Abstract
Preventing the formation of dysfunctional telomeres is essential for genomic stability. In most organisms, the ribo-nucleoprotein reverse transcriptase telomerase is responsible for telomere GT-strand elongation. However, in telomerase-negative cells, low-frequency recombination mechanisms can avert lethality by elongating critically short telomeres. This study focuses on the involvement of the budding yeast Mre11 in telomere recombination and homeostasis. We have identified a novel allele of MRE11, mre11-A470T, that, in telomerase-positive cells, confers a semidominant decrease in telomere size and a recessive defect in telomere healing. In addition, mutant cells lack normal telomere size homeostasis. Telomerase-negative mre11-A470T cells display a Rad51-dependent bypass of replicative senescence via induction of a highly efficient type I-related recombination pathway termed type IA. The type IA pathway involves an amplification of subtelomeric Y′ elements, coupled with elongated and more heterogeneous telomere tracts relative to the short telomere size of type I survivors. The data have led us to propose the involvement of break-induced replication in telomere expansion. The differing phenotypes elicited by the mre11-A470T mutants in telomerase-positive and telomerase-negative cells have also led us to speculate that the telomere end structure may be modified differentially in mre11-A470T cells, directing the telomere into specific pathways.
A major function of DNA damage response pathways at the telomere, the nucleoprotein complexes present at chromosomal termini, is to recognize and repair functional or structural defects in telomeres. Such damage includes constitutive single-stranded 3′ overhangs (Polotnianka et al. 1998), short dysfunctional telomeres (Lundblad and Szostak 1989), and promiscuous homologous and nonhomologous telomeric recombination (Pennaneach and Kolodner 2004). A failure to protect against these activities can lead to end-to-end fusions and more complex rearrangements that compromise genomic stability (Pennaneach and Kolodner 2004).
The two major DNA damage response checkpoints in Saccharomyces cerevisiae are the Tel1(ATM)-Mre11 (TM) and the Mec1(ATR)-Ddc1 pathways (Usui et al. 2001). These checkpoint pathways are not necessarily coregulated. For example, under some conditions (e.g., overproduction of Tel1), phosphorylation of Rad53 can be Tel1-dependent and Mec1-independent (Clerici et al. 2001).
Both upstream and downstream points of the TM pathway also control telomere homeostasis. Upstream, the Mre11/Rad50/Xrs2 complex (MRX) and Tel1 bind to telomeric DNA, with Tel1 having a preference for shorter telomeres (Hector et al. 2007; Seidel et al. 2008). MRX is the yeast ortholog of the vertebrate MRN complex. The preference for short telomeres permits the subsequent elongation of telomeres mediated by Tel1 kinase phosphorylation of the single-stranded telomere binding protein Cdc13 (Tseng et al. 2006; Hector et al. 2007), thereby stimulating the association of telomerase components (Goudsouzian et al. 2006). Concurrently, activation of Exo1 nuclease through phosphorylation leads to checkpoint arrest (Morin et al. 2008). In the absence of Exo1, double-strand break processing, at least in nontelomeric ends, takes place through the step-wise MRX initiation of rescission and Sae2 nuclease activity (Zhu et al. 2008). Furthermore, Mre11 is necessary for constitutive 3′ overhang formation (Larrivee et al. 2004). Superimposed on this process is the cell-cycle-regulated phosphorylation of Cdc13 by the cyclin-dependent CDK1 gene that also acts to stabilize telomerase (Li et al. 2009).
The primary pathway for telomere addition in most organisms is the ribo-nucleoprotein reverse transcriptase, telomerase, that uses its RNA component as a template for simple sequence addition. Telomerase also has a binding preference for short telomeres (Bianchi and Shore 2007). However, limited amounts or the absence of telomerase leads to an attrition of telomere tracts that ultimately provokes a Mec1-dependent G2/M arrest (Enomoto et al. 2002; Ijpma and Greider 2003).
Telomerase-negative, but recombination-proficient, yeast cells are able to overcome cell cycle growth arrest predominantly through one of two low-frequency recombination pathways. The type I pathway requires Rad51, Rad52, Rad54, and Rad57 (Lundblad and Blackburn 1993; Le et al. 1999; Teng et al. 2000; Chen et al. 2001; Tsai et al. 2002; Davis and Symington 2004). In haploid cells, this scenario involves break-induced replication (BIR). A single strand of one of two misaligned sister chromatids invades Y′ subtelomeric sequences of either the sister chromatid or a nonhomolog. Subsequent DNA replication to the end of the elongated chromosome completes BIR. The best evidence for this mechanism is the dependence of the type I pathway on the replication factor Pol32 (Lydeard et al. 2007). It is not yet known whether telomeric tracts can undergo the same form of BIR. The RAD52 gene must be continuously present (Lundblad and Blackburn 1993; Teng and Zakian 1999) to maintain type I survivors, an indication of tract loss associated with inviability. Presumably, type I survivors display Y′ amplification and the dual process of regeneration and telomere loss that may, as a consequence, lead to the observed slow growth (Lundblad and Blackburn 1993; Teng and Zakian 1999). Other factors may also be involved in the type I slow-growth phenotype.
The type II pathway in yeast produces a range of elongated telomeres, with sizes up to or >20 kb (Lundblad and Blackburn 1993; Le et al. 1999; Teng et al. 2000) and is the consequence of recombination between telomere sequences per se (Natarajan and McEachern 2002; Groff-Vindman et al. 2005; Cesare et al. 2008). This process may be mediated either through rolling circle replication of telomere circles, possibly excised from t-loops, or through rolling loop replication of the t-loop (Lustig 2003; Tomaska et al. 2004; Cesare et al. 2008). Type II survivors are also likely to be mediated by BIR, given their dependence on Pol32 (Lydeard et al. 2007). Once again, in the absence of the continued presence of Rad52, telomeres begin to senesce (Lundblad and Blackburn 1993; Teng and Zakian 1999).
The type II pathway is also dependent on the Srs2 and Sgs1 helicases that form a complex with Mre11 (Cohen and Sinclair 2001; Signon et al. 2001; Chiolo et al. 2005; Azam et al. 2006). The Sgs1 helicase may have multiple effects on recombination, as measured at nontelomeric sites. Among the roles of Sgs1 is the inhibition of recombination between inexact (homeologous) sequences, possibly including the irregular TG (1-3) yeast telomere repeat (Spell and Jinks-Robertson 2004). However, Sgs1 and Srs2 also influence the accumulation of recombination intermediates and Rad51 filament formation (Krejci et al. 2003; Wu and Hickson 2003; Sugawara et al. 2004; Azam et al. 2006; Schmidt and Kolodner 2006; Seidel et al. 2008).
Mre11 is considered to play an exclusive role in generating type II survivors (Teng et al. 2000). However, most studies have used null alleles of MRE11 that fail to form MRX. These alleles cannot distinguish between the function of MRX and functions specific to Mre11. Indeed, Mre11 influences the intrachromatid telomere rapid deletion process (TRD) in a highly allele-specific fashion (Williams et al. 2005). The specific roles of Mre11 in the regulation of telomere recombination are largely unknown.
Most telomerase-negative cancers utilize an alternative (ALT) pathway that bears striking structural and genetic similarities to the type II pathway. In both yeast and human cells, Southern blots display telomere signals up to at least 20 kb in size. In yeast, type II recombination is dependent on Rad52, the Rad52 paralogue Rad59, MRX, and Tel1 (Chen et al. 2001; Tsai et al. 2002). ALT appears to require at least Rad52, Rad51, and the MRN complex (Wu et al. 2003; Muntoni and Reddel 2005). However, whether the mechanism involves BIR between unequal telomere sister-chromatid exchange, a rolling circle pathway (Nittis et al. 2008), or an as-yet-undiscovered mechanism remains unclear (Morrish and Greider 2009).
We have identified a gain-of-function missense mre11 allele that confers a multitude of phenotypes, including the presence of short telomeres, a loss of telomere homeostasis, and an abrogation of telomere healing. Furthermore, mre11-A470T cells confer the ability to bypass senescence efficiently in a telomerase-negative background. The cause of this bypass appears to be the induction of both a variant of the type I pathway, in which the telomere tracts undergo recombinational elongation, and the type II pathway.
MATERIALS AND METHODS
Strains used in the characterization of mre11-A470T:
All strains used in this study were isogenic to W303. The JPY704 genetic background is identical to W303, with the exception that it lacks a weak hypomorphic rad5 allele present in W303. The mre11-A470T mutant was originally isolated by EMS mutagenesis of the diploid strain BL38 (Li and Lustig 1996), marked with ADE2 and URA3 at the left arm of chromosome VII (VIIL) and the right arm of chromosome V (VR), respectively, and carrying 50% of telomeres in an elongated state. This screen took advantage of TRD as an assay for telomere recombination, while excluding mutants influencing ADE2 expression. Specifically, mutants that decreased telomere size but did not affect telomere silencing were selected. Four isolates were characterized. Of these, only a single nuclear mutation segregated 2:2 after sporulation of heterozygous diploids. This mutation was originally termed telomeric rapid deletion 1 (trd1). Given the tight linkage of both trd1 and mre11 mutations to yKu, we sequenced the trd1 mutation and found it to be an allele (mre11-A470T) of MRE11. A spore colony from a heterozygous TRD1/trd1 diploid strain was backcrossed to W303 three times to yield spore colonies trd1-9c and trd1-2b. The strain trd1-9c (MATa) was mated with W303α, and the resulting diploid was disrupted by one-step gene disruption of one of the two TLC1 genes (Li and Lustig 1996) to create IJ2. The same method was used to construct heterozygous diploids of RAD51, RAD52, LIG4, and SGS1 using PCR-amplified cassettes of yeast strains carrying the rad52 Δ∷kanMX, rad51 Δ∷kanMX, lig4 Δ∷kanMX, or sgs1 Δ∷kanMX mutations. These disruptions gave rise to the heterozygous diploids IJ2rad52, IJ2rad51, IJ2lig4, and IJ2sgs1, respectively. The deletion cassettes were constructed by the Saccharomyces Genome Deletion Project (http://www-sequence.stanford.edu/group/yeast_deletion_project) and purchased from Research Genetics.
The trd1-9c spore colony was used for most mre11-A470T experiments. MRE11/MRE11 and mre11-A470T/mre11-A470T strains were obtained by crossing opposite mating types of W303 and by crossing trd1-9c with trd1-2b, respectively.
Primers:
PCR primers used to identify the MRE11 or mre11-A470T alleles were Mre11-Forward: 5′- GTC AGA GTT CAC AAG CAA GC-3′ and Mre11-Reverse: 5′-CCA TGG TCT CCT CTA TAT CTC-3′. The primer used for sequencing the 470A region of MRE11 was 5′-CGATAAAGATGCTACGTC-3′. The primers used for testing the disruption of TLC1 were TLC Forward: 5′-ACT CGA TGG TGA AGA GAT AGT GTC-3′ and TLC Reverse: 5′-AAA TAT TAA GAG GCA TAC CTC CGC-3′. Primers described in the Saccharomyces deletion database were used to amplify disrupted genes from null alleles generated in the Saccharomyces Genome Deletion Project (http://www-sequence.stanford.edu/group/yeast_deletion_project) for SGS1, LIG4 and RAD52. The KAN B primer was 5′-ATG CAG CGA GGA GCC GTA AT-3′. The chromosome III forward primer, 5′- TTA CTA CCA CCA ACC CAC CGT-3′, and the chromosome III reverse primer, 5′-TCG TTC ATA TAT TCG TTA GCA-3′, were used to amplify a 1053-bp sequence in the subtelomeric region of chromosome IIIL.
Growth assays:
Growth rate:
Freshly dissected spore colonies were grown on YPAD solid medium for 48 hr (s0) and then inoculated into 5 ml YPAD liquid medium and grown for an additional 24 hr. Cells were counted using a hemocytometer, diluted to 1 × 105 cells/ml in 5 ml YPAD, and grown for 24 hr. We performed this serial subculturing daily for 6–9 days after the initial inoculation. The number of cells from each spore colony at the end of each 24-hr cycle was plotted relative to the subculture number. The growth pattern of mre11-A470T tlc1Δ was carried out in four experiments derived from three tetrads at 30°. All of the patterns were qualitatively identical.
Growth capacity:
At the end of each 24-hr subculture, 2 × 107 cells were plated onto 60- × 15-mm YPAD plates. Six hours after plating, random frames of the plates for each spore colony were photographed microscopically using a ×40 objective. The initial number of colonies with greater or fewer than four cells was counted to compare cell division potentials among different genotypes. The number of colonies with more than four cells after 6 hr of growth was plotted relative to the subculture number. The experiment was carried out in two complete tetrads with similar results.
Telomere healing assays:
Telomere healing assays were performed as previously described (Bhattacharyya et al. 2008). The strains used for these experiments were modified by the integration of a GAL-HO gene and by the construction of an HO cut site at a VIIL∷ADE2-marked telomere, as described in Diede and Gottschling (1999). To measure complementation of telomere healing, an MRE11 centromeric plasmid (Sikorski and Hieter 1989) was introduced into trd1-9c cells. Telomere healing was then assayed in both cases.
Data quantification:
We quantified scanned tiff files of Southern and Western blot autoradiograms using both Total Lab (Nonlinear USA) and Quantity One (Bio-Rad) software. The two methods gave identical results. Species were normalized using the first subculturing set arbitrarily at the value of 1. For quantification of Y′ elements, the blots were initially probed with a LEU2 fragment to generate loading and normalization controls. The blot was then stripped and reprobed with nick-translated poly d(GT)/poly d(CA). The intensity of the 6.7 and 5.2 kb Y′ tandem repeats was quantified relative to the intensity of the LEU2 species. In some experiments, we used phosphorimager analysis to quantify heterogeneity.
RESULTS
A mutation of the highly conserved mre11-A470 residue confers a semidominant defect in telomere length:
Through a screen for telomeric recombination mutations (see materials and methods), we identified a novel mutation within the MRE11 gene. This mutation alters an alanine to threonine at amino acid 470 (mre11-A470T). This missense allele did not cause a change in growth rate (data not shown). A phylogenetic comparison of this region demonstrates the ubiquitous presence of the A residue within the context of a 13-amino-acid conserved motif in all species except for dipterans (Figure S1). On the basis of these comparisons, we derived a consensus sequence for the conserved motif.
We found that the telomere length of the mre11-A470T haploid strain is ∼150 bp compared to 300 bp in MRE11 cells. Homozygous mre11-A470T/mre11-A470T diploids recapitulated the short telomere phenotype. Telomere sizes from MRE11/mre11-A470T heterozygotes, however, migrated at a position between mre11-A470T and MRE11 cells, indicating a partial dominance (Figure 1, compare lanes 6 and 7 to lanes 3 and 10). These data suggest the presence of a gain- or separation-of-function allele with respect to telomere size, as opposed to a hypomorph.
Figure 1.—
mre11-A470T is semidominant. XhoI digests of DNA from haploid MRE11 cells, mre11-A470T cells, or diploids created by crosses between the strains and subsequently subjected to Southern analysis using poly d(GT)/poly d(CA) as the probe. The genotypes in lanes 1–12 are given on the left. Lanes 1 and 5, 2 and 3, and 6 and 7 were duplicate samples. Horizontal bars on the left indicate the position of size markers in the following order (top to bottom): 5, 4, 3, 2, 1.5, and 1.0 kb. The map below depicts a standard Y′-class telomere showing the position of the conserved XhoI site relative to the junction of the telomeric tract (0.87 kb) and the variable size of the telomeric fragment (0.87+).
Telomere heterogeneity in mre11-A470T cells:
Another barometer of telomere structure in wild-type and mutant cells is telomere homeostasis, that is, the degree by which telomeres are maintained around a specific mean size. The extent of telomeric repeat tract variability in yeast has been correlated to its mean length. Cells with larger average repeat tract sizes have variances around that size that are larger than cells with smaller tract sizes (e.g., tel1, tel2, wt, rif1, rif1rif2, and rap1-17, in ascending order of mean size and heterogeneity) (Lustig and Petes 1986; Kyrion et al. 1992; Wotton and Shore 1997). Since the average telomeric repeat tract size in mre11-A470T cells is about half that of MRE11 cells, we expected a decrease in heterogeneity. However, this correlation did not hold in mre11-A470T cells. Despite the significant difference in average size between MRE11 and mre11-A470T cells, the range of telomere sizes are similarly dispersed around the mean. Specifically, Southern analysis of the heterogeneity of telomere sizes in a large sample of independent cultures revealed that the median heterogeneity of the telomere Y′ class is comparable in mre11-A470T cells (325 ± 48, n = 36) compared to wild type (300 ± 59 bp, n = 28), although the mean telomere size is one-half that of wild-type telomeres. Hence, mre11-A470T alters the precision of telomere size homeostasis.
mre11-A470T abrogates telomere healing:
We also found that mre11-A470T cells abrogate telomere healing in nocodazole-arrested cells, an assay for telomerase healing activity (Diede and Gottschling 1999) (Figure 2). In MRE11 cells, after generation of HO cleavage by galactose induction of a plasmid-borne HO gene and growth in nocodazole for 20 hr to attain a G2/M arrest, most of the telomeric seed sequences (TSS) had served as substrate for telomerase. This loss of the ability to extend the telomeric seeds is a recessive trait. The mre11-A470T mutant transformed with plasmid-borne MRE11 also displayed wild-type healing (data not shown). In contrast, HO breaks in the presence of telomere seed sequences cannot serve as substrate for the addition of telomere sequences in mre11Δ or mre11-A470T cells. These data suggest that cells harboring the mre11-A470T allele have lost the ability either to detect or to process freshly exposed telomere seed sequences.
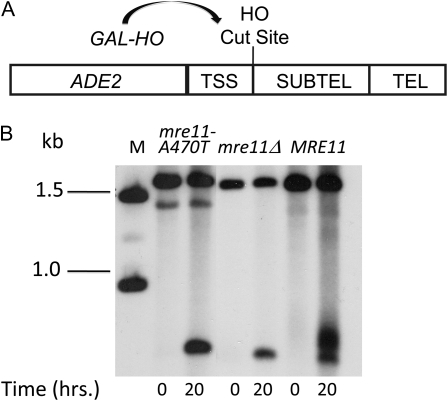
Figure 2.—
Telomere healing is abrogated in mre11-A470T cells. (A) Strains designed as described in materials and methods contain (telomere distal to proximal) the ADE2 marker, a telomere seed sequence (TSS), an HO cut site, subtelomeric sequences (SUBTEL), and telomeric sequences (TEL) (Diede and Gottschling 1999). In these cells, the HO-encoded endonuclease is under the control of a galactose-regulated HO endonuclease promoter (GAL-HO). (B) Southern blotting of SpeI-digested DNA using ADE2 as a probe was used to determine the cleavage of the recognition site and the addition of telomere repeats to the telomeric seed sequence. Southern blots were performed for strains that carried mre11-A470T, mre11Δ, or MRE11 alleles as shown on the top of the gel. M refers to the size marker lane. The image is derived from one gel that is spliced between the A470T-20 hr and mre11Δ-0 hr. The cells were synchronized in nocodazole and shifted to galactose for the time indicated on the bottom of the gel (in hr). Only MRE11 and mre11-A470T cells transformed with plasmid-borne MRE11 (data not shown) displayed full healing activity.
mre11-A470T bypasses tlc1Δ-induced senescence:
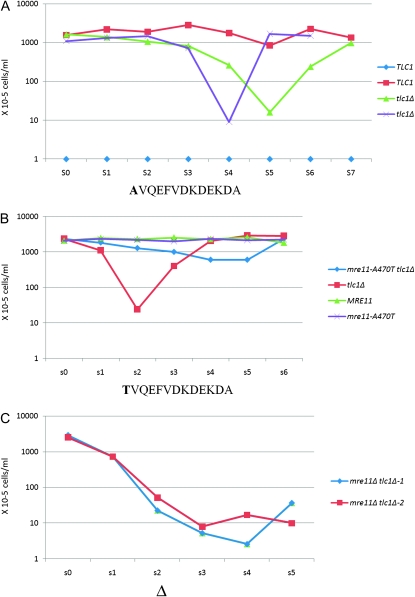
To test the association between mre11-A470T and the telomerase pathway, we analyzed mre11-A470T growth characteristics in the absence of telomerase RNA (TLC1). Specifically, we compared the phenotypes of MRE11, tlc1Δ, mre11-A470T tlc1Δ, and mre11-A470T cells. Growth characteristics were analyzed by three established methods. First, we determined the growth rate during repetitive liquid subculturing for specified periods after sporulation of the relevant diploid. Individual tetrads were analyzed because the spore products displayed a more highly reproducible pattern than between random spore colonies. However, all tetrads and tlc1Δ spore colonies displayed a qualitatively, although not a quantitatively, identical pattern of growth. Second, we ascertained the number of single cells capable of continued growth beyond 6 hr after each subculture (“growth capacity”). Finally, we verified these findings by clonal subculturing of average-sized colonies on solid medium (Figure S2) (Lustig and Petes 1986).
The liquid subculturing growth assay of mre11-A470T tlc1Δ cells displayed only a small decrease in growth rate relative to MRE11 cells (Figure 3B). This finding differs from the replicative senescence of tlc1Δ cells after 40–50 population doublings (Figure 3A). Further rounds of subculturing led to an increase in the growth rate of the tlc1Δ single mutant due to the accumulation of type I and type II recombinant survivors (Lundblad and Blackburn 1993) (Figure 3, A and B). Growth capacity assays revealed that MRE11 tlc1Δ colonies formed a high percentage of cells that were incapable of further growth beyond the four-cell stage as senescence progressed (data not shown). In contrast, the mre11-A470T tlc1 double mutants displayed a greater number of cells that were capable of growth beyond the four-cell stage. Similarly, we observed only a small decrease in growth rate relative to wild-type cells in solid subculturing of mre11-A470T (Figure S2), even at the minimal point of tlc1Δ growth. Thus, by all three criteria, the mre11-A470T mutation displayed a bypass of tlc1Δ senescence. This is not due to a loss of Mre11 function because mre11Δ tlc1Δ strains underwent the typical slow loss of growth previously observed in rad50 tlc1Δ mutants (Figure 3C) (Le et al. 1999).
Figure 3.—
Bypass of senescence in mre11-A470T tlc1Δ cells. (A) Liquid subculturing of the TLC1 and tlc1Δ spore colonies from a representative tetrad derived from a diploid heterozygous for tlc1Δ. Liquid subculturing was carried out for seven rounds (s0–s7). Three tlc1Δ spore colonies were examined. Two are shown in A and one in B. (B) Six rounds of liquid subculturing (s0–s6) of the MRE11, mre11-A470T tlc1Δ, mre11-A470T, and tlc1Δ spore colonies of a representative tetrad of IJ2 (a diploid strain heterozygous for mre11-A470T and tlc1Δ). Four mre11-A470T tlc1Δ trials from independent tetrads were analyzed. (C) Liquid subculturing of an mre11Δ tlc1Δ strain was conducted for five rounds (s0–s5), displaying the typical slow senescent pattern observed previously in this strain. Two spore colonies were analyzed from independent tetrads. Each subculturing represents 24 hr of growth. To reduce artifacts of stochastic variation in the kinetics of cells approaching senescence, only the products of a single complete tetrad are shown in A and B. The x-axis refers to the rounds of subculturing, with each round representing 24 hr of growth.
Suppression of senescence in mre11-A470T tlc1Δ cells is correlated with increased telomere tract size and heterogeneity:
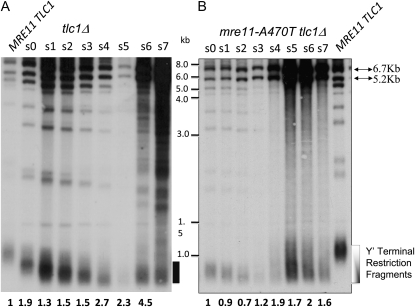
In tlc1Δ strains proficient for recombination, survivors of tlc1Δ senescence undergo type I and type II recombination between subtelomeric and telomeric regions, respectively. As noted, type I cells grow slowly and are ultimately overtaken by type II recombinants (Lundblad and Blackburn 1993). To test whether suppression of senescence and the ensuing telomere maintenance in mre11-A470T tlc1Δ cells may be the consequence of type I and/or type II recombination mechanisms, we conducted Southern analysis of DNA derived from liquid-subcutured mre11-A470T tlc1Δ cells. These experiments revealed the presence of a recombinant, termed type IA, that bears similarities to type I recombinants, with the exception of a heterogeneous slower migrating telomeric pattern (Figure 4). Type IA recombinants displayed a modest two- to eightfold amplification of Y′ elements, but did not display the exclusively short telomeres associated with type I recombination. Rather, we observed an increase in median telomere tract size and an expanded distribution of sizes around the median, with telomere tracts reaching MRE11 (300 bp) length (Figure 4B). These data suggest that homeostatic regulation of telomere size has been altered in type IA recombinants.
Figure 4.—
A novel recombinant is associated with the bypass of senescence. (A) Southern analysis of DNA isolated from subcultured tlc1Δ or an MRE11 TLC1 control strain after digestion with XhoI and hybridization, using poly d(GT)/d(CA) as the telomeric probe. The degree of subculturing (s0–s7) is shown above the relevant lanes of the gel. (B) Southern analysis of DNA isolated from a subcultured (s0–s7) mre11-A470T tlc1Δ spore colony or an MRE11 TLC1 control strain carried out as described above. A size ladder (in kilobases) is at the left of the gel. Drawing to the right of each gel depicts the distributions exhibited by type 1 (A) and type IA (B). The numbers below the blots reflect the ratio of Y′ repeat hybridization signal to a single-copy LEU2 probe normalized to a value of 1.0 for the first subculture. Arrows on the right in B indicate the positions of the subtelomeric Y′ repeats.
Type IA telomere tract expansion may be restricted to Y′ class telomeres, or it may reflect a more general phenomenon influencing all telomeres. To distinguish between these possibilities, we examined whether the chromosome III left arm (cIIIL) non-Y′ telomere became larger and more heterogeneous in mre11-A470T tlc1Δ cells during liquid subculturing (Figure S3). Of the three individual clones analyzed, one displayed a diffuse cIIIL telomere tract distribution as shown, with the others involved in more complex rearrangements. These data suggest that telomere heterogeneity in mutant cells is not restricted to Y′ subtelomeric elements.
Several lines of evidence argue that the diffuse telomeric distribution and increase in mean size in type IA events are related mechanistically to type I recombination. First, we were unable to identify any cultures containing a Y′ amplification that did not also contain a diffuse telomeric distribution in mre11-A470T tlc1Δ cells. Second, the two-to-eight fold amplification of Y′ elements, typical of strain W303, coincides temporally with heterogeneous telomeres during liquid subculturing (Figure 4B). This is not an artifactual consequence of variations in the amount of DNA loaded because even underloaded samples displayed a clearly diffuse distribution (Figure 4B, s4). Third, cells containing these recombinants grow at wild-type rates. This finding is consistent with the presence of an equilibrium between functional and dysfunctional telomeres near the minimal functional telomere size in type I survivors, an effect that could be substantially reduced in type IA recombinants (Lundblad and Blackburn 1993; Teng and Zakian 1999).
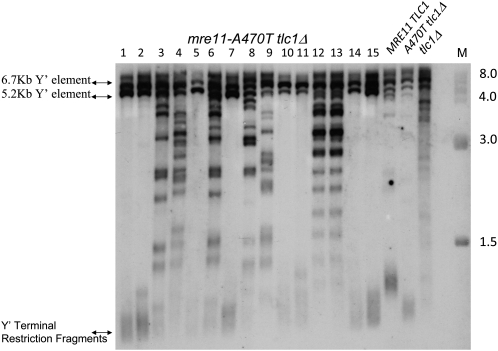
Southern analysis of DNA derived from type IA recombinants revealed a diffuse background, suggesting the simultaneous presence of type II recombinants. To test for the presence of type II recombinants in mre11-A470T tlc1Δ cells, we plated cells from cultures late in subculturing. Of 15 cells plated from such subcultures (Figure 5, lanes 1–15), eight colonies displayed the type IA pattern (lanes 1, 2, 5, 7, 10, 11, 14, 15), while seven colonies (lanes 3, 4, 6, 8, 9, 12, 13) were typical of type II survivors (Figure 5). The ratio of type IA to type II differs from the ratio of type I to type II survivors of MRE11 tlc1Δ senescing cells. In the latter case, type I survivors are overtaken by low-frequency, but rapidly growing, type II survivors.
Figure 5.—
Clonal products of liquid-subcultured type IA cells. Southern blot of DNA isolated from 15 subclones of an mre11-A470T tlc1Δ spore colony subcultured for four rounds were plated onto YPD medium, and probed with poly d(GT)/poly d(CA). MRE11 TLC1, A470T tlc1Δ, and tlc1Δ were used as control strains. The strain or subclone number is presented above the blot and size markers are presented to the right. The distribution of telomere sizes normally found in this strain and the positions of the Y′ elements are shown on the left.
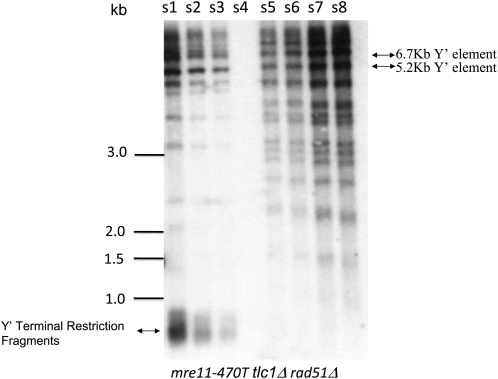
To assess more clearly the link between type I and type IA, we analyzed the products generated during senescence of mre11-A470T tlc1Δ mutants in a rad51Δ background (Figure 6). The presence of a similar mechanism leading to Y′ amplification and telomere heterogeneity predicts a common dependence on Rad51. We found two distinct phenotypes in rad51Δ mre11-A470T tlc1Δ cells in the spore products of the three tetrads that were tested. In spore colonies from two tetrads, rad51Δ mre11-A470T tlc1Δ spore colonies gave rise to type II, but not type IA, recombinants following senescence (e.g., Figure 6). In the rad51Δ mre11-A470T tlc1Δ spore colony from a third tetrad, growth of the spore colony led to an irreversible loss of viability (data not shown). Similarly, the diffuse telomere distribution was not present in the short-lived mre11-A470T rad52 mutants (data not shown). The data demonstrate that both Rad51 and Rad52 are required for the generation of the type IA species and reaffirm that these forms are the consequence of homologous recombination. Consistent with the exclusive role of homologous recombination in the bypass phenomenon, a null mutation in the LIG4 gene encoding the DNA ligase required for nonhomologous end joining has no effect on the formation of survivors (data not shown).
Figure 6.—
Formation of the type IA recombinant is Rad51-dependent. Southern analysis of DNA isolated from mre11-A470T tlc1Δ rad51Δ spore colonies derived from the diploid IJ2rad51 that were subcultured in liquid media for seven rounds (s1–s8) and probed with the telomeric probe poly d(GT)/poly d(CA). Cells senesced, accounting for the low level of DNA in lane s4, followed by the accumulation of type II survivors. Size markers and position of the telomeres are shown to the left of the blot. The positions of Y′ repeat fragments are shown on the right.
The effect of Sgs1 on telomere heterogeneity in mre11-A470T tlc1Δ cells:
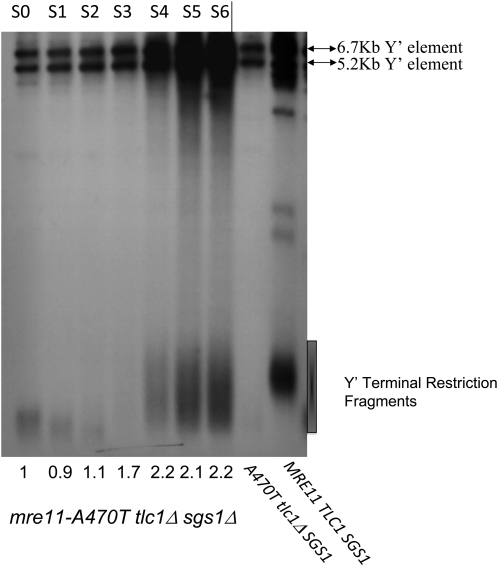
The Sgs1 helicase is involved in recombination as (a) an effector capable of attenuating the resolution of replication intermediates (Lee et al. 2007), (b) a facilitator of Rad51 filament formation (Krejci et al. 2003), and (c) an inhibitor of recombination between imperfect repeats (Spell and Jinks-Robertson 2004). Such imperfect repeats are, of course, present in the irregular telomere tracts of S. cerevisiae. Sgs1 is also required for type II recombination. Indeed, a role in mismatch repair and in telomere processing may be related (Cohen and Sinclair 2001; Azam et al. 2006; Bonetti et al. 2009). The triple mutant mre11-A470T sgs1Δ tlc1Δ conferred type IA events that display a small but uniform increase in heterogeneity (Figure 7) relative to mre11-A470T tlc1Δ cells, with some telomeres exceeding wild-type size. These data raise the possibility of a negative role for Sgs1 in type IA recombination.
Figure 7.—
Variability of telomere size increase in mre11-A470T strains lacking Sgs1. Southern analysis of DNA isolated either from mre11-A470T tlc1Δ sgs1Δ spore colonies after six rounds (s0–s6) of subculturing or from A470T tlc1Δ SGS1 and MRE11 TLC1 SGS1 control strains and probed with poly d(GT)/poly d(CA). Arrows on the right indicate the position of the Y′ repeats. Cells accumulated type IA survivors at high frequencies with uniform increases in heterogeneity greater than those found in mre11-A470T cells. The drawing to the right of the gel represents the distribution of telomere sizes normally found in this strain. The numbers below the blots reflect the ratio of the Y′ repeat hybridization signal to a single-copy LEU2 probe normalized to a value of 1.0 for the first subculture.
DISCUSSION
In this article, we report multiple striking effects of the mre11-A470T allele. These phenotypes provide an initial insight into the recombinational mechanism(s) of Mre11. To summarize our findings on telomerase-positive cells, we observed the following: (a) short telomeric tracts, (b) a failure to maintain the coupled relationship between telomere tract length and heterogeneity, and (c) the abrogation of telomerase-dependent telomere healing. In contrast, in telomerase-negative cells, we observed an efficient bypass of senescence and a concomitant efficient Rad51-dependent recombinational expansion of the telomeric repeat tracts. This amplification is coupled with a high efficiency of type II recombination. These type II events take place even in the presence of a mutant allele (mre11-A470T) of MRE11. The distribution of type IA telomeres appears to be expanded further in a null allele of SGS1.
The short telomere phenotype is unlikely to reflect a loss of function identical to the loss in the mre11Δ allele. The phenotype of the latter allele would not explain the gain of function resulting in shorter telomeres that was observed in the mre11-A470T allele. These data suggest two possibilities: (a) a deactivation or interference with telomerase or (b) a process that may be characteristic of mre11-A470T, e.g., the activation of a recombination-based pathway for telomere elongation. Although a telomerase-based mechanism is likely, we cannot yet rule out the activation of a second pathway related to the activity of the telomerase-negative phenotypes.
The alteration in the length of the telomere, relative to the distribution of the heterogeneous signal that flanks the mean size, is more curious. These data now provide the opportunity to separate these two functions of telomere sizing. The genetic separation of functions regulating the mean and the pattern of heterogeneity will allow us to selectively influence the heterogeneity pathway and determine the role, if any, of recombination in this process.
Telomere healing is dependent on Tel1 (Frank et al. 2006; Bhattacharyya et al. 2008) and abrogated by the mre11-A470T allele. The mutant mre11-A470T could act at an upstream point in substrate recognition. More likely, however, the known resection initiation activity of Mre11 in conjunction with Sae2 at internal sites (Zhu et al. 2008) may be involved in CA-strand resection. These activities may produce sufficient single-strand sequence for efficient telomerase binding and activity.
Current data on Rad53 phosphorylation in response to MMS damage indicate that the vast majority of phosphorylation in MRE11 and mre11-A470T is dependent on Mec1 (data not shown). Experiments that measure telomeric damage specifically are necessary to draw any further conclusions.
One unifying explanation for these various phenotypes is the presence of potential differences in terminal end structure in mre11-A470T in telomerase-positive and telomerase-negative cells. For example, the resection activity of Mre11 that precedes the nuclease action of Sae2 in exo1Δ cells may vary in mre11-A470T cells (if telomeres have similar characteristics as internal breaks) (Zhu et al. 2008). Indeed, loss of the endonuclease Exo1 lessens the impact of telomerase loss and increases the efficiency of both type I and type II recombination (Bertuch and Lundblad 2004). Further analysis is required to determine whether exo1 mutations act synergistically with mre11-A470T on rescuing tlc1 senescence. Alternatively, mre11-A470T may exert its action through a change in the length of the terminal single strand in mre11-A470T cells (Larrivee et al. 2004). Although it is known that short telomere sequences improve the efficiency of telomerase in yeast (Bianchi and Shore 2007; Hector et al. 2007) and recombination in vertebrates (Morrish and Greider 2009), any preferential recombinational end structure produced by mre11-A470T should normally be limiting or absent in telomerase-plus cells.
In the telomerase-negative cells, it is highly likely that the mre11-A470T-induced bypass of senescence is due to an efficient type IA recombination pathway that increases the heterogeneity of the Y′ (or chromosome IIIL) telomere tracts. This probably explains retention of the wild-type growth rate in the mre11-A470T telomerase-negative cells. Classic type I amplification is the consequence of Rad51-dependent break-induced replication, requiring both unidirectional strand invasion and replicative expansion to the chromosomal terminus (Davis and Symington 2004; Lydeard et al. 2007). We propose that the expansion of telomere tracts in type IA recombination similarly is due to break-induced replication within the telomere tract itself. We note that the presence of subtelomeric Y′ elements is not necessary for this process, strengthening the argument for recombination between telomeres per se. After several rounds of unequal sister-chromatid alignment, single-strand invasion and replication should produce a significant population of elongated telomeres. The apparent absence of type IA recombinants during normal telomere attrition suggests that MRE11 normally restricts this pathway. This expansion of the telomere tract may involve release of constraints placed on homeologous strand exchange, which is consistent with the effect of Sgs1 loss in cells carrying the mre11-A470T allele. Sgs1 has been noted, at least at internal sites, to act as a repressor of homeologous recombination, raising the possibility that both mre11-A470T and sgs1Δ (or mutations in mismatch repair pathways) allow recombination under conditions of lower homology.
One potentially unique characteristic of BIR-mediated recombination is the ability to test telomere sizes relative to one another, leading to an overall telomere homeostasis. This is particularly true of interactions among nonhomologous chromosomes. Type IA BIR may play an ancillary role, even in telomerase-positive cells, in compensating for changes in the sizes of individual telomeres by replicating to the end of heterologous chromosomes.
An alternative to BIR-mediated recombination is the ability of simple-sequence tracts to recombine at short regions of tract homology. Regions of homology could exist through the presence of adjacent conserved Rap1 binding sites. Such a short tract homology system has been demonstrated in yeast (Ira and Haber 2002). Additionally, we cannot formally exclude the presence of unequal reciprocal exchanges in mre11-A470T cells.
The unexpected finding of multiple recombinant forms in clonal subcultures (i.e., type IA and type II) supports the assertion that a single defect allows high efficiencies for both recombinant type I and type II. Indeed, these data also suggest that different termini in a single cell can carry out either type IA or type II recombination. It remains to be seen whether the enhancement of recombination is due to a global or telomeric increase in BIR and whether BIR activity may act through unequal sister-chromatid recombination to amplify repeats in higher eukaryotic ALT cells (Muntoni and Reddel 2005).
Acknowledgments
We thank Prescott Deininger, Titia de Lange, Raymund Wellinger, Kametra Matthews, Melanie Cross, and E. B. Hoffman for critical reading of the manuscript and Mark Tidwell, Greg McCarty, and Kametra Matthews for technical assistance. These studies were funded by National Institutes of Health grant R01 GM069943 (to A.J.L.) and the Louisiana Cancer Research Consortium (to M.K.B., A.K., and A.J.L.).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.117598/DC1.
References
- Azam, M., J. Y. Lee, V. Abraham, R. Chanoux, K. A. Schoenly et al., 2006. Evidence that the S. cerevisiae Sgs1 protein facilitates recombinational repair of telomeres during senescence. Nucleic Acids Res. 34 506–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertuch, A. A., and V. Lundblad, 2004. EXO1 contributes to telomere maintenance in both telomerase-proficient and telomerase-deficient Saccharomyces cerevisiae. Genetics 166 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya, M. K., K. M. Matthews and A. J. Lustig, 2008. Mre11 nuclease and C-terminal tail-mediated DDR functions are required for initiating yeast telomere healing. Chromosoma 117 357–366. [DOI] [PubMed] [Google Scholar]
- Bianchi, A., and D. Shore, 2007. Increased association of telomerase with short telomeres in yeast. Genes Dev. 21 1726–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti, D., M. Martina, M. Clerici, G. Lucchini and M. P. Longhese, 2009. Multiple pathways regulate 3′ overhang generation at S. cerevisiae telomeres. Mol. Cell 35 70–81. [DOI] [PubMed] [Google Scholar]
- Cesare, A. J., C. Groff-Vindman, S. A. Compton, M. J. McEachern and J. D. Griffith, 2008. Telomere loops and homologous recombination-dependent telomeric circles in a Kluyveromyces lactis telomere mutant strain. Mol. Cell. Biol. 28 20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Q., A. Ijpma and C. W. Greider, 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21 1819–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo, I., W. Carotenuto, G. Maffioletti, J. H. Petrini, M. Foiani et al., 2005. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol. Cell. Biol 25 5738–5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici, M., V. Paciotti, V. Baldo, M. Romano, G. Lucchini et al., 2001. Hyperactivation of the yeast DNA damage checkpoint by TEL1 and DDC2 overexpression. EMBO J. 20 6485–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, H., and D. A. Sinclair, 2001. Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc. Natl. Acad. Sci. USA 98 3174–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, A. P., and L. S. Symington, 2004. RAD51-dependent break-induced replication in yeast. Mol. Cell. Biol. 24 2344–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede, S. J., and D. E. Gottschling, 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99 723–733. [DOI] [PubMed] [Google Scholar]
- Enomoto, S., L. Glowczewski and J. Berman, 2002. MEC3, MEC1, and DDC2 are essential components of a telomere checkpoint pathway required for cell cycle arrest during senescence in Saccharomyces cerevisiae. Mol. Biol. Cell 13 2626–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, C. J., M. Hyde and C. W. Greider, 2006. Regulation of telomere elongation by the cyclin-dependent kinase CDK1. Mol. Cell 24 423–432. [DOI] [PubMed] [Google Scholar]
- Goudsouzian, L. K., C. T. Tuzon and V. A. Zakian, 2006. S. cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol. Cell 24 603–610. [DOI] [PubMed] [Google Scholar]
- Groff-Vindman, C., A. J. Cesare, S. Natarajan, J. D. Griffith and M. J. McEachern, 2005. Recombination at long mutant telomeres produces tiny single- and double-stranded telomeric circles. Mol. Cell. Biol. 25 4406–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector, R. E., R. L. Shtofman, A. Ray, B. R. Chen, T. Nyun et al., 2007. Tel1p preferentially associates with short telomeres to stimulate their elongation. Mol. Cell 27 851–858. [DOI] [PubMed] [Google Scholar]
- Ijpma, A., and C. W. Greider, 2003. Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol. Biol. Cell 14 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira, G., and J. E. Haber, 2002. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 22 6384–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci, L., S. Van Komen, Y. Li, J. Villemain, M. S. Reddy et al., 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423 305–309. [DOI] [PubMed] [Google Scholar]
- Kyrion, G., K. A. Boakye and A. J. Lustig, 1992. C-terminal truncation of RAP1 results in the deregulation of telomere size, stability, and function in Saccharomyces cerevisiae. Mol. Cell. Biol. 12 5159–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrivee, M., C. LeBel and R. J. Wellinger, 2004. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 18 1391–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, S., J. K. Moore, J. E. Haber and C. W. Greider, 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. Y., M. Kozak, J. D. Martin, E. Pennock and F. B. Johnson, 2007. Evidence that a RecQ helicase slows senescence by resolving recombining telomeres. PLoS Biol. 5 e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B., and A. J. Lustig, 1996. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 10 1310–1326. [DOI] [PubMed] [Google Scholar]
- Li, S., S. Makovets, T. Matsuguchi, J. D. Blethrow, K. M. Shokat et al., 2009. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell 136 50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad, V., and E. H. Blackburn, 1993. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell 73 347–360. [DOI] [PubMed] [Google Scholar]
- Lundblad, V., and J. W. Szostak, 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57 633–643. [DOI] [PubMed] [Google Scholar]
- Lustig, A. J., 2003. Clues to catastrophic telomere loss in mammals from yeast telomere rapid deletion. Nat. Rev. Genet. 9 916–923. [DOI] [PubMed] [Google Scholar]
- Lustig, A. J., and T. D. Petes, 1986. Identification of yeast mutants with altered telomere structure. Proc. Natl. Acad. Sci. USA 83 1398–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydeard, J. R., S. Jain, M. Yamaguchi and J. E. Haber, 2007. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448 820–823. [DOI] [PubMed] [Google Scholar]
- Morin, I., H. P. Ngo, A. Greenall, M. K. Zubko, N. Morrice et al., 2008. Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J. 27 2400–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrish, T. A., and C. W. Greider, 2009. Short telomeres initiate telomere recombination in primary and tumor cells. PLoS Genet. 5 e1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntoni, A., and R. R. Reddel, 2005. The first molecular details of ALT in human tumor cells. Hum. Mol. Genet. 14 Spec No. 2: R191–R196. [DOI] [PubMed] [Google Scholar]
- Natarajan, S., and M. J. McEachern, 2002. Recombinational telomere elongation promoted by DNA circles. Mol. Cell. Biol. 22 4512–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nittis, T., L. Guittat and S. A. Stewart, 2008. Alternative lengthening of telomeres (ALT) and chromatin: Is there a connection? Biochimie 90 5–12. [DOI] [PubMed] [Google Scholar]
- Pennaneach, V., and R. D. Kolodner, 2004. Recombination and the Tel1 and Mec1 checkpoints differentially effect genome rearrangements driven by telomere dysfunction in yeast. Nat. Genet. 36 612–617. [DOI] [PubMed] [Google Scholar]
- Polotnianka, R. M., J. Li and A. J. Lustig, 1998. The yeast Ku heterodimer is essential for protection of the telomere against nucleolytic and recombinational activities. Curr. Biol. 8 831–834. [DOI] [PubMed] [Google Scholar]
- Schmidt, K. H., and R. D. Kolodner, 2006. Suppression of spontaneous genome rearrangements in yeast DNA helicase mutants. Proc. Natl. Acad. Sci. USA 103 18196–18201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel, J. J., C. M. Anderson and E. H. Blackburn, 2008. A novel Tel1/ATM N-terminal motif, TAN, is essential for telomere length maintenance and a DNA damage response. Mol. Cell. Biol. 28 5736–5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signon, L., A. Malkova, M. L. Naylor, H. Klein and J. E. Haber, 2001. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol. 21 2048–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spell, R. M., and S. Jinks-Robertson, 2004. Examination of the roles of Sgs1 and Srs2 helicases in the enforcement of recombination fidelity in Saccharomyces cerevisiae. Genetics 168 1855–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara, N., T. Goldfarb, B. Studamire, E. Alani and J. E. Haber, 2004. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc. Natl. Acad. Sci. USA 101 9315–9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, S. C., and V. A. Zakian, 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19 8083–8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, S., J. Chang, B. McCowan and V. A. Zakian, 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, rif-inhibited recombinational process. Mol. Cell 6 947–952. [DOI] [PubMed] [Google Scholar]
- Tomaska, L., M. J. McEachern and J. Nosek, 2004. Alternatives to telomerase: keeping linear chromosomes via telomeric circles. FEBS Lett. 567 142–146. [DOI] [PubMed] [Google Scholar]
- Tsai, Y. L., S. F. Tseng, S. H. Chang, C. C. Lin and S. C. Teng, 2002. Involvement of replicative polymerases, Tel1p, Mec1p, Cdc13p, and the Ku complex in telomere-telomere recombination. Mol. Cell. Biol. 22 5679–5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng, S. F., J. J. Lin and S. C. Teng, 2006. The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res. 34 6327–6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui, T., H. Ogawa and J. H. Petrini, 2001. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7 1255–1266. [DOI] [PubMed] [Google Scholar]
- Williams, B., M. K. Bhattacharyya and A. J. Lustig, 2005. Mre11p nuclease activity is dispensable for telomeric rapid deletion. DNA Repair (Amst.) 4 994–1005. [DOI] [PubMed] [Google Scholar]
- Wotton, D., and D. Shore, 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11 748–760. [DOI] [PubMed] [Google Scholar]
- Wu, G., X. Jiang, W. H. Lee and P. L. Chen, 2003. Assembly of functional ALT-associated promyelocytic leukemia bodies requires Nijmegen breakage syndrome 1. Cancer Res. 63 2589–2595. [PubMed] [Google Scholar]
- Wu, L., and I. D. Hickson, 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426 870–874. [DOI] [PubMed] [Google Scholar]
- Zhu, Z., W. H. Chung, E. Y. Shim, S. E. Lee and G. Ira, 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]