Abstract
Autophagy is an evolutionarily conserved degradative pathway that has been implicated in a number of physiological events important for human health. This process was originally identified as a response to nutrient deprivation and is thought to serve in a recycling capacity during periods of nutritional stress. Autophagy activity appears to be highly regulated and multiple signaling pathways are known to target a complex of proteins that contains the Atg1 protein kinase. The data here extend these observations and identify a particular phosphorylation event on Atg1 as a potential control point within the autophagy pathway in Saccharomyces cerevisiae. This phosphorylation occurs at a threonine residue, T226, within the Atg1 activation loop that is conserved in all Atg1 orthologs. Replacing this threonine with a nonphosphorylatable residue resulted in a loss of Atg1 protein kinase activity and a failure to induce autophagy. This phosphorylation required the presence of a functional Atg1 kinase domain and two known regulators of Atg1 activity, Atg13 and Atg17. Interestingly, the levels of this modification were found to increase dramatically upon exposure to conditions that induce autophagy. In addition, T226 phosphorylation was associated with an autophosphorylated form of Atg1 that was found specifically in cells undergoing the autophagy process. In all, these data suggest that autophosphorylation within the Atg1 activation loop may represent a point of regulatory control for this degradative process.
MACROAUTOPHAGY (hereafter referred to as autophagy) is a highly conserved process of self-degradation that is essential for cell survival during periods of nutrient limitation (Tsukada and Ohsumi 1993). During autophagy, a double membrane grows out from a specific nucleation site, known as the pre-autophagosomal structure, or PAS, in Saccharomyces cerevisiae and the phagophore assembly site in mammals (Suzuki and Ohsumi 2007). This membrane encapsulates bulk protein and other constituents of the cytoplasm and ultimately targets this material to the vacuole/lysosome for degradation (Xie and Klionsky 2007). Recent studies have linked this pathway to a number of processes important for human health, including tumor suppression, innate immunity, and neurological disorders, like Huntington's disease (Rubinsztein et al. 2007; Levine and Kroemer 2008). Determining how this pathway is regulated is therefore important for our understanding of these processes and our attempts to manipulate autophagy in clinically beneficial ways.
Most of the molecular components of the autophagy pathway were initially characterized in the budding yeast, S. cerevisiae, but orthologs of many of these Atg proteins have since been found in other eukaryotes (Tsukada and Ohsumi 1993; Meijer et al. 2007). A complex of proteins that contains the Atg1 protein kinase is of special interest and appears to be a key point of regulatory control within this pathway (Kamada et al. 2000; Budovskaya et al. 2005; He and Klionsky 2009; Stephan et al. 2009). In S. cerevisiae, genetic and biochemical data indicate that this complex is targeted by at least three different signaling pathways. Two of these pathways, involving the Tor and cAMP-dependent protein kinases, inhibit this process, whereas the AMP-activated protein kinase is needed for the full induction of autophagy (Noda and Ohsumi 1998; Wang et al. 2001; Budovskaya et al. 2004; Stephan and Herman 2006; Kamada et al. 2010). The manner in which these signaling pathways regulate Atg1 activity and the precise role of this kinase in the autophagy process are presently matters of intense scrutiny.
Although Atg1 kinase activity is required for the induction of autophagy, relatively little is known about how this enzyme is regulated in vivo. Two proteins associated with Atg1, Atg13 and Atg17, have been shown to be required for full Atg1 kinase activity both in vitro and in vivo (Kamada et al. 2000; Stephan et al. 2009). The roles of these proteins appear to be conserved through evolution as functional homologs of both have been identified in fruit flies and/or mammals (Hara et al. 2008; Chan et al. 2009; Chang and Neufeld 2009; Ganley et al. 2009; Hosokawa et al. 2009; Jung et al. 2009; Mercer et al. 2009). However, it is not yet clear precisely how these proteins stimulate Atg1 activity. In this study, we show that Atg1 is autophosphorylated within the activation loop and that this phosphorylation is required for both Atg1 kinase activity and the induction of autophagy. The activation loop is a structurally conserved element within the kinase domain and phosphorylation within this loop is often a necessary prerequisite for efficient substrate binding and/or phosphotransfer in the catalytic site (Johnson et al. 1996; Nolen et al. 2004). This loop generally corresponds to the sequence between two signature elements within the core kinase domain, the DFG and APE motifs (Hanks and Hunter 1995). Phosphorylation within this loop tends to result in a more ordered structure for this region and the proper positioning of key elements within the catalytic core of the kinase domain (Knighton et al. 1991; Johnson and O'reilly 1996; Huse and Kuriyan 2002). We found that Atg1 activation loop phosphorylation was correlated with the onset of autophagy and that replacing the site of phosphorylation with a phosphomimetic residue led to constitutive Atg1 autophosphorylation in vivo. In all, the data here suggest that Atg1 phosphorylation within its activation loop may be an important point of regulation within the autophagy pathway and models that discuss these data are presented.
MATERIALS AND METHODS
Strains and growth media:
Standard Escherichia coli growth conditions and media were used throughout this study. The yeast rich-growth medium, YPAD, consists of 1% yeast extract, 2% Bacto-peptone, 500 mg/liter adenine–HCl, and 2% glucose. The yeast YM glucose and SC glucose minimal growth media have been described (Kaiser et al. 1994; Chang et al. 2001). The nitrogen starvation medium, SD-N, consists of 0.17% yeast nitrogen base lacking amino acids and ammonium sulfate, and 2% glucose. Growth media reagents were from Difco (Detroit). Standard yeast genetic methods were used in this study. The yeast strains used were TN125 (MATa ade2 his3 leu2 lys2 trp1 ura3 pho8∷pho8Δ60), YYK126 (TN125 atg1Δ∷LEU2), YYK130 (TN125 atg13Δ∷TRP1), PHY3687 (TN125 atg1Δ∷LEU2 atg13Δ∷kanMX), BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), PHY4167 (BY4741 atg1Δ∷kanMX), PHY4177 (BY4741 atg13Δ∷kanMX), and PHY4181 (BY4741 atg17Δ∷kanMX) (Noda et al. 1995; Kamada et al. 2000; Giaever et al. 2002).
Plasmid constructions:
Site-directed mutageneses were performed with the Clontech Transformer kit on the ATG1-containing plasmids, pPHY1115 and pPHY2376, to generate the variants (K54A, D211A, T226A, and T226E) used herein. Plasmid pPHY1115 was originally named pRS316-3xmycATG1 and was generously provided by Dr. Yoshinori Ohsumi. Plasmid pPHY2376 is a pRS423-based construct in which Atg1 has three copies of the HA epitope at its N terminus; this plasmid was originally provided by Dr. Daniel Klionsky. Variants in the YFP–Atg1 fusion background were generated similarly with the wild-type plasmid, pPHY2297, as the starting vehicle (Budovskaya et al. 2005). For the rAtg1 variants, fragments encoding residues 1–420 were PCR amplified from the appropriate yeast plasmids and cloned into the pET21a expression vector. The plasmid, pPHY2427, encoded an HA epitope-tagged version of Atg13 under the control of the promoter from the copper-inducible CUP1 gene. The ATG13 locus was cloned from the plasmid, pRS316–ATG13, that was kindly provided by Dr. Takeshi Noda. A site-directed mutagenesis was performed to insert an XmaI site immediately following the start codon of ATG13. The ATG13 coding sequences and transcriptional terminator were then cloned as a 3.1 kb XmaI–NotI fragment into pPHY2203 to form pPHY2427. The pPHY2203 plasmid was described previously and contains the CUP1 promoter and the HA epitope (Deminoff et al. 2006).
Western blotting and immunoprecipitation:
Protein samples for Western blotting were prepared as described (Budovskaya et al. 2005). The proteins were separated on SDS–polyacrylamide gels, transferred to nitrocellulose membranes and the membranes were probed with the appropriate primary and secondary antibodies. The Supersignal chemiluminescent substrate (Pierce Chemical, Rockford, IL) or Odyssey infrared imaging system (LI-COR Biosciences) was subsequently used to detect the reactive bands or fluorescence intensities. Immunoprecipitations were performed as described (Budovskaya et al. 2005; Deminoff et al. 2009). HA-tagged proteins were generally precipitated on anti-HA matrix beads (Roche) whereas myc-tagged proteins were immunoprecipitated with a monoclonal anti-myc antibody and then collected on Protein A–Sepharose (GE Healthcare) as described (Budovskaya et al. 2005; Deminoff et al. 2006). The amount of immunoprecipitated and co-immunoprecipitated proteins were subsequently assessed by Western immunoblotting with the appropriate antibodies. To assess phosphorylation of position T226 within Atg1, a polyclonal antiserum was generated in rabbits against a phosphorylated peptide, FLPNTSLAE(pT)LCGSPLY, that corresponds to the sequence of the Atg1 activation loop (see Figure 1A). The peptide synthesis, and antibody generation and purification was performed by Lampire Biological Laboratories. Western blots with this antibody were generally performed with cells overexpressing Atg1 from a 2μ plasmid.
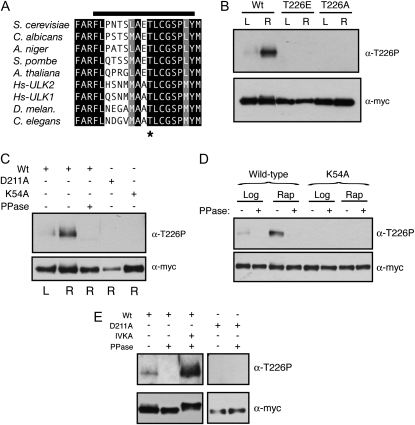
Figure 1.—
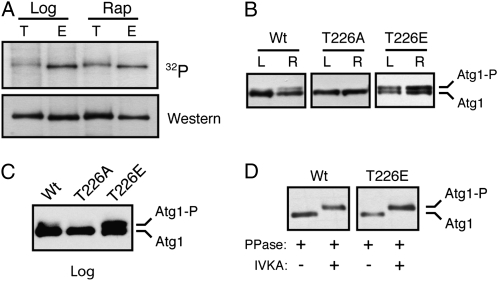
T226 within the activation loop was a site of Atg1 autophosphorylation. (A) The activation loop sequences for the Atg1 orthologs from the indicated organisms are shown. Residues identical in all the orthologs are shown on a black background and those residues that are chemically similar are shown on a gray background. The T226 residue is indicated by an asterisk and the bar at the top indicates the peptide used to generate the phosphospecific antibody. Hs, Homo sapiens. (B) Phosphorylation at position T226 within the Atg1 activation loop increased upon rapamycin treatment. Note that the Atg1T226A and Atg1T226E variants were not recognized by the anti-T226P phosphospecific antibody. In B–E, the indicated Atg1 proteins were immunoprecipitated from cell extracts with an antibody directed against the indicated epitope, separated on SDS-polyacrylamide gels, and then analyzed by Western blotting with the indicated antibodies. The cell extracts were prepared from either log-phase (L) or rapamycin-treated (R) cultures. The protein signals were assessed by autoradiography following chemiluminescence illumination. (C and D) Atg1 recognition by the T226-P phosphospecific antibody was lost upon phosphatase treatment and was absent from cells that contained kinase-defective variants of Atg1. Where indicated, the precipitated Atg1 proteins were treated with λ phosphatase (PPase) prior to SDS-polyacrylamide gel electrophoresis. K54A and D211A refer to kinase-defective variants of Atg1. (E) The T226 phosphorylation signal was regenerated in an in vitro kinase assay. The indicated Atg1 proteins were precipitated from yeast cells that had been treated with rapamycin, and the relative level of T226 phosphorylation was assessed by Western blotting with the phosphospecific antibody. Where indicated, the precipitated Atg1 proteins were treated with λ phosphatase (PPase) and then subjected to an in vitro kinase autophosphorylation assay (IVKA).
In vitro kinase assays (IVKAs):
The immunoprecipitated Atg1 proteins, or purified recombinant Atg1 fragments, were incubated for 30–60 min at 30° with 10 μCi [γ-32P]ATP in a 40-μl reaction (50 mm potassium phosphate, 5 mm NaF, 10 mm MgCl2, 4.5 mm DTT, protease inhibitors, and phosphatase inhibitors) with or without 10 μg of myelin basic protein (MBP) (Sigma, St. Louis). The reactions products were separated on SDS–polyacrylamide gels and the gels were fixed, dried, and analyzed either by autoradiography or by phosphorimaging with a Typhoon Trio (GE Healthcare).
Recombinant protein purification:
E. coli cells were grown to an OD600 of ∼0.5/ml and IPTG was added to a final concentration of 1 mm to induce expression of the rAtg1 proteins. The induction was carried out for 4 hr. The cells were collected by centrifugation and lysed by sonication in lysis buffer (50 mm sodium phosphate, 500 mm NaCl, and protease inhibitors). Clarified cell lysates were then incubated with 1 ml Ni–NTA agarose beads (Qiagen) at 4° overnight. The beads were collected by centrifugation and washed three times with PBS containing 20 mm imidazole, and the recombinant proteins were eluted with PBS containing 250 mm imidazole.
Autophagy assays:
Autophagy activity was assessed with several different assays that have been described previously. In general, autophagy was induced by treating mid-log-phase cells with 200 ng/ml rapamycin for the indicated times. The ALP-based assay measures the delivery and subsequent activation in the vacuole of an altered form of the Pho8 alkaline phosphatase, known as Pho8Δ60 (Noda et al. 1995; Noda and Klionsky 2008). The relative level of autophagy was indicated by the difference in alkaline phosphatase activity detected between extracts prepared from log-phase and rapamycin-treated cells. The data presented represent the average of at least three independent experiments where the autophagy levels generally varied by less than 15% between the different experimental replicates. In the GFP–Atg8 assay, the relative level of free GFP generated by proteolysis in the vacuole is used as an indicator of autophagy activity (Shintani and Klionsky 2004). The GFP–Atg8 fusion protein and the free GFP were detected by Western blotting with an anti-GFP antibody. The final assay assesses the processing of overexpressed aminopeptidase I, or Ape1, that occurs via the autophagy pathway (Budovskaya et al. 2004). In this assay, the cells express Ape1 at a level that saturated the cytoplasm-to-vacuole, or Cvt, transport system. Previous work has shown that the precursor Ape1 protein in these cells is processed by the autophagy pathway following either rapamycin treatment or nitrogen starvation (Budovskaya et al. 2004). The Ape1 proteins were detected by Western blotting with an anti-Ape1 antibody provided by Dr. Daniel Klionsky.
Fluorescence microscopy:
The CFP–Atg11 and YFP–Atg1 fusions were under the control of the inducible promoter from the yeast CUP1 gene. Expression of the fusion proteins was induced by the addition of 100 μm CuSO4 for 2 hr at 30°, and the samples were imaged as described (Budovskaya et al. 2005).
RESULTS
Residue T226 within the activation loop was a site of Atg1 autophosphorylation:
The activation loop of the S. cerevisiae Atg1 contains a candidate site of phosphorylation at threonine-226 (T226). This position, and much of the surrounding sequence, is conserved in Atg1 orthologs (Figure 1A). To test whether T226 was indeed a site of phosphorylation, we generated an antibody that specifically recognized the phosphorylated form of a peptide whose sequence corresponded to that of the Atg1 activation loop (Figure 1A; see materials and methods). This antibody recognized the wild-type Atg1 protein but not the Atg1T226A and Atg1T226E variants that had substitutions at position T226 (Figure 1B). Interestingly, the signal with this antibody was found to increase upon exposure to conditions that lead to an induction of autophagy, such as rapamycin treatment (Figure 1B; see below). The recognition by this antibody was lost upon phosphatase treatment of the immunoprecipitated Atg1 and was absent from cells expressing only the kinase-defective variants, Atg1K54A or Atg1D211A (Figure 1, C and D). The positions altered in these variants, K54 and D211, are highly conserved in protein kinases and are important for the proper positioning of ATP within the active site (Zoller et al. 1981; Hanks et al. 1988; Adams 2001). These results suggested that the recognition by this antibody required the presence of a phosphate group at position T226 and that the addition of this phosphate might be catalyzed by Atg1 itself. To examine this latter possibility further, we attempted to regenerate the phosphorylation signal at T226 in vitro with immunoprecipitated Atg1 proteins that had been pretreated with λ phosphatase. We found that the T226 phosphorylation was restored with the wild-type Atg1 but not with kinase-defective variants (Figure 1E; data not shown). Altogether, these data suggested that position T226 within the activation loop was a site of Atg1 autophosphorylation and that the presence of this modification was coincident with the induction of autophagy.
Phosphorylation at position T226 within the activation loop was required for Atg1 protein kinase activity:
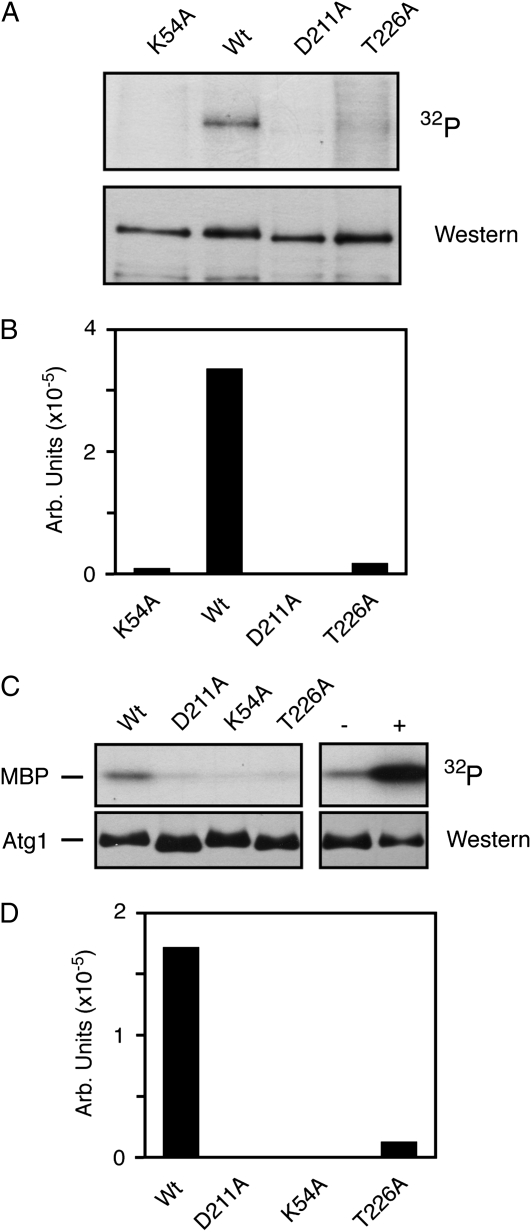
To assess the importance of activation loop phosphorylation for Atg1 function, we replaced T226 with a nonphosphorylatable alanine residue and assessed Atg1 kinase activity both in vivo and in vitro. The in vitro assays examined Atg1 autophosphorylation and the phosphorylation of a nonphysiological substrate, the MBP (Matsuura et al. 1997; Kamada et al. 2000). In each case, the Atg1 proteins examined were precipitated from rapamycin-treated cells. We found that this Atg1T226A variant exhibited significantly diminished activity, relative to the wild-type enzyme, in both of these assays (Figure 2, A–D). Two kinase-defective versions of Atg1, Atg1K54A and Atg1D211A, were used as negative controls in these experiments and the level of kinase activity associated with Atg1T226A was similar to that observed with these control proteins. We also found that a pre-incubation of the precipitated Atg1 with ATP resulted in elevated kinase activity toward MBP (Figure 2C). This stimulation required the presence of the kinase domain and suggested that Atg1 autophosphorylation might increase the specific activity of this enzyme (supporting information, Figure S1, A and B). This autophosphorylation could occur at position T226 and/or other positions within the Atg1 protein.
Figure 2.—
The presence of T226 within the Atg1 activation loop was required for protein kinase activity. Wt, wild-type. (A) Replacing T226 with a nonphosphorylatable residue resulted in a loss of Atg1 autophosphorylation. Atg1 proteins with the indicated alterations were immunoprecipitated from rapamycin-treated cells and subjected to an in vitro autophosphorylation assay as described in the materials and methods. The Atg1K54A and Atg1D211A variants were kinase-defective controls. In both A and B, the Atg1 proteins were tagged at the N terminus with three copies of the myc epitope. (B) Quantification of the phosphorimager data shown in A. (C) An in vitro kinase assay, using the MBP as substrate, was performed with the indicated Atg1 proteins isolated from rapamycin-treated cells. In the two right-hand lanes, the wild-type Atg1 protein was immunoprecipitated and either mock treated (−) or pre-incubated with 3 mM ATP for 60 min at 30°C (+). The samples were subsequently washed to remove the excess ATP and an in vitro kinase assay was performed with [γ-32P]ATP and MBP. (D) Quantification of the phosphorimager analysis data in C.
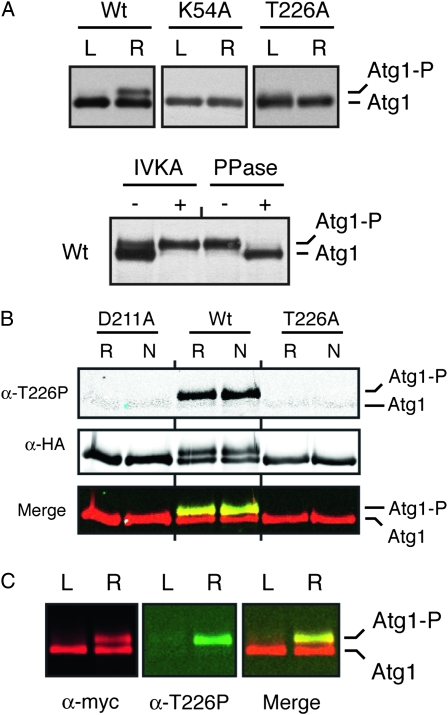
To assess Atg1 kinase activity in vivo, we took advantage of previous observations indicating that Atg1 autophosphorylation results in the production of a slower-migrating form of this protein on SDS-polyacrylamide gels (Atg1-P in Figure 3A) (Kamada et al. 2000; Budovskaya et al. 2005; Stephan et al. 2009). Our data indicate that the site of autophosphorylation responsible for this shift is distinct from T226 (see below and our unpublished observations). The relative level of this second, upper band can serve as a measure of the Atg1 kinase activity present in cells (Stephan et al. 2009). Consistent with the in vitro data above, we found significantly less of the Atg1T226A variant, relative to the wild type, in the slower-migrating form following either rapamycin treatment or nitrogen starvation, two conditions that are known to induce an autophagy response (Figure 3, A and B). The relative activity of the Atg1T226A variant again resembled the kinase-defective controls, Atg1K54A and Atg1D211A. Interestingly, we found that the α-T226P antibody specifically recognized this slower-migrating form of Atg1 on Western blots (Figure 3, B and C). In all, these data suggested that T226 phosphorylation within the activation loop was required for Atg1 kinase activity and was associated with an autophosphorylated form of Atg1 found specifically in cells undergoing autophagy.
Figure 3.—
The Atg1T226A variant was defective for Atg1 autophosphorylation in vivo. (A) The presence of the T226A alteration resulted in a loss of Atg1 autophosphorylation in vivo. Western blots were performed with extracts prepared from either log-phase (L) or rapamycin-treated (R) cells with the indicated Atg1 proteins. In the bottom part, immunoprecipitated wild-type Atg1 was either mock treated (IVKA−) or subjected to an in vitro autophosphorylation reaction with 3 mM ATP (IVKA+). An aliquot of this latter sample was then split into two and either mock treated (PPase−) or incubated with λ phosphatase (PPase+). PPase, λ phosphatase; IVKA, in vitro kinase assay; Atg1-P, autophosphorylated form of Atg1 that exhibits an altered mobility on SDS-polyacrylamide gels. (B) Atg1 activation loop phosphorylation was elevated in response to nitrogen starvation and required the presence of a functional Atg1 kinase domain. In B and C, the indicated Atg1 proteins were immunoprecipitated from cell extracts with an antibody directed against the indicated epitope, separated on SDS-polyacrylamide gels, and then analyzed by Western blotting with the indicated antibodies. The cell extracts were prepared from either log-phase (L), rapamycin-treated (R) or nitrogen starved (N) cultures. The protein signals were detected with a LI-COR Odessey infrared detection system. For nitrogen starvation, log-phase cells were transferred to SD-N medium for 6 hr. D211A, a kinase-defective variant of Atg1. (C) The anti-T226P phosphospecific antibody (α-T226P) recognized a slower-migrating, autophosphorylated form of Atg1 that appeared upon rapamycin treatment. The protein signals were detected with a LI-COR Odessey infrared detection system and the right-most part shows a merged image for the α-myc and α-T226P signals.
Activation loop phosphorylation was required for the induction of autophagy:
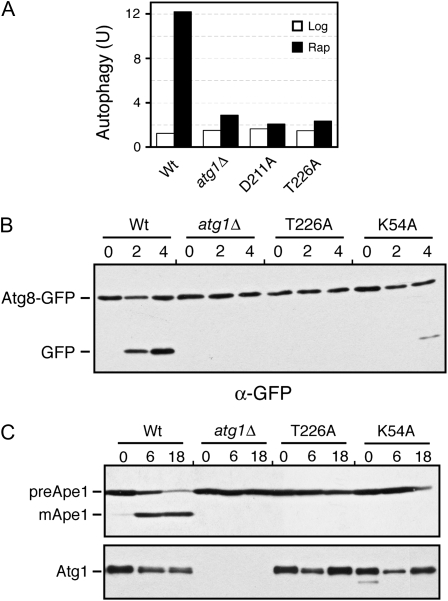
We used three different assays to test whether phosphorylation within the Atg1 activation loop was required for the induction of autophagy. The first assessed the vacuolar delivery and processing of an altered form of the Pho8 alkaline phosphatase, known as Pho8Δ60 (Noda et al. 1995). This truncated protein lacks a membrane-spanning domain and the targeting information necessary for normal delivery through the secretory pathway (Klionsky and Emr 1989; Noda et al. 1995). As a result, this protein can be delivered to the vacuole, and therein activated, only by the autophagy pathway. Using this assay, we found that no significant autophagy activity was detectable in cells carrying only the Atg1T226A variant (Figure 4A). The levels of autophagy detected were similar to those associated with the atg1Δ control strain.
Figure 4.—
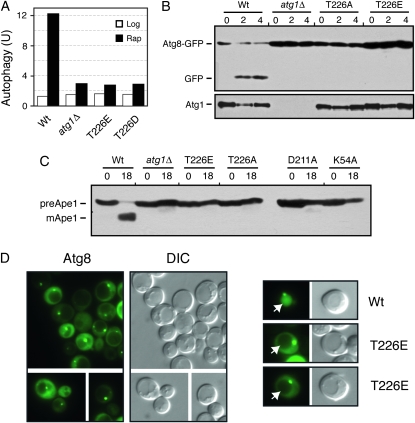
The Atg1T226A variant was defective for autophagy. (A) Autophagy activity was assessed with the ALP Pho8Δ60-based assay using an atg1Δ strain expressing the indicated Atg1 proteins. The atg1Δ control strain contained a vector plasmid. Extracts were prepared from cells that had been treated with 200 ng/ml rapamycin for either 0 (Log) or 4 hr (Rap). The data shown are the average of two independent experiments where the values obtained varied by less than 15% between replicates. (B) Autophagy activity was assessed with a GFP–Atg8 processing assay as described in materials and methods. The level of free GFP was an indicator of the relative autophagy activity present. (C) Autophagy activity was assessed with an assay measuring the vacuolar processing of overexpressed Ape1 as described in materials and methods. The presence of the mature, processed Ape1 protein (mApe1) was an indicator of autophagy activity. preApe1, precursor Ape1. The bottom part shows the relative levels of the different myc-tagged Atg1 proteins examined. For B and C, cells were treated with 200 ng/ml rapamycin for the hours indicated.
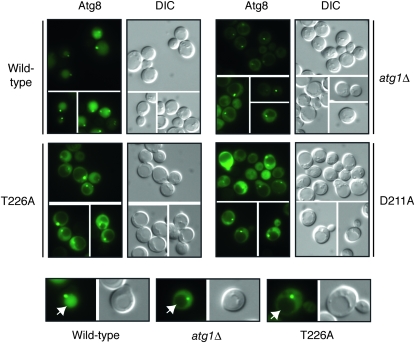
The second assay measured the processing of a GFP–Atg8 fusion protein (Shintani and Klionsky 2004). Atg8 is normally incorporated into the autophagosome and delivered into the vacuolar lumen (Kim et al. 2001; Suzuki et al. 2001; Suzuki and Ohsumi 2007). The Atg8 is then degraded by vacuolar hydrolases, thereby releasing the more stable GFP protein (Shintani and Klionsky 2004). The relative level of free GFP can be used as a measure of the autophagy activity present. As with the ALP-based assay, no significant processing of the GFP–Atg8 fusion was detected in cells with the Atg1T226A variant (Figure 4B). This autophagy defect was corroborated by an examination of the subcellular localization of the GFP fluorescence in these cells. In wild-type cells, the GFP–Atg8 fusion is delivered to the vacuole and the free GFP accumulates within the lumen of this organelle (Figure 5). In contrast, very little, if any, GFP fluorescence is detected in the vacuoles of mutants defective for autophagy (see atg1Δ in Figure 4) (Suzuki et al. 2001). Consistent with the above processing defects, we found no significant GFP fluorescence within the vacuoles of atg1Δ cells carrying the Atg1T226A variant (Figure 5). It should be noted that this GFP–Atg8 fusion was normally localized to the PAS in these cells (Figure 5).
Figure 5.—
An GFP-Atg8 fusion protein was not delivered to the vacuole in cells containing the Atg1T226A variant. Yeast strains carrying the indicated Atg1 proteins were treated for 2 hr with 200 ng/ml rapamycin and the subcellular localization of an GFP–Atg8 fusion protein was then assessed by fluorescence microscopy. Atg8 is normally incorporated into the autophagosome and transported to the vacuolar compartment. This transport results in the presence of GFP fluorescence in the vacuoles of cells with a functional autophagy pathway. The vacuoles of the representative cells shown in the bottom row are indicated by the white arrows. Note that the GFP–Atg8 fusion was still found at the PAS in cells containing the Atg1T226A variant.
Finally, we assessed the autophagy-dependent maturation of theApe1 protein. Ape1 is a normal cargo for the cytoplasm-to-vacuole, or Cvt, transport pathway that is responsible for delivering a set of proteins to the vacuole during log-phase growth (Klionsky et al. 1992; Khalfan and Klionsky 2002). Overexpression of Ape1 saturates the Cvt system and results in the cytoplasmic accumulation of an unprocessed, precursor form of this protein (see Figure 4C) (Klionsky et al. 1992). Upon the induction of autophagy, this protein is efficiently delivered to the vacuole and processed to its mature, active form (Scott et al. 1996; Budovskaya et al. 2004). We found that the Atg1T226A variant was defective for this autophagy-mediated processing and that the Ape1 protein was present only in its precursor form in these cells (Figure 4C). Altogether, these data indicated that Atg1 autophosphorylation at T226 was necessary for the induction of the autophagy pathway in S. cerevisiae.
The Atg1T226E variant exhibited constitutive autophosphorylation activity:
To test whether the presence of T226 phosphorylation was sufficient for Atg1 autophosphorylation, we replaced this position with the potential phosphomimetic residues, glutamic and aspartic acid. Similar results were obtained with both substitutions, and we focus here on the Atg1T226E variant. Interestingly, we found that this Atg1T226E variant was competent for autophosphorylation in vitro when isolated from rapamycin-treated cells (Figure S2, A). However, unlike the wild-type protein, Atg1T226E also exhibited elevated levels of autophosphorylation in log-phase cells that were typically not undergoing autophagy. This constitutive, or unregulated, autophosphorylation activity was observed in both in vitro and in vivo assays (Figure 6, A–C). For example, in an in vitro assay, elevated levels of radioactivity, relative to the wild-type protein, were incorporated into the Atg1T226E variant isolated from log-phase cells (Figure 6A). In addition, the fraction of this variant found in the slower-migrating form on SDS-polyacrylamide gels was elevated in log-phase cell extracts (Figure 6, B and C). In both assays, the overall level of autophosphorylation was slightly elevated with the Atg1T226E variant. These in vivo results with the Atg1T226E protein supported the above contention that the presence of the slower-migrating form of Atg1 was due to autophosphorylation at a site distinct from T226. In particular, we found that the upper band associated with the Atg1T226E protein was lost upon phosphatase treatment and was regenerated in a subsequent incubation with ATP (Figure 6D and Figure S2, B).
Figure 6.—
The Atg1T226E variant was a constitutively active protein kinase. (A) The Atg1T226E variant exhibited elevated protein kinase activity in log-phase cell extracts. Atg1 was immunoprecipitated from either log-phase (Log) or rapamycin-treated (Rap) cells and subjected to an in vitro autophosphorylation assay with [γ-32P]ATP. T, wild-type Atg1; E, Atg1T226E variant. The top is the autoradiograph image and the bottom is the Western blot control performed with an anti-myc antibody. For A–D, the Atg1 proteins were tagged with three copies of the myc epitope. (B) Atg1T226E exhibited constitutive autophosphorylation in vivo. Western blots were performed with extracts prepared from either log-phase (L) or rapamycin-treated (R) cultures of an atg1Δ strain with the indicated Atg1 proteins. (C) The relative level of the slower-migrating form of the Atg1T226E variant was elevated in log-phase cells. Western blotting was performed with log-phase extracts prepared from atg1Δ cells carrying the indicated Atg1 proteins. (D) The slower-migrating form of Atg1 was the result of an autophosphorylation at a position distinct from T226. The wild-type Atg1 and Atg1T226E proteins were immunoprecipitated from rapamycin-treated cells and incubated with λ phosphatase. The sample was then split into two aliquots and either mock treated with the reaction buffer or incubated with 3 mM ATP. PPase, λ phosphatase; IVKA, in vitro kinase assay.
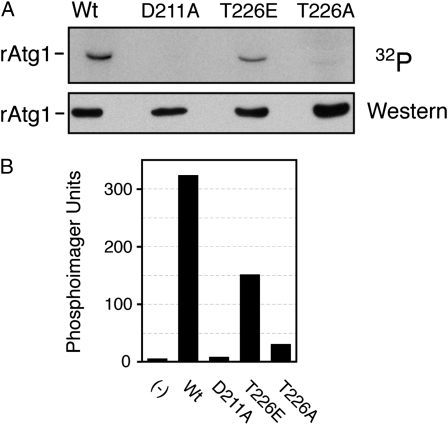
Finally, we found that a recombinant fragment of Atg1 produced in E. coli (rAtg1) was also capable of catalyzing an autophosphorylation reaction (Figure 7). This fragment contained residues 1–420 of the wild-type protein and included the entire protein kinase domain located at the N terminus of Atg1. This autophosphorylation was also dependent upon the presence of T226 as replacing this position with an alanine residue resulted in significantly diminished activity (Figure 7). This rAtg1T226A variant also exhibited decreased activity in assays with MBP as substrate (Figure S3). In contrast, a recombinant protein with glutamic acid replacing T226 was capable of significant autophosphorylation (Figure 7). This result was similar to that obtained above with the wild-type protein and together these observations suggested that it was the phosphorylation at position T226 that was important for this Atg1 autophosphorylation. Moreover, the presence of a phosphomimetic at position 226 appeared to disrupt the normal control of Atg1 and resulted in constitutive autophosphorylation.
Figure 7.—
Autophosphorylation of a recombinant Atg1 protein required the presence of position T226, the site of activation loop phosphorylation. (A) Recombinant fragments of Atg1 containing residues 1–420 were partially purified from E. coli cell extracts and incubated with [γ-32P]ATP to assess autophosphorylation in vitro. The relative level of label incorporation was then assessed with a phosphorimager and the image from this analysis is shown at the top. The bottom shows the Western blot control performed with an antibody specific to the His6-tag present on these recombinant proteins. (B) Quantification of the phosphorimager analysis shown in A.
The Atg1T226E variant was nonfunctional for autophagy:
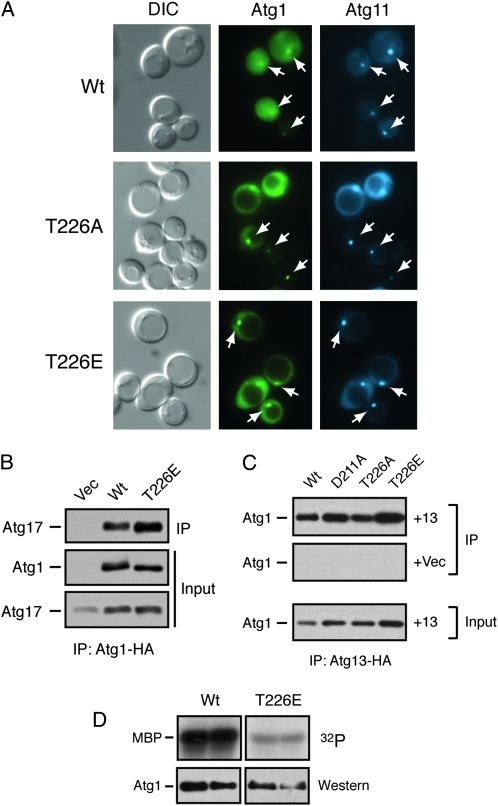
We also tested whether the Atg1T226E and Atg1T226D variants were functional for autophagy. Using the three processing assays described above, we found little, if any, autophagy activity in atg1Δ cells carrying these variants (Figure 8, A–C). Moreover, we found that the GFP–Atg8 fusion protein was not transported efficiently into the lumen of the vacuole in atg1–T226E cells (Figure 8D). This latter defect is consistent with a failure to carry out the normal autophagy process. Therefore, although these variants exhibited constitutive autophosphorylation activity, neither was capable of replacing the wild-type protein in the autophagy process. These variants also failed to induce autophagy in log-phase cells and had little detrimental effect upon autophagy when overexpressed in wild-type cells (Figure 8A and Figure S4). The autophagy defects did not appear to be due simply to gross misfolding as the Atg1T226E protein was present at normal steady-state levels in yeast cells, was localized normally to the PAS, and interacted with both Atg13 and Atg17, two key regulators of Atg1 kinase activity (Figure 9, A–C) (Kamada et al. 2000; Kabeya et al. 2005). However, we did find that the Atg1T226E variant was less effective than the wild-type protein at phosphorylating the nonphysiological substrate, MBP, in an in vitro assay (Figure 9D). Therefore, it is possible that the autophagy defects observed with this variant might be due to a failure to phosphorylate an important, but as yet unknown, substrate in vivo.
Figure 8.—
The Atg1T226E variant was defective for autophagy. (A) The ALP-based assay was used to assess autophagy activity in an atg1Δ strain expressing the indicated Atg1 proteins. Extracts were prepared from cells that had been treated with 200 ng/ml rapamycin for either 0 (Log) or 4 hr (Rap). The data shown are the average of two independent experiments where the values obtained varied by less than 15% between replicates. The atg1Δ control strain contained a vector plasmid. (B and C) Autophagy activity was assessed with either the GFP–Atg8 (B) or the Ape1 (C) processing assays as described in materials and methods. In each case, the strains were treated with 200 ng/ml rapamycin for the indicated time (hours). B, bottom, shows the relative levels of the different Atg1 proteins examined. (D) A GFP–Atg8 fusion protein was not delivered to the vacuole in cells expressing only Atg1T226E. The subcellular localization of an GFP–Atg8 fusion protein was assessed by fluorescence microscopy in atg1Δ cells carrying the indicated Atg1 proteins. The cells were treated with 200 ng/ml rapamycin for 2 hr prior to examination. Note that the GFP–Atg8 fusion was localized to the PAS in cells containing the Atg1T226E variant.
Figure 9.—
The Atg1T226E variant was localized to the PAS and interacted with Atg13 and Atg17. (A) The Atg1T226A and Atg1T226E proteins were localized normally to the PAS. The subcellular localization of YFP fusions to the indicated Atg1 proteins was assessed in an atg1Δ strain by fluorescence microscopy. An Atg11–CFP fusion protein was used as a marker for the PAS. The cells were all treated with 200 ng/ml rapamycin for 2 hr before examination. The white arrows indicate the location of the PAS in the respective cells. (B) The Atg1T226E variant interacted with Atg17. The indicated HA-tagged Atg1 proteins were precipitated from cell extracts and the relative amount of associated Atg17 protein was assessed by Western blotting with an anti-myc antibody. (C) The Atg1T226E variant interacted with Atg13. HA-tagged Atg13 was precipitated from cell extracts and the relative amounts of the associated Atg1 proteins were assessed by Western blotting with an anti-myc antibody. +13, cells containing the HA-tagged Atg13 protein; +Vec, cells containing a vector control and thus lacking the tagged Atg13 protein. (D) An in vitro kinase assay using MBP as substrate was performed with the wild-type Atg1 and the Atg1T226E variant. The indicated Atg1 proteins were immunoprecipitated and incubated with [γ-32P]ATP and the nonphysiological substrate, MBP; each sample was performed in duplicate. The bottom parts show the amount of Atg1 protein present in the in vitro reactions.
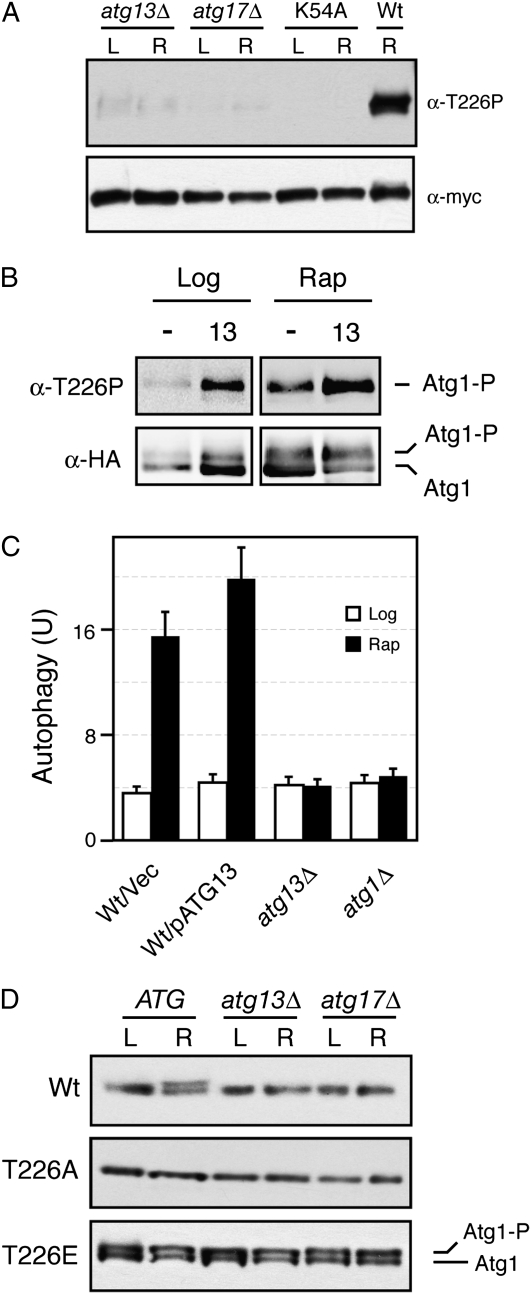
Atg13 and Atg17 were required for T226 phosphorylation:
The precise roles of Atg13 and Atg17 in the regulation of Atg1 kinase activity have not yet been defined. Here, we tested whether these regulators were required for the phosphorylation of Atg1 within its activation loop. Indeed, we found that T226 phosphorylation was greatly diminished in both atg13Δ and atg17Δ cells (Figure 10A). The residue levels remaining in these mutants were higher than those observed in strains carrying only a kinase-defective version of Atg1 (Figure S5). This low level of T226 phosphorylation might be important for the suppression of atg13 defects observed upon the overexpression of Atg1 in these mutants (Funakoshi et al. 1997). We also found that the overexpression of Atg13 resulted in an elevated level of T226 phosphorylation in both log-phase and rapamycin-treated cells (Figure 10B). Consistent with this observation, the overexpression of Atg13 also led to slightly increased levels of autophagy in rapamycin-treated cultures (Figure 10C). Finally, we tested whether the autophosphorylation associated with the Atg1T226E variant required either Atg13 or Atg17. We found that the slower-migrating form of Atg1T226E that was indicative of Atg1 kinase activity in vivo was still present in both atg13Δ and atg17Δ cells (Figure 10D). Therefore, the presence of a phosphomimetic within the Atg1 activation loop bypassed the normal requirement for these regulators for Atg1 autophosphorylation.
Figure 10.—
Atg13 and Atg17 were required for Atg1 autophosphorylation within the activation loop. (A) T226 phosphorylation in atg13Δ and atg17Δ mutants. The wild-type Atg1 was precipitated from atg13Δ, atg17Δ, or wild-type cells and analyzed by Western blotting with the anti-T226P phosphospecific antibody. The Atg1K54A variant (K54A) was used as a kinase-defective control. L, log phase; R, rapamycin-treated. (B) Overexpression of Atg13 resulted in an increased level of T226 phosphorylation. The relative level of T226 phosphorylation was assessed in both log-phase and rapamycin-treated wild-type cells carrying either a vector control (−) or an ATG13 overexpression plasmid. The wild-type Atg1 was immunoprecipitated from these cells with an antibody specific for the HA epitope present on this protein. (C) Overexpression of Atg13 resulted in elevated levels of autophagy. A plasmid overexpressing Atg13 or a vector control were introduced into wild-type cells, and autophagy levels were assessed with the Pho8Δ60 ALP-based assay. The cells were treated with 200 ng/ml rapamycin for 0 (Log) or 4 hr (Rap) before analysis. (D) Atg13 and Atg17 were not required for the autophosphorylation of Atg1T226E in vivo. Cell extracts were prepared from either log-phase (L) or rapamycin-treated (R) cultures of wild-type, atg13Δ, and atg17Δ cells carrying the indicated Atg1 proteins. The relative levels of the slower-migrating, autophosphorylated form of Atg1 were assessed by Western blotting with an anti-myc antibody. The Atg1 proteins assayed were all tagged with three copies of the myc epitope.
DISCUSSION
The autophagy pathway has been implicated in a wide variety of phenomena important for human health and has been suggested as a potential therapeutic target for a number of disease conditions (Levine 2005; Rubinsztein et al. 2007; Levine and Kroemer 2008). To manipulate autophagy in a productive way, it is critical that we develop a thorough understanding of how this pathway is regulated in vivo. Toward this end, the work here identifies a specific phosphorylation event within the activation loop of Atg1 as a potential point of control for autophagy in S. cerevisiae. Several observations point to the importance of this phosphorylation at position T226 for the autophagy process. First, the substitution of a nonphosphorylatable alanine residue for the threonine normally present resulted in a loss of Atg1 kinase activity and a failure to induce the autophagy process. Second, a phosphospecific antibody, prepared against a phosphopeptide corresponding to the Atg1 activation loop, recognized the wild-type Atg1 but not the Atg1T226A variant. Third, the relative level of T226 phosphorylation was found to increase specifically upon exposure to conditions that induce the autophagy process. Fourth, this antibody specifically recognized an autophosphorylated form of Atg1 that was present in cells undergoing autophagy. Fifth, we found that replacing the T226 position with an acidic residue, either glutamic or aspartic acid, resulted in an enzyme that exhibited constitutive activity with respect to autophosphorylation. This latter observation is important as it indicates that the presence of a potential phosphomimetic at position 226 in the activation loop results in the production of an active enzyme. Taken altogether, these data support the contention that Atg1 is phosphorylated within its activation loop and that this phosphorylation is important for the normal induction of the autophagy process. Since this site has been evolutionarily conserved in Atg1 orthologs, it is possible that this phosphorylation will be a point of control for autophagy in other eukaryotes.
Many protein kinases require phosphorylation within their activation loop for full activity and this modification can be a critical event in the activation of these enzymes (Nolen et al. 2004). This phosphorylation can be catalyzed by a second enzyme or can be the consequence of an autophosphorylation reaction (Lochhead et al. 2005; Imajo et al. 2006; Lochhead et al. 2006; Pirruccello et al. 2006; Oliver et al. 2007). For Atg1, the data here suggest that this modification is self-catalyzed. A functional Atg1 kinase domain was required for T226 phosphorylation in vivo and this modification occurred in vitro with Atg1 immunoprecipitates. This phosphorylation was also shown to require the presence of Atg13 and Atg17, two previously identified regulators of Atg1 kinase activity. Interestingly, replacing position T226 with a phosphomimetic, such as glutamic acid, resulted in an enzyme that no longer required Atg13 or Atg17 for autophosphorylation in vivo. This bypass of the requirement for these regulators may suggest that facilitating Atg1 autophosphorylation within the activation loop is one of the functions of the Atg13 and Atg17 proteins during the autophagy process.
Although the data here suggest that T226 phosphorylation is required for the induction of autophagy, neither the Atg1T226E nor the Atg1T226D variant was capable of supporting this degradative process in vivo. The underlying basis for this defect is not yet known, but there are a number of potential explanations that can be considered. On one extreme is the possibility that it is the threonine itself at position 226, and not its phosphorylation, that is important for Atg1 function. We do not favor this possibility given the strong correlation between T226 phosphorylation and both the induction of autophagy and the presence of Atg1 kinase activity, as measured by autophosphorylation. In addition, this model is difficult to reconcile with the observation that the presence of a phosphomimetic at position 226 led to constitutive Atg1 autophosphorylation. Therefore, we feel that the results obtained here with the Atg1T226E and Atg1T226D variants require a different explanation. One possibility is that the phosphomimetic does not fully restore the functionality of a phosphorylated threonine and therefore results in an enzyme with only partial activity. This could explain why the Atg1T226E protein is competent for autophosphorylation but not the phosphorylation of an exogenous substrate, like MBP. Alternatively, this failure to phosphorylate MBP might reflect some inherent aspect of Atg1 biology. For example, Atg1 might normally participate in a negative feedback loop that is permanently engaged in the Atg1T226E variant. Another possibility is that the autophagy defects might be due to the inability of these variants to exist in a kinase-inactive form. Previous work has indicated that Atg1 has both kinase-dependent and kinase-independent activities in the autophagy process (Abeliovich et al. 2003; Cheong et al. 2008; Kawamata et al. 2008). Atg1 may therefore need to oscillate between kinase-active and kinase-inactive states to stimulate autophagy. Clearly, further work will be necessary to distinguish between these and other potential explanations. However, we feel that the work here in its totality identifies the autophosphorylation of Atg1 within its activation loop as an important event in the normal induction of autophagy. Since Atg1 and its regulators have been conserved through evolution, a continued analysis of this modification is likely to provide additional insights into the general control of this degradative process.
Acknowledgments
We thank Daniel Klionsky, Yoshiaki Kamada, Takeshi Noda, and Yoshinori Ohsumi for reagents used in this study, Yoshiaki Kamada for helpful discussions, and Venkat Gopalan, Mark Parthun, and members of the Herman lab for comments on the manuscript. This work was supported by a grant from the National Institutes of Health (GM65227) to P.K.H.
Supporting information available at http://www.genetics.org/cgi/content/full/genetics.110.116566/DC1.
References
- Abeliovich, H., C. Zhang, W. A. Dunn, Jr., K. M. Shokat and D. J. Klionsky, 2003. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol. Biol. Cell 14 477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, J. A., 2001. Kinetic and catalytic mechanisms of protein kinases. Chem. Rev. 101 2271–2290. [DOI] [PubMed] [Google Scholar]
- Budovskaya, Y. V., J. S. Stephan, F. Reggiori, D. J. Klionsky and P. K. Herman, 2004. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 279 20663–20671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya, Y. V., J. S. Stephan, S. J. Deminoff and P. K. Herman, 2005. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 102 13933–13938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, E. Y., A. Longatti, N. C. McKnight and S. A. Tooze, 2009. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Cell. Biol. 29 157–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y. W., S. C. Howard, Y. V. Budovskaya, J. Rine and P. K. Herman, 2001. The rye mutants identify a role for Ssn/Srb proteins of the RNA polymerase II holoenzyme during stationary phase entry in Saccharomyces cerevisiae. Genetics 157 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, Y. Y., and T. P. Neufeld, 2009. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol. Biol. Cell 20 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong, H., U. Nair, J. Geng and D. J. Klionsky, 2008. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 19 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deminoff, S. J., S. C. Howard, A. Hester, S. Warner and P. K. Herman, 2006. Using substrate-binding variants of the cAMP-dependent protein kinase to identify novel targets and a kinase domain important for substrate interactions in Saccharomyces cerevisiae. Genetics 173 1909–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deminoff, S. J., V. Ramachandran and P. K. Herman, 2009. Distal recognition sites in substrates are required for efficient phosphorylation by the cAMP-dependent protein kinase. Genetics 182 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi, T., A. Matsuura, T. Noda and Y. Ohsumi, 1997. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene 192 207–213. [DOI] [PubMed] [Google Scholar]
- Ganley, I. G., D. H. Lam, J. Wang, X. Ding, S. Chen et al., 2009. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284 12297–12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles et al., 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418 387–391. [DOI] [PubMed] [Google Scholar]
- Hanks, S. K., and T. Hunter, 1995. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9 576–596. [PubMed] [Google Scholar]
- Hanks, S. K., A. M. Quinn and T. Hunter, 1988. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241 42–52. [DOI] [PubMed] [Google Scholar]
- Hara, T., A. Takamura, C. Kishi, S. Iemura, T. Natsume et al., 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181 497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, C., and D. J. Klionsky, 2009. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. [DOI] [PMC free article] [PubMed]
- Hosokawa, N., T. Hara, T. Kaizuka, C. Kishi, A. Takamura et al., 2009. Nutrient-dependent mTORC1 association with the ULK1-Atg13–FIP200 complex required for autophagy. Mol. Biol. Cell 20 1981–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse, M., and J. Kuriyan, 2002. The conformational plasticity of protein kinases. Cell 109 275–282. [DOI] [PubMed] [Google Scholar]
- Imajo, M., Y. Tsuchiya and E. Nishida, 2006. Regulatory mechanisms and functions of MAP kinase signaling pathways. IUBMB Life 58 312–317. [DOI] [PubMed] [Google Scholar]
- Johnson, L. N., and M. O'Reilly, 1996. Control by phosphorylation. Curr. Opin. Struct. Biol. 6 762–769. [DOI] [PubMed] [Google Scholar]
- Johnson, L. N., M. E. Noble and D. J. Owen, 1996. Active and inactive protein kinases: structural basis for regulation. Cell 85 149–158. [DOI] [PubMed] [Google Scholar]
- Jung, C. H., C. B. Jun, S. H. Ro, Y. M. Kim, N. M. Otto et al., 2009. ULK-Atg13–FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20 1992–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya, Y., Y. Kamada, M. Baba, H. Takikawa, M. Sasaki et al., 2005. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell 16 2544–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, C., S. Michaelis and A. Mitchell, 1994. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Kamada, Y., T. Funakoshi, T. Shintani, K. Nagano, M. Ohsumi et al., 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150 1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada, Y., K. Yoshino, C. Kondo, T. Kawamata, N. Oshiro et al., 2010. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol. Cell. Biol. 30 1049–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata, T., Y. Kamada, Y. Kabeya, T. Sekito and Y. Ohsumi, 2008. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol. Biol. Cell 19 2039–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalfan, W. A., and D. J. Klionsky, 2002. Molecular machinery required for autophagy and the cytoplasm to vacuole targeting (Cvt) pathway in S. cerevisiae. Curr. Opin. Cell Biol. 14 468–475. [DOI] [PubMed] [Google Scholar]
- Kim, J., W. P. Huang and D. J. Klionsky, 2001. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J. Cell Biol. 152 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D. J., and S. D. Emr, 1989. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 8 2241–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D. J., R. Cueva and D. S. Yaver, 1992. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 119 287–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knighton, D. R., J. H. Zheng, L. F. Ten Eyck, V. A. Ashford, N. H. Xuong et al., 1991. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253 407–414. [DOI] [PubMed] [Google Scholar]
- Levine, B., 2005. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 120 159–162. [DOI] [PubMed] [Google Scholar]
- Levine, B., and G. Kroemer, 2008. Autophagy in the pathogenesis of disease. Cell 132 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochhead, P. A., G. Sibbet, N. Morrice and V. Cleghon, 2005. Activation-loop autophosphorylation is mediated by a novel transitional intermediate form of DYRKs. Cell 121 925–936. [DOI] [PubMed] [Google Scholar]
- Lochhead, P. A., R. Kinstrie, G. Sibbet, T. Rawjee, N. Morrice et al., 2006. A chaperone-dependent GSK3beta transitional intermediate mediates activation-loop autophosphorylation. Mol. Cell 24 627–633. [DOI] [PubMed] [Google Scholar]
- Matsuura, A., M. Tsukada, Y. Wada and Y. Ohsumi, 1997. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192 245–250. [DOI] [PubMed] [Google Scholar]
- Meijer, W. H., I. J. van der Klei, M. Veenhuis and J. A. Kiel, 2007. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy 3 106–116. [DOI] [PubMed] [Google Scholar]
- Mercer, C. A., A. Kaliappan and P. B. Dennis, 2009. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 5 649–662. [DOI] [PubMed] [Google Scholar]
- Noda, T., and D. J. Klionsky, 2008. The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol. 451 33–42. [DOI] [PubMed] [Google Scholar]
- Noda, T., and Y. Ohsumi, 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273 3963–3966. [DOI] [PubMed] [Google Scholar]
- Noda, T., A. Matsuura, Y. Wada and Y. Ohsumi, 1995. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 210 126–132. [DOI] [PubMed] [Google Scholar]
- Nolen, B., S. Taylor and G. Ghosh, 2004. Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell 15 661–675. [DOI] [PubMed] [Google Scholar]
- Oliver, A. W., S. Knapp and L. H. Pearl, 2007. Activation segment exchange: A common mechanism of kinase autophosphorylation? Trends Biochem. Sci. 32 351–356. [DOI] [PubMed] [Google Scholar]
- Pirruccello, M., H. Sondermann, J. G. Pelton, P. Pellicena, A. Hoelz et al., 2006. A dimeric kinase assembly underlying autophosphorylation in the p21 activated kinases. J. Mol. Biol. 361 312–326. [DOI] [PubMed] [Google Scholar]
- Rubinsztein, D. C., J. E. Gestwicki, L. O. Murphy and D. J. Klionsky, 2007. Potential therapeutic applications of autophagy. Nat. Rev. Drug Discov. 6 304–312. [DOI] [PubMed] [Google Scholar]
- Scott, S. V., A. Hefner-Gravink, K. A. Morano, T. Noda, Y. Ohsumi et al., 1996. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc. Natl. Acad. Sci. USA 93 12304–12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani, T., and D. J. Klionsky, 2004. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279 29889–29894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan, J. S., and P. K. Herman, 2006. The regulation of autophagy in eukaryotic cells: Do all roads pass through Atg1? Autophagy 2 146–148. [DOI] [PubMed] [Google Scholar]
- Stephan, J. S., Y. Y. Yeh, V. Ramachandran, S. J. Deminoff and P. K. Herman, 2009. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc. Natl. Acad. Sci. USA 106 17049–17054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., and Y. Ohsumi, 2007. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 581 2156–2161. [DOI] [PubMed] [Google Scholar]
- Suzuki, K., T. Kirisako, Y. Kamada, N. Mizushima, T. Noda et al., 2001. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20 5971–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada, M., and Y. Ohsumi, 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333 169–174. [DOI] [PubMed] [Google Scholar]
- Wang, Z., W. A. Wilson, M. A. Fujino and P. J. Roach, 2001. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol. Cell. Biol. 21 5742–5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Z., and D. J. Klionsky, 2007. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 9 1102–1109. [DOI] [PubMed] [Google Scholar]
- Zoller, M. J., N. C. Nelson and S. S. Taylor, 1981. Affinity labeling of cAMP-dependent protein kinase with p-fluorosulfonylbenzoyl adenosine: covalent modification of lysine 71. J. Biol. Chem. 256 10837–10842. [PubMed] [Google Scholar]