT-rich, and the intergenic regions and introns are even more AT-biased. The reason for the AT bias is unknown, but the resultant codon usage bias greatly facilitated the prediction of coding regions. Also, most genes contain only one or two introns, and they are usually very small: ∼100–200 bp. The genome sequence is maintained and annotated at dictyBase (http://dictybase.org/), which also runs the Dictyostelium Stock Center. Also, a large-scale EST project has generated cDNAs representing ∼55% of the estimated genes, and many cDNAs are available as full-length inserts (Urushihara et al. 2006).
Although there is a sexual cycle, it is not experimentally useful and, until the era of gene cloning, only para-sexual gene mapping of mutants generated using chemical mutagenesis could be applied to the organism. Fortunately, Dictyostelium is readily transformable and now supports a relatively sophisticated set of molecular genetic techniques. Key among these is gene disruption via the homologous recombination of a targeting construct. This works with an efficiency that is sometimes as high as 90%, and serial rounds of gene disruption are possible using the Cre-loxP system to recycle mutants (Faix et al. 2004). This can be invaluable when studying gene families and has been used, for example, to knock out sequentially in one strain all five of the type 1 phosphoinositide 3-kinase genes (Hoeller and Kay 2007).
Because the organism is haploid, gene disruption is not applicable in those situations where the target gene is essential for growth. In such a case, a target gene's activity can be inhibited by using an antisense or a dominant negative construct under the control of a regulatable promoter, e.g., the tetracycline “off” system (Blaauw et al. 2000). More sophisticated manipulations such as gene “knock-ins” are also routinely used to modify genes in the genome. Using this technology, Dictyostelium became the first eukaryotic organism in which individual gene transcription events were seen in living cells (Chubb et al. 2006). This follows a long tradition of technical advances developed using the organism; it was, for example, the first organism in which univalent antibodies were used to study cell–cell adhesion (Beug et al. 1970).
Although large numbers of interesting and informative mutants were generated using chemical mutagenesis, attempts to identify the cognate genes by molecular complementation using libraries of genomic DNA have thus far been unsuccessful. Fortunately, an alternative method of gene isolation, termed Restriction Enzyme-Mediated Integration (REMI), was devised by Kuspa and Loomis (1992). They modified a technique, originally developed in yeast recombination studies, to randomly insert a DNA “tag” into the Dictyostelium genome. Insertion of the REMI tag will usually inactivate the tagged gene, and the cellular consequences of growth and/or development can be determined. The tagged gene can then either be isolated by cloning in Escherichia coli or, more usually since the advent of the genome sequence, directly amplified, sequenced, and mapped onto the genome. This technique has been one of the mainstays of Dictyostelium molecular genetics and has even been used to generate informative suppressor mutations (Souza et al. 1998; Tekinay et al. 2003). In a good example of cross-model fertilization, REMI was further modified by the Xenopus community as the method for inserting transgenes into sperm, the key to successful DNA transformation of the germ line (Kroll and Amaya 1996).
DICTYOSTELIUM BRIDGES AN EVOLUTIONARY GAP
Dictyostelium is an excellent test bed for developing new techniques, but its various experimental attributes would be of little general importance were it not also a “relevant” model system. Although this term is often taken to mean relevant to higher organisms such as humans, it is worth noting that Entamoeba histolytica is also an amoebozoan, that infection by E. histolytica is the second most common cause of death from parasitic disease, and that there is an urgent need for basic information on the organism (Stanley and Samuel 2003). There is an E. histolytica genome sequence, and the organism supports some gene manipulation techniques. However, these are relatively limited and Dictyostelium has become the model amoebozoan, to which E. histolytica and other members of this diverse phyllum are compared. Also, Dictyostelium acts as a technical trailblazer for researchers using other amoebozoan species, suggesting molecular genetic approaches that are likely to succeed with their organism.
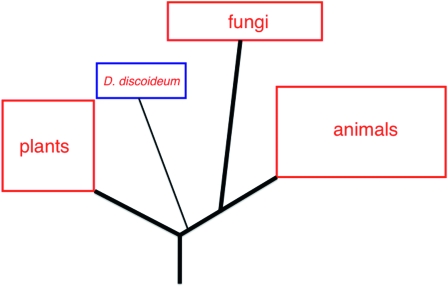
The interspecies comparison of thousands of individual protein sequences, made possible by the genome sequence, has allowed Dictyostelium's evolutionary relationship to higher organisms to be reassessed (Eichinger et al. 2005). This reassessment confirmed earlier work, suggesting that the ancestor of Dictyostelium diverged from the ancestors of animals and fungi at some time after the divergence of ancestral plants (Loomis and Smith 1990; Baldauf and Doolittle 1997). Thus Dictyostelium is more closely related to present-day animals than are plants (Figure 1). The position of Dictyostelium with respect to fungi and animals should perhaps be clarified. Although orthologous animal and yeast proteins will generally show a higher degree of sequence similarity to each other than to the Dictyostelium counterpart, Dictyostelium has many more genes held in common with animals than do either of the two yeasts. The simple interpretation of these facts is that, at some time after the divergence of fungi from animals, there was massive gene loss during fungal evolution. What this means in practice is that Dictyostelium offers access to many protein classes that are not represented in the yeasts. The SH2 domain containing proteins provides a good example of this “horses for courses” principle.
Figure 1.—
The evolutionary relationships of D. discoideum. This schematic, deduced from sequence comparisons, displays the evolutionary position of D. discoideum relative to other selected phyla.
SH2 domains are regulatable protein–protein interaction domains that bind to a phosphotyrosine-containing recognition sequence on an interaction partner (Pawson et al. 2001). They were originally believed to be confined to the animal kingdom, but we now know that Dictyostelium has 13 SH2 domain proteins (Eichinger et al. 2005). This is still a much smaller set than mouse or human, which both possess 110 SH2 domain proteins (Liu et al. 2006), but the only organism other than animals in which SH2 domains have been functionally validated. The four best characterized are STAT proteins a–d (Williams 2003). Analysis of STATc revealed a functional interaction with CblA, another of the 13 SH2 domain proteins and an ortholog of the metazoan Cbl proteins (Langenick et al. 2008). The presence of STAT and Cbl orthologs in Dictyostelium fits the general pattern discussed above, whereby genes previously considered characteristic of animal cells are often found to be also present in Dictyostelium.
Cross-kingdom homology is sometimes sufficiently strong for a Dictyostelium ortholog to provide information directly relevant to its metazoan counterparts. STAT proteins dimerize and become transcriptionally activated by reciprocal SH2 domain:phosphotyrosine interactions (Bromberg and Darnell 2000). The structure of dimeric Dictyostelium STATa, serendipitously crystallized without a DNA ligand (Soler-Lopez et al. 2004), provided an important general insight into the mechanism of STAT action by suggesting that binding to the DNA ligand causes a major conformational change in the STAT protein dimer.
DEVELOPMENT AS A READOUT OF GENE FUNCTION
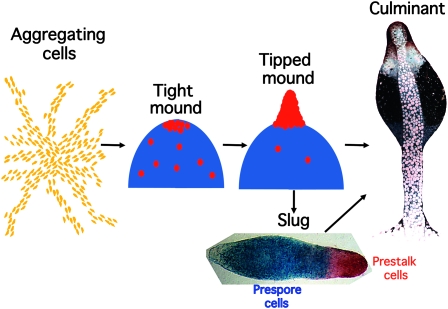
During normal multicellular development, up to 100,000 Dictyostelium cells aggregate together to form a mound (Figure 2). At about this stage, cells differentiate as either prestalk or prespore cells. Prespore cells are induced to differentiate by extracellular cAMP signaling, and they synthesize and release another small signaling molecule, the polyketide DIF-1, that directs a proportion of the remaining, uncommitted cells to become prestalk cells (Kay et al. 1999). The prestalk cells are initially scattered throughout the mound, but they move to the apex where they intercalate and form themselves into a tip. This elongates to form a standing slug that then either forms a fruiting body in situ or moves away as a motile slug, seeking out an appropriate environment in which to culminate. Slug formation constitutes a remarkable behavioral transition. In just 12 hr, thousands of cells exchange a solitary, foraging lifestyle to become one cog in a multicellular organism in which cells communicate, cooperate, and display a division of labor.
Figure 2.—
The Dictyostelium life cycle. Development in Dictyostelium is highly regulative and, depending on the local cell density, from 100 to 100,000 cells may aggregate together in response to cAMP signals emanating from the center of an aggregation territory. At the end of aggregation a mound of cells is formed and prespore cell differentiation, represented by blue shading, is induced by cAMP signaling. A subset of the uncommitted cells become prestalk cells (red circles) and migrate to the apex of the mound. There these cells form themselves into a nipple-shaped structure that extends to form an upright slug-shaped structure. Depending on environmental conditions, this structure may enter culmination to form a fruiting body immediately or may migrate for a time before completing development. The slug displayed was transformed with an ecmA-to-lacZ promoter fusion that is expressed in all prestalk cells (red) and a pspA-to-gus promoter fusion that is expressed in all prespore cells (blue). The two expression patterns were revealed by double enzymatic, gus–gal staining. The fruiting body is an electron microscopy image (copyright by M. J. Grimson and R. L. Blanton) that is false colored to show the position of the prestalk cells (red), upper cup cells (white), and spore cell precursors (blue).
Dictyostelium is unusual in that cell division and multicellular development are uncoupled. As one mark of this dichotomy, very many mutant strains grow normally but are defective in some aspect of development. Development acts as a stringent biological filter, so that assaying the developmental competence of a mutant strain can often be an invaluable tool for studying and defining a gene's function. This is nicely exemplified by myoA, the gene encoding myosin II, the first Dictyostelium gene to be inactivated using molecular genetics. It was inactivated by homologous gene disruption and, in a parallel study, by expression of an antisense transcript (De Lozanne and Spudich 1987; Knecht and Loomis 1987). Surprisingly, the myosin II null and knock-down cells have only a partial defect in cell movement and cytokinesis, but they completely arrest their development at the mound stage. It seems that the myosin-null prestalk cells are in some way defective in their movement and cannot squeeze themselves through the multicellular milieu of the mound (Knecht and Shelden 1995).
DEVELOPMENTAL MARKERS AND PRESTALK AND STALK CELL SUBTYPES
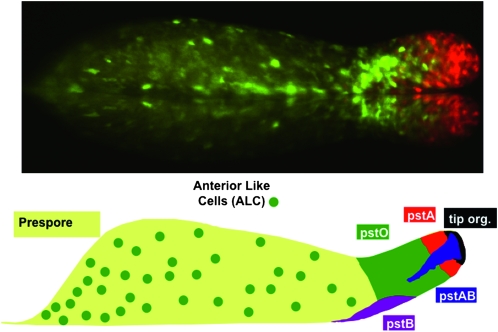
Not all developmental defects are as dramatic as that of the myoA null. The development of many mutants appears outwardly normal but, by using appropriate markers, cryptic differentiation defects or defects in morphogenetic cell movement can be detected. This possibility greatly increases the utility of development as a phenotypic readout. During development past the mound stage, to form at first a slug and then a culminant, prestalk cells undertake highly coordinated morphogenetic movements before completing their differentiation into vacuolated stalk cells. These different movement patterns are possible because there are multiple prestalk cell subtypes that yield, by their distinct movement patterns and further cellular differentiation, multiple stalk cell types that are located in different parts of the culminant (Figure 3; Gaudet et al. 2008; http://dictybase.org/Dicty_Info/dicty_anatomy_ontology.html).
Figure 3.—
Prestalk and stalk cell heterogeneity. (Top) The slug displayed was transformed with two different ecmA promoter-to-GFP fusions that are expressed in pstA cells (red), pstO cells, or anterior-like cells (ALCs) (green) (D. Dormann, N. Zhukovskaya, J. G. Williams and C. J. Weijer, unpublished results). (Bottom) A representation of the different prestalk cell populations present in the slug. The anterior prestalk region contains four partially overlapping populations (Gaudet et al. 2008; http://dictybase.org/Dicty_Info/dicty_anatomy_ontology.html). The pstA and pstO cells, respectively, occupy the front and rear halves and are identified by their ability to use different parts of the promoter of the ecmA gene. The pstAB cells are pstA cells that prematurely and sporadically express the ecmB gene. This is the same transitional event that occurs continuously during culmination. The pstAB cells sporadically drop backward and fall out the back of the migrating slug. At culmination, they form the inner part of the basal disc. The tip organizer cells are characterized by their ability to use a specific region of the promoter of the cudA gene. They control the migration and maintain the integrity of the slug. The pstB cells express ecmB at a higher level than ecmA and move backward and forward as a group along the anterior–posterior axis of the ventral surface of the prespore region. At culmination, they form the outer part of the basal disc. The ALCs are a mixed group of cells that are the direct precursors of the upper and lower cups of the culminant.
The routinely used markers for the major three prestalk cell subtypes—pstA, pstO, and pstB cells—derive from the promoters of two genes that encode extracellular matrix proteins of the slug: ecmA and ecmB. They have been used to delineate the developmental lesion in many mutant strains. For example, mutants in DIF-1 signaling are defective in the differentiation of pstO and pstB cells but are unaffected in pstA cell differentiation (Thompson and Kay 2000; Thompson et al. 2004; Fukuzawa et al. 2006; Keller and Thompson 2008; Saito et al. 2008). The signaling molecule that induces pstA differentiation is unknown, but it seems likely that it is another polyketide (Saito et al. 2006).
A recent study has revealed a further, unexpected, and quite remarkable anatomical feature of the slug (Chen et al. 2007). The study identified cells scattered within the slug, the Sentinel cells (S cells), that phagocytose bacteria and sequester toxins. Because they are periodically shed from the slug, they function in much the same way as phagocytic scavenger cells of the metazoan innate immune system. The S cells may also be relevant to the evolution of animal innate immunity because they selectively express a gene, tlrA, that contains a domain with sequence similarity to the metazoan Toll/interleukin-1 receptor. Moreover, null mutants for tlrA are hypersensitive to infection by a Legionella strain that is normally avirulent. This observation, of another radical division of labor in the slug cell population, reinforces the notion of Dictyostelium as a facultatively multicellular organism of unexpected sophistication.
GENOME SEQUENCES FROM OTHER DICTYOSTELIDS OFFER NEW INSIGHTS AND OPPORTUNITIES
Thus far the focus has been on D. discoideum, the organism employed in the vast majority of studies. However, there is great diversity in the Dictyostelids. All share the same survival and dispersal strategy of lifting resistant spores off the substratum on a stalk, but there are major differences in their fruiting body morphologies and developmental game plans. In Acytostelium subglobosum, a spore mass is supported by an acellular stalk (Figure 4). At the other end of the complexity spectrum Polysphonylium pallidum forms a beautiful fruiting body, composed of delicately branching whorled stalks, each supporting a spore mass (Figure 4). A comparison of small subunit ribosomal RNA sequences and α-tubulin sequences across almost all of the known Dictyostelids has produced a molecular evolutionary tree (Schaap et al. 2006). It contains four subdivisions and is significantly different from the traditional taxonomy based solely on morphological characteristics. For example, despite their radically different morphological complexities, both A. subglobosum and P. pallidum are in group 2.
Figure 4.—
Degrees of anatomical complexity within the Dictyostelids. These are images of three of the species for which a complete genome sequence is available or soon will be available. Both A. subglobosum and P. pallidum are in group 2 of the molecular phylogeny of Schaap et al. (2006) while D. discoideum is in group 4. Note the differences in size, indicated by the scale bars. These images were kindly provided by F. Spiegel.
A key task now is to determine how morphological diversification evolved within and between the subgroups. This is being addressed by whole-genome sequence analysis of at least one representative from each of the four groups and by parallel studies of genes known to be key to the development of D. discoideum (Kawabe et al. 2002, 2009). The new sequences, from A. subglobosum (http://acyto.sequence.info/), D. purpurem (http://dictybase.org/), P. pallidum (http://sacgb.fli-leibniz.de/), Dictyostelium fasciculatum (http://sacgb.fli-leibniz.de/), and D. lacteum (P. Schaap, personal communication), will also be extremely valuable in other, more general ways. When studying a gene's structure and function, it will be possible to compare a series of orthologs of graded divergence and so identify conserved and potentially important residues. The promoters of orthologous, coregulated genes can also be compared to identify conserved sequence elements.
ALTRUISM, THE STALK–SPORE RATIO, AND CHEATER MUTANTS
One of the features of D. discoideum development that is extremely useful to the developmental biologist interested in pattern formation is the approximate correspondence between the number of prestalk cells and the number of stalk cells. During development ∼20% of cells are diverted from prespore to prestalk differentiation, and the normal fate of a prestalk cell is to further differentiate and die as a stalk cell. An often-ignored fact is that in many other Dictyostelids there is little or no prestalk region; most or all of the stalk cells are formed by the trans-differentiation of prespore cells. This occurs positionally, at the entrance to the stalk tube, and stalk formation is continuous throughout slug migration. Nonetheless, the decision to differentiate and die as a stalk cell, whether made by a “half-way house” prestalk cell or a “turncoat” prespore cell, raises the same evolutionary paradox. What do the individual cells that opt for this sterile fate gain by their decision? And what is to stop the appearance, over evolutionary time, of “cheaters”—mutants that decline to die as stalk cells and instead opt for survival as spore cells?
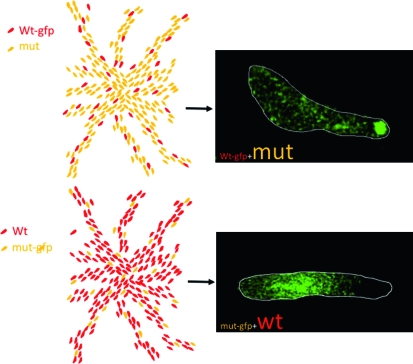
The first cheater strain was a field sample of Dictyostelium mucoroides, a species that produces a stalk continuously during slug migration (Buss 1982). It represents the extreme example of cheating because it formed only spores. More recently, there has been a major resurgence of interest in the cheating phenomenon but using D. discoideum and studying much subtler cheating mutants. Cheating behavior can be easily assessed in mixing experiments in which a minority of mutant cells, labeled genetically or with a fluorescent dye, are co-developed with parental cells, and their position within the slug and their relative contribution to the spore population is determined (Figure 5).
Figure 5.—
A typical synergy experiment as used in cheater assays. The drawings at the left represent aggregation territories formed by (top) a minority (usually 5–10%) of GFP-labeled parental cells mixed with a large excess of GFP-labeled cheater mutant cells and (bottom) a majority of unlabeled parental cells mixed with a minority of GFP-labeled cheater mutant cells. The images at the right are from such an experiment using a mutant defective in several aspects of late development (C. Sugden and J. Williams, unpublished data). In both variants of the mixing protocol, the mutant cells are selectively excluded from the prestalk zone.
Serial selections of REMI mutagenized cells have allowed the identification of genes that, when inactivated, lead to a cheater phenotype. The most frequently occurring and best characterized of these, fbxA, encodes an F box protein (Ennis et al. 2000; Nelson et al. 2000). The F box family of ubiquitin ligases targets proteins for ubiquitination and degradation. Genetic and biochemical analyses suggest that FbxA functions by regulating the degradation of PKA, the cAMP-dependent protein kinase (Mohanty et al. 2001; Tekinay et al. 2003). A larger-scale REMI mutant selection, incorporating a screen to exclude mutants like fbxA that fail to culminate when developing alone, identified >100 cheater mutations (Santorelli et al. 2008). The genes that were identified encode a very mixed bag of proteins, with no marked enrichment for any one functional class.
In the laboratory, therefore, cheating is a very well-established phenomenon that has kindled the interest of evolutionary biologists. But how relevant is it to life on the forest floor? Three questions need answering: (i) Do cheaters co-exist with non-cheaters in natural populations? (ii) Does cheating ever really occur in nature? (iii) If so, what prevents cheaters from coming to dominate over evolutionary time? In answer to the first questions, a field study has shown that naturally occurring strains that behave as cheaters under laboratory conditions do coexist in close proximity with non-cheaters (Strassmann et al. 2000). There is therefore the potential for cheating. The second question is of course extremely difficult to address directly for natural populations. Students of cheating, therefore, must assume that it does happen and concern themselves principally with addressing the third question: the evolutionary containment of cheater mutations. One theoretical possibility is that of a developmental cost to cheating, and there is evidence for this (Foster et al. 2004). DimB is a DIF-1-activated b-Zip transcription factor that is required for DIF-1 responsiveness (Huang et al. 2006; Zhukovskaya et al. 2006). Since the dimB null strain is a DIF-1 signal reception mutant, it would be expected to behave as a cheater, but it forms very few viable spores when synergized with wild-type cells (Foster et al. 2004).
Formerly, the favored explanation for the evolutionary survival of altruism in the Dictyostelids was that it reflects the pressure of intense kin selection. By lifting up the spore head and aiding spore dispersal, stalk cells increase the survival chances of the gene complement represented within that particular fruiting body. In the laboratory, a clonal plaque of fruiting bodies can be generated by depositing a single spore on a plate spread with bacteria. If clonal propagation were also the usual scenario in nature, then the altruistic behavior of stalk cells would be readily explicable: when all cells within a fruiting body are identical, then the self-sacrificial stalk cell is simply promoting the survival of copies of its own genome. A recent field study, in which spores were picked from individual fruiting bodies and their relatedness was determined by genomic analysis, gives very strong support to just such a mechanism (Gilbert et al. 2007). It revealed a remarkably high degree of relatedness, sufficient to prevent the spread of cheaters. It would seem therefore that the study of cheaters is yielding fascinating insights into cellular cooperation and competition but carries the proviso that it may well be a laboratory rather than a natural phenomenon.
DICTYOSTELIUM IN BIOMEDICAL RESEARCH
The genome sequence has revealed orthologs of many genes implicated in human disease that are present in Dictyostelium but not in the two commonly studied yeasts (Eichinger et al. 2005). Several of these have already been the subject of functional studies (Lee et al. 1998; Harris et al. 2002; Wessels et al. 2006; Langenick et al. 2008), and this seems likely to be a major future use of the organism. Also, Dictyostelium has been used to help identify targets for drugs used to treat human diseases: cis-platin, an anticancer agent (Li et al. 2000); lithium used to treat bipolar disorder (Williams et al. 1999); and bisphosphonates used to treat osteoporosis (Grove et al. 2000). These studies are reviewed elsewhere (Williams et al. 2006), so I will concentrate on just one recent application that well illustrates the power of the system: investigating the infection mechanisms of clinically important bacteria, particularly Legionella pneumophila.
Legionella, the causative agent of Legionnaires' disease, infects and parasitizes amoebozoan species present in stagnant water sources such as cooling towers. It does so by subverting the normal phagocytic feeding mechanisms to avoid digestion. Once ingested, Legionella replicates within membrane-bound vesicles the Legionella-containing vacuoles (LCVs) that prevent fusion with lysosomes. If elderly or immunocompromised individuals inhale an aerosol containing Legionella, it can infect their macrophages and cause a pneumonia that is often fatal. Because they survive by ingesting bacteria, Dictyostelium cells are highly active in phagocytosis, and the process appears to be very similar to that occurring in mammalian cells (Bozzaro et al. 2008; Cosson and Soldati 2008).
As a “professional” phagocyte, with well-founded cell and molecular biology, Dictyostelium provides a model both for the natural protozoan host and for the alternate host: the human macrophage. This is now a major field of research that is comprehensively reviewed elsewhere (Steinert and Heuner 2005; Bozzaro et al. 2008; Bruhn and Steinert 2008; Cosson and Soldati 2008; Clarke 2010) so I will give only an overview. Dictyostelium is susceptible to Legionella infection, LCVs are formed, and some Legionella mutant strains that are avirulent for humans are avirulent for Dictyostelium (Solomon et al. 2000). A number of Dictyostelium mutants that affect the cytoskeleton have been shown to differ in their relative susceptibility to Legionella infection (reviewed by Bruhn and Steinert 2008). For example, a null mutant in coronin, an actin cytoskeleton-associated protein required for optimal phagocytosis, is excessively permissive for intracellular Legionella growth (Solomon et al. 2000).
In animal cells, mitochondria become associated with the LCVs, and Dictyostelium strains with altered levels of expression of AMP-activated protein kinase, a known regulator of mitochondrial function, show altered capacities to replicate Legionella (Francione et al. 2009). The complementary, more global approach, of identifying cellular components that change in response to Legionella infection, has also been informative. Expression profiling reveals changes in the expression of ∼5% of the genes represented on the array, and many of these encoded components that might be expected to be regulated in response to infection (Farbrother et al. 2006). Purification of Legionella containing phagosomes from cells infected with Legionella and analysis of their proteome have given further insights into the infection process (Shevchuk et al. 2009).
Thus far I have emphasized the similarities between Legionella infection in Dictyostelium and animal cells but there are, as might be expected, some differences. For example, certain Legionella mutant strains that are virulent for humans are avirulent for Dictyostelium (Bruhn and Steinert 2008). Analysis of the mitochondria of Legionella-infected Dictyostelium cells has revealed another major difference: Legionella infection causes degradation of the Dictyostelium mitochondrial messenger RNAs and three specific cleavages of the large subunit of the mitochondrial ribosomal RNA (Zhang and Kuspa 2009). These effects do not occur in human cells nor, interestingly, do they occur in Acanthamoeba, a natural host for Legionella.
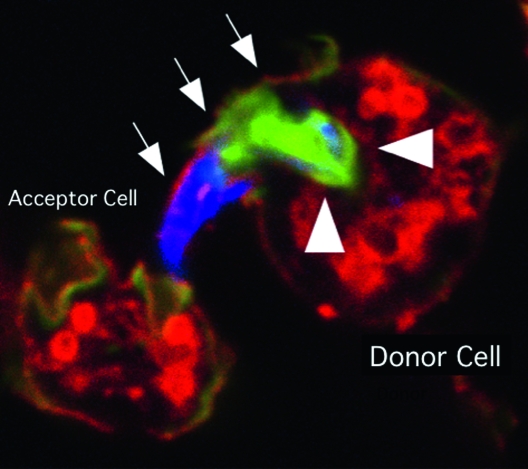
Other medically important infectious agents have also been studied in Dictyostelium to good effect. Pseudomonas aeruginosa, an opportunistic pathogen, replicates in Dictyostelium, and comparative expression profiling of cells infected with two bacterial strains with different virulences has identified genes whose expression correlates with higher virulence (Cosson et al. 2002; Carilla-Latorre et al. 2008). Mycobacterium tuberculosis also infects Dictyostelium cells, and an entirely novel bacteria-induced structure was recently discovered using Dictyostelium as a model: the ejectosome, an actin-based structure responsible for nonlytic spreading of the mycobacterium (Figure 6; Skriwan et al. 2002; Hagedorn et al. 2009).
Figure 6.—
An ejectosome produced by a Dictyostelium cell infected with Mycobacterium marinum. A bundle of mycobacteria is being ejected from a donor cell via an ejectosome (Hagedorn et al. 2009). The plasma membrane bulge (arrows) is ruptured at the tip, where it contacts the acceptor cell. Actin tails, stained green with phalloidin (arrowheads), are polarized at the posterior of the bacteria, which are stained blue. This image was kindly provided by T. Soldati.
THE BROADER PICTURE AND A WISH LIST
This review has illustrated the utility of Dictyostelium by describing two radically different modeling roles: as an example of altruism in sociobiological research and as an investigative tool for human bacterial pathogenesis. However, this is but a small part of a much larger picture; many other biological processes are being vigorously investigated. In addition to those mentioned, cell movement, chemotaxis, phagocytosis, processes such as cytokinesis, stress responses, non-apoptotic cell death, cell–cell signaling, signal transduction, and pattern formation are all under active study. A major part of the organism's appeal is the ability to rapidly, cheaply, and cleanly knock out individual “known” genes and entire gene families. The identification of “unknown” genes involved in a particular biological process using REMI is also a very powerful approach. A systematic REMI project designed to provide a null strain for every targetable gene has been combined with high-throughput time-lapse analysis to provide a devlopmental phenotype for thousands of strains (Sawai et al. 2007). However, gaps remain in the genetic armory.
An extremely rapid method of gene expression knock down, akin to the genome-wide RNA interference (RNAi)-based methods used in other organisms, would be high on one's wish list. While less definitive than a total gene disruptant, such an approach would facilitate high-throughput screening and primary gene identification. Both antisense and RNAi inhibition have been used to knock down expression of individual genes, but success has been sporadic, especially for developmental genes (reviewed by Kuhlmann et al. 2006). This problem may be surmountable by a more mechanistically informed approach, as evidenced by the recent demonstration of the greatly improved frequency of knock-down strains in a null mutant for a dicer-related helicase (Popova et al. 2006).
Another approach is gene complementation using a total genomic DNA library, a key method in yeast molecular genetics. Although valuable results have been obtained using a Dictyostelium cDNA expression library (Robinson and Spudich 2000; Shimada and Kawata 2007; Shimada et al. 2008), the library is very unlikely to contain full-length sequences representing all the less highly transcribed genes. The alternative approach of genomic DNA complementation has never been made to work, probably because of the difficulty of amplifying representative genomic Dictyostelium DNA libraries in E. coli. New cloning hosts for genomic DNA or the use of gridded, complete Dictyostelium cDNA expression libraries might offer ways around these problems. That would open up the study of the vast range of classically generated primary mutants and also allow the isolation of multi-copy suppressor genes. This would permit the identification of entirely new components of signaling pathways genetically and would broaden the organism's horizons even further.
References
- Baldauf, S. L., and W. F. Doolittle, 1997. Origin and evolution of the slime molds (Mycetozoa). Proc. Natl. Acad. Sci. USA 94 12007–12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beug, H., G. Gerisch, S. Kempff, V. Riedel and G. Cremer, 1970. Specific inhibition of cell contact formation in Dictyostelium by univalent antibodies. Exp. Cell Res. 63 147–158. [DOI] [PubMed] [Google Scholar]
- Blaauw, M., M. H. K. Linskens and P. J. M. van Haastert, 2000. Efficient control of gene expression by a tetracycline-dependent transactivator in single Dictyostelium discoideum cells. Gene 252 71–82. [DOI] [PubMed] [Google Scholar]
- Bozzaro, S., C. Bucci and M. Steinert, 2008. Phagocytosis and host-pathogen interactions in Dictyostelium with a look at macrophages. Int. Rev. Cell Mol. Biol. 271 253–300. [DOI] [PubMed] [Google Scholar]
- Bromberg, J., and J. E. Darnell, Jr., 2000. The role of STATs in transcriptional control and their impact on cellular function. Oncogene 19 2468–2473. [DOI] [PubMed] [Google Scholar]
- Bruhn, H., and M. Steinert, 2008. Dictyostelium, a tractable model host organism for Legionella, pp. 125–152 in Legionella: Molecular Microbiology, edited by K. A. S. Heuner and M. Caister. Academic Press, San Diego.
- Buss, L. W., 1982. Somatic cell parasitism and the evolution of somatic tissue compatibility. Proc. Natl. Acad. Sci. USA 79 5337–5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carilla-Latorre, S., J. Calvo-Garrido, G. Bloomfield, J. Skelton, R. R. Kay et al., 2008. Dictyostelium transcriptional responses to Pseudomonas aeruginosa: common and specific effects from PAO1 and PA14 strains. BMC Microbiol. 8 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G., O. Zhuchenko and A. Kuspa, 2007. Immune-like phagocyte activity in the social amoeba. Science 317 678–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb, J. R., T. Trcek, S. M. Shenoy and R. H. Singer, 2006. Transcriptional pulsing of a developmental gene. Curr. Biol. 16 1018–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, M., 2010. Recent insights into host-pathogen interactions from Dictyostelium. Cell Microbiol. 12 283–291. [DOI] [PubMed] [Google Scholar]
- Cosson, P., and T. Soldati, 2008. Eat, kill or die: when amoeba meets bacteria. Curr. Opin. Microbiol. 11 271–276. [DOI] [PubMed] [Google Scholar]
- Cosson, P., L. Zulianello, O. Join-Lambert, F. Faurisson, L. Gebbie et al., 2002. Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J. Bacteriol. 184 3027–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lozanne, A., and J. A. Spudich, 1987. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science 236 1086–1091. [DOI] [PubMed] [Google Scholar]
- Eichinger, L., J. A. Pachebat, G. Glockner, M. A. Rajandream, R. Sucgang et al., 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435 43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennis, H. L., D. N. Dao, S. U. Pukatzki and R. H. Kessin, 2000. Dictyostelium amoebae lacking an F-box protein form spores rather than stalk in chimeras with wild type. Proc. Natl. Acad. Sci. USA 97 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix, J., L. Kreppel, G. Shaulsky, M. Schleicher and A. R. Kimmel, 2004. A rapid and efficient method to generate multiple gene disruptions in Dictyostelium discoideum using a single selectable marker and the Cre-loxP system. Nucleic Acids Res. 32 e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farbrother, P., C. Wagner, J. Na, B. Tunggal, T. Morio et al., 2006. Dictyostelium transcriptional host cell response upon infection with Legionella. Cell Microbiol. 8 438–456. [DOI] [PubMed] [Google Scholar]
- Foster, K. R., G. Shaulsky, J. E. Strassmann, D. C. Queller and C. R. Thompson, 2004. Pleiotropy as a mechanism to stabilize cooperation. Nature 431 693–696. [DOI] [PubMed] [Google Scholar]
- Franca-Koh, J., Y. Kamimura and P. Devreotes, 2006. Navigating signaling networks: chemotaxis in Dictyostelium discoideum. Curr. Opin. Genet. Dev. 16 333–338. [DOI] [PubMed] [Google Scholar]
- Francione, L., P. K. Smith, S. L. Accari, P. E. Taylor, P. B. Bokko et al., 2009. Legionella pneumophila multiplication is enhanced by chronic AMPK signalling in mitochondrially diseased Dictyostelium cells. Dis. Model. Mech. 2 479–489. [DOI] [PubMed] [Google Scholar]
- Fukuzawa, M., N. V. Zhukovskaya, Y. Yamada, T Araki and J. G. Williams, 2006. Regulation of Dictyostelium prestalk-specific gene expression by a SHAQKY family MYB transcription factor. Development 133 1715–1724. [DOI] [PubMed] [Google Scholar]
- Gaudet, P., J. G. Williams, P. Fey and R. L. Chisholm, 2008. An anatomy ontology to represent biological knowledge in Dictyostelium discoideum. BMC Genomics 9 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, O. M., K. R. Foster, N. J. Mehdiabadi, J. E. Strassmann and D. C. Queller, 2007. High relatedness maintains multicellular cooperation in a social amoeba by controlling cheater mutants. Proc. Natl. Acad. Sci. USA 104 8913–8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove, J. E., R. J. Brown and D. J. Watts, 2000. The intracellular target for the antiresorptive aminobisphosphonate drugs in Dictyostelium discoideum is the enzyme farnesyl diphosphate synthase. J. Bone Miner. Res. 15 971–981. [DOI] [PubMed] [Google Scholar]
- Hagedorn, M., K. H. Rohde, D. G. Russell and T. Soldati, 2009. Infection by tubercular mycobacteria is spread by nonlytic ejection from their amoeba hosts. Science 323 1729–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, M. W., M. V. Han and S. G. Han, 2007. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 3 e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E., N. Wang, W. Wu, A. Weatherford, A. De Lozanne et al., 2002. Dictyostelium LvsB mutants model the lysosomal defects associated with Chediak-Higashi syndrome. Mol. Biol. Cell 13 656–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeller, O., and R. R. Kay, 2007. Chemotaxis in the absence of PIP3 gradients. Curr. Biol. 17 813–817. [DOI] [PubMed] [Google Scholar]
- Huang, E. Y., S. L. Blagg, T. Keller, M. Katoh, G. Shaulsky et al., 2006. bZlP transcription factor interactions regulate DIF responses in Dictyostelium. Development 133 449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe, Y., H. Kuwayama, T. Morio, H. Urushihara and Y. Tanaka, 2002. A putative serpentine receptor gene tasA required for normal morphogenesis of primary stalk and branch structure in Polysphondylium pallidum. Gene 285 291–299. [DOI] [PubMed] [Google Scholar]
- Kawabe, Y., T. Morio, J. L. James, A. R. Prescott, Y. Tanaka et al., 2009. Activated cAMP receptors switch encystation into sporulation. Proc. Natl. Acad. Sci. USA 106 7089–7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay, R. R., P. Flatman and C. R. L. Thompson, 1999. DIF signalling and cell fate. Sem. Cell Dev. Biol. 10 577–585. [DOI] [PubMed] [Google Scholar]
- Kay, R. R., P. Langridge, D. Traynor and O. Hoeller, 2008. Changing directions in the study of chemotaxis. Nat. Rev. Mol. Cell Biol. 9 455–463. [DOI] [PubMed] [Google Scholar]
- Keller, T., and C. R. Thompson, 2008. Cell type specificity of a diffusible inducer is determined by a GATA family transcription factor. Development 135 1635–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht, D. A., and W. F. Loomis, 1987. Antisense RNA inactivation of myosin heavy chain gene expression in Dictyostelium discoideum. Science 236 1081–1086. [DOI] [PubMed] [Google Scholar]
- Knecht, D. A., and E. Shelden, 1995. Three-dimensional localization of wild-type and myosin II mutant cells during morphogenesis of Dictyostelium. Dev. Biol. 170 434–444. [DOI] [PubMed] [Google Scholar]
- Kolsch, V., P. G. Charest and R. A. Firtel, 2008. The regulation of cell motility and chemotaxis by phospholipid signaling. J. Cell Sci. 121 551–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll, K. L., and E. Amaya, 1996. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development 122 3173–3183. [DOI] [PubMed] [Google Scholar]
- Kuhlmann, M., B. Popova and W. Nellen, 2006. RNA interference and antisense-mediated gene silencing in Dictyostelium. Methods Mol. Biol. 346 211–226. [DOI] [PubMed] [Google Scholar]
- Kumar, A., P. M. Harrison, K. H. Cheung, N. Lan, N. Echols et al., 2002. An integrated approach for finding overlooked genes in yeast. Nat. Biotechnol. 20 58–63. [DOI] [PubMed] [Google Scholar]
- Kuspa, A., and W. F. Loomis, 1992. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc. Natl. Acad. Sci. USA 89 8803–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenick, J., T. Araki, Y. Yamada and J. G. Williams, 2008. A Dictyostelium homologue of the metazoan Cbl proteins regulates STAT signalling. J. Cell Sci. 200 3524–3530. [DOI] [PubMed] [Google Scholar]
- Lee, S. K., S. L. Yu, H. Alexander and S. Alexander, 1998. A mutation in repB, the Dictyostelium homolog of the human xeroderma pigmentosum B gene, has increased sensitivity to UV-light but normal morphogenesis. Biochim. Biophys. Acta 1399 161–172. [DOI] [PubMed] [Google Scholar]
- Li, G., H. Alexander, N. Schneider and S. Alexander, 2000. Molecular basis for resistance to the anticancer drug cisplatin in Dictyostelium. Microbiology 146 2219–2227. [DOI] [PubMed] [Google Scholar]
- Liu, B. A., K. Jablonowski, M. Raina, M. Arce, T. Pawson et al., 2006. The human and mouse complement of SH2 domain proteins: establishing the boundaries of phosphotyrosine signaling. Mol. Cell 22 851–868. [DOI] [PubMed] [Google Scholar]
- Loomis, W. F., and D. W. Smith, 1990. Molecular phylogeny of Dictyostelium dscoideum by protein sequence comparison. Proc. Natl. Acad. Sci. USA 87 9093–9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty, S., S. Lee, N. Yadava, M. J. Dealy, R. S. Johnson et al., 2001. Regulated protein degradation controls PKA function and cell-type differentiation in Dictyostelium. Genes Dev. 15 1435–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, M. K., A. Clark, T. Abe, A. Nomura, N. Yadava et al., 2000. An F-Box/WD40 repeat-containing protein important for Dictyostelium cell-type proportioning, slug behaviour, and culmination. Dev. Biol. 224 42–59. [DOI] [PubMed] [Google Scholar]
- Olsen, R. M., 2005. How many protein encoding genes does Dictyostelium discoideum have? pp. 265–278 in Dictyostelium Genomics, edited by W. F. Loomis and A. Kuspa. Horizon Bioscience.
- Pawson, T., G. D. Gish and P. Nash, 2001. SH2 domains, interaction modules and cellular wiring. Trends Cell Biol. 11 504–511. [DOI] [PubMed] [Google Scholar]
- Popova, B., M. Kuhlmann, A. Hinas, F. Soderbom and W. Nellen, 2006. HeIF, a putative RNA helicase acts as a nuclear suppressor of RNAi but not antisense mediated gene silencing. Nucleic Acids Res. 34 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, D. N., and J. A. Spudich, 2000. Dynacortin, a genetic link between equatorial contractility and global shape control discovered by library complementation of a Dictyostelium discoideum cytokinesis mutant. J. Cell Biol. 150 823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, T., G. W. Taylor, J. C. Yang, D. Neuhaus, D. Stetsenko et al., 2006. Identification of new differentiation inducing factors from Dictyostelium discoideum. Biochim. Biophys. Acta 1760 754–761. [DOI] [PubMed] [Google Scholar]
- Saito, T., A. Kato and R. R. Kay, 2008. DIF-1 induces the basal disc of the Dictyostelium fruiting body. Dev. Biol. 317 444–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santorelli, L. A., C. R. Thompson, E. Villegas, J. Svetz, C. Dinh et al., 2008. Facultative cheater mutants reveal the genetic complexity of cooperation in social amoebae. Nature 451 1107–1110. [DOI] [PubMed] [Google Scholar]
- Sawai, S., X. J. Guan, A. Kuspa and E. C. Cox, 2007. High-throughput analysis of spatio-temporal dynamics in Dictyostelium. Genome Biol. 8 R144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaap, P., T. Winckler, M. Nelson, E. Alvarez-Curto, B. Elgie et al., 2006. Molecular phylogeny and evolution of morphology in the social amoebas. Science 314 661–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchuk, O., C. Batzilla, S. Hagele, H. Kusch, S. Engelmann et al., 2009. Proteomic analysis of Legionella-containing phagosomes isolated from Dictyostelium. Int. J. Med. Microbiol. 299 489–508. [DOI] [PubMed] [Google Scholar]
- Shimada, N., and T. Kawata, 2007. Evidence that noncoding RNA dutA is a multicopy suppressor of Dictyostelium discoideum STAT protein Dd-STATa. Eukaryot. Cell 6 1030–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, N., N. Kanno-Tanabe, K. Minemura and T. Kawata, 2008. GBF-dependent family genes morphologically suppress the partially active Dictyostelium STATa strain. Dev. Genes Evol. 218 55–68. [DOI] [PubMed] [Google Scholar]
- Skriwan, C., M. Fajardo, S. Hagele, M. Horn, M. Wagner et al., 2002. Various bacterial pathogens and symbionts infect the amoeba Dictyostelium discoideum. Int. J. Med. Microbiol. 291 615–624. [DOI] [PubMed] [Google Scholar]
- Soler-Lopez, M., C. Petosa, M. Fukuzawa, R. Ravelli, J. G. Williams et al., 2004. Structure of an activated Dictyostelium STAT in its DNA-unbound form. Mol. Cell 13 791–804. [DOI] [PubMed] [Google Scholar]
- Solomon, J. M., A. Rupper, J. A. Cardelli and R. R. Isberg, 2000. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect. Immunol. 68 2939–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza, G. M., S. Lu and A. Kuspa, 1998. YakA, a protein kinase required for the transition from growth to development in Dictyostelium. Development 125 2291–2302. [DOI] [PubMed] [Google Scholar]
- Stanley, J. R., and L. Samuel, 2003. Amoebiasis. Lancet 361 1025–1034. [DOI] [PubMed] [Google Scholar]
- Steinert, M., and K. Heuner, 2005. Dictyostelium as host model for pathogenesis. Cell. Microbiol. 7 307–314. [DOI] [PubMed] [Google Scholar]
- Strassmann, J. E., Y. Zhu and D. C. Queller, 2000. Altruism and social cheating in the social amoeba Dictyostelium discoideum. Nature 408 965–967. [DOI] [PubMed] [Google Scholar]
- Tekinay, T., H. L. Ennis, M. Y. Wu, M. Nelson, R. H. Kessin et al., 2003. Genetic interactions of the E3 ubiquitin ligase component FbxA with cyclic AMP metabolism and a histidine kinase signaling pathway during Dictyostelium discoideum development. Eukaryot. Cell 2 618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, C. R. L., and R. R. Kay, 2000. The role of DIF-1 signaling in Dictyostelium development. Mol. Cell 6 1509–1514. [DOI] [PubMed] [Google Scholar]
- Thompson, C. R. L., Q. Fu, C. Buhay, R. R. Kay and G. Shaulsky, 2004. A bZIP/bRLZ transcription factor required for DIF signaling in Dictyostelium. Development 131 513–523. [DOI] [PubMed] [Google Scholar]
- Urushihara, H., T. Morio and Y. Tanaka, 2006. The cDNA sequencing project. Methods Mol. Biol. 346 31–49. [DOI] [PubMed] [Google Scholar]
- Van Haastert, P. J., and D. M. Veltman, 2007. Chemotaxis: navigating by multiple signaling pathways. Sci. STKE 2007 pe40. [DOI] [PubMed] [Google Scholar]
- Wessels, D., T. Srikantha, S. Yi, S. Kuhl, L. Aravind et al., 2006. The Shwachman-Bodian-Diamond syndrome gene encodes an RNA-binding protein that localizes to the pseudopod of Dictyostelium amoebae during chemotaxis. J. Cell Sci. 119 370–379. [DOI] [PubMed] [Google Scholar]
- Williams, J. G., 2003. The STAT proteins of Dictyostelium, pp. 105–121 in Signal Transducers and Activators of Transcription (STATs): Activation and Biology, edited by P. Segal, D. Levy, and T. Hirano, Kluwer Academic Publishers, Dordecht, The Netherlands.
- Williams, R. S. B., M. Eames, W. J. Ryves, J. Viggars and A. J. Harwood, 1999. Loss of a prolyl oligopeptidase confers resistance to lithium by elevation of inositol (1,4,5) trisphosphate. EMBO J. 18 2734–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, R. S., K. Boeckeler, R. Graf, A. Muller-Taubenberger, Z. Li et al., 2006. Towards a molecular understanding of human diseases using Dictyostelium discoideum. Trends Mol. Med. 12 415–424. [DOI] [PubMed] [Google Scholar]
- Zhang, C., and A. Kuspa, 2009. Transcriptional down-regulation and rRNA cleavage in Dictyostelium discoideum mitochondria during Legionella pneumophila infection. PLoS One 4 e5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhukovskaya, N. V., M. Fukuzawa, Y. Yamada, T. Araki and J. G. Williams, 2006. The Dictyostelium bZIP transcription factor DimB regulates prestalk-specific gene expression. Development 133 439–448. [DOI] [PubMed] [Google Scholar]