Abstract
Natural populations of forest trees exhibit striking phenotypic adaptations to diverse environmental gradients, thereby making them appealing subjects for the study of genes underlying ecologically relevant phenotypes. Here, we use a genome-wide data set of single nucleotide polymorphisms genotyped across 3059 functional genes to study patterns of population structure and identify loci associated with aridity across the natural range of loblolly pine (Pinus taeda L.). Overall patterns of population structure, as inferred using principal components and Bayesian cluster analyses, were consistent with three genetic clusters likely resulting from expansions out of Pleistocene refugia located in Mexico and Florida. A novel application of association analysis, which removes the confounding effects of shared ancestry on correlations between genetic and environmental variation, identified five loci correlated with aridity. These loci were primarily involved with abiotic stress response to temperature and drought. A unique set of 24 loci was identified as FST outliers on the basis of the genetic clusters identified previously and after accounting for expansions out of Pleistocene refugia. These loci were involved with a diversity of physiological processes. Identification of nonoverlapping sets of loci highlights the fundamental differences implicit in the use of either method and suggests a pluralistic, yet complementary, approach to the identification of genes underlying ecologically relevant phenotypes.
ENVIRONMENTAL heterogeneity at multiple spatial scales influences the distribution of genetic variation across plant populations. Correlations between genetic variation and environmental gradients have been identified in a variety of plant species (Antonovics and Bradshaw 1970; Antonovics 1971; Hamrick and Allard 1972; Westfall and Conkle 1992; Linhart and Grant 1996; Mitton 1997; Savolainen et al. 2007), and such associations are often interpreted as evidence of natural selection (Allard et al. 1972; Dudley 1996a,b; Gram and Sork 2001; Vasemägi and Primmer 2005; Parisod and Christin 2008). Renewed interest in identifying correlations between environmental and genetic variation has emerged as high-throughput sequencing and genotyping platforms are applied to functional genetic variation within natural populations of non-model species (cf. Joost et al. 2007; Namroud et al. 2008).
Forest trees illustrate clear phenotypic adaptations to environmental gradients at multiple spatial scales (Morgenstern 1996; Savolainen et al. 2007 and references therein). An extensive history of provenance, common garden, and genecological studies has established the highly polygenic basis of these adaptive traits (Langlet 1971; Namkoong 1979). One of the major abiotic stressors for conifers is water availability. Traits related to water-deficit stress (WDS) have a genetic basis for many conifers and variation at these traits is adaptive (Zhang and Marshall 1994; Aitken et al. 1995; Johnsen et al. 1999; Olivas-Garcia et al. 2000; Brendel et al. 2002; González-Martínez et al. 2007; Baltunis et al. 2008). Physiological responses to drought involve many different cellular and molecular pathways (Newton et al. 1991; Ingram and Bartels 1996), and functional and gene expression studies in Arabidopsis have implicated several gene networks in WDS responses, as well as interactions between drought and cold-hardiness traits (Shinozaki and Yamaguchi-Shinozaki 2000, 2007; Bray 2004). Population genetic studies, moreover, have identified functionally diverse genes with skewed site-frequency spectra (González-Martínez et al. 2006), extreme allele-frequency differences across populations (Eveno et al. 2008), altered gene expression (Dubos and Plomion 2003; Watkinson et al. 2003; Yang and Loopstra 2005), or significant associations with WDS (González-Martínez et al. 2008) for multiple pine species distributed across strong precipitation gradients.
This study investigates the potential association between genetic variation at individual loci and aridity gradients for loblolly pine (Pinus taeda L.). The predominant abiotic gradient across the range of loblolly pine is water availability. Water use efficiency is a WDS-related trait and in loblolly pine is heritable, but displays significant non-additive variance as well as strong environmental influences (Warren et al. 2001) and genotype-by-environment interactions (Baltunis et al. 2008). The canonical approach to searching for environmental associations would be to correlate measures of genetic diversity (e.g., allele frequencies, heterozygosity) to environmental variation related to drought stress (Linhart and Grant 1996; Mitton 1997; Mitton et al. 1998; Vasemägi and Primmer 2005). One limitation to this approach is that environment is likely to be confounded with geography and, by proxy, overall genetic structure. Correction for population structure is common in association studies that aim at identifying markers contributing to phenotypes (cf. Yu et al. 2006). An obvious solution to the confounding of genetic structure with environmental variation, therefore, is to apply existing association approaches to environmental data, treating the environment methodologically as a phenotype.
Here, we use two genome-wide marker data sets to address the following questions: (1) What are the patterns of population structure across the range of loblolly pine? (2) What is the degree of confounding between environmental variation and population structure due to geography? (3) Which loci are associated with water availability across the range of loblolly pine? (4) Are loci with strong genotypic correlations with aridity also those with extreme allele-frequency differences among populations? In answering these questions, we highlight the need for further integration of environmental and genetic data in genome scans for loci subject to natural selection, as well as the promise of combining complementary approaches for the identification of functionally important genetic variation within natural populations (cf. Vasemägi and Primmer 2005).
MATERIALS AND METHODS
Focal species:
Loblolly pine (Pinus taeda L.) is distributed throughout the southeastern United States, ranging from Texas to Delaware. Its 370,000-km2 range is divided primarily by the Mississippi River Valley, with 60% of the distribution range located east of the Mississippi River (Al-Rabab'ah and Williams 2002). Isozyme and nuclear simple sequence repeat (SSR) loci illustrate moderate genetic differentiation between populations located to the east and west of the Mississippi River Valley, as well putative population contraction in the westernmost populations (Wells et al. 1991; Schmidtling et al. 1999; Al-Rabab'ah and Williams 2002, 2004; Xu et al. 2008). A review of phylogeographical patterns in unglaciated eastern North America identified six major patterns, of which loblolly pine conforms to the Mississippi River discontinuity (Soltis et al. 2006). The structure of this discontinuity is consistent with a dual Pleistocene refugial model, which has also been used to explain differential growth abilities, disease resistance, and concentrations of secondary metabolites among families located across this discontinuity (Wells and Wakeley 1966; Squillace and Wells 1981; Schmidtling 2003).
Sampling:
Needle tissue was collected from 907 largely unrelated trees sampled across the natural range of loblolly pine (Figure 1). Seven hundred of these trees are first-generation selections (i.e., trees grown from wild-collected seed) with known source localities. These samples are geo-referenced by county (ncounties = 188). The average number of sampled trees per county was 4 ± 6 (range: 1–67). The remaining 207 are second-generation selections (i.e., trees resulting from crosses between the first-generation selections). These trees also comprise two experimental populations currently being used for association mapping: Weyerhaeuser (cf. González-Martínez et al. 2007) and North Carolina State University (cf. P. Cumbie, A. Eckert, J. Wegrzyn, R. Whetten, D. Neale and B. Goldfarb, unpublished results). Total genomic DNA was isolated from each sample at the U.S. Department of Agriculture National Forest Genetics Laboratory (Placerville, CA) using DNeasy plant kits (Qiagen, Valencia, CA) following the manufacturer's protocol.
Figure 1.—
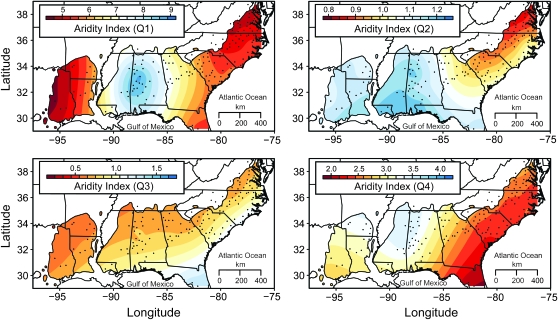
The distribution of loblolly pine, sampling localities (black points) and aridity gradients (color gradients) used to assess patterns of population structure and environmental associations. The distribution of loblolly pine is available as a GIS layer from the U. S. Geological Survey (http://esp.cr.usgs.gov/data/atlas/little/). Aridity gradients were smoothed with kriging using algorithms in the fields package available in R (R Development Core Team 2007). Aridity gradients are shown by annual quarter (Q1: January–March; Q2: April–June; Q3: July–September; Q4: October–December).
Marker discovery and genotyping:
We utilized two sets of molecular markers. The first set comprises 23 unlinked nuclear SSR markers selected from the PtTX marker set for medium to high polymorphism rate and full coverage of the linkage map (Auckland et al. 2002; González-Martínez et al. 2006, 2007). The second set comprises ∼23,000 single nucleotide polymorphism (SNP) markers, of which we chose 7216 for genotyping, that were identified through the resequencing of 7535 uniquely expressed sequence tag (EST) contigs in 18 loblolly pine haploid megagametophytes. These SNPs cover the entire linkage map for loblolly pine (TG accession: TG091; http://dendrome.ucdavis.edu/cmap/), with 117 ± 18 SNPs mapped on average per linkage group for a total of 1635 mapped SNPs with an average distance of 1.2 ± 1.1 cM between SNPs. Selection of SNPs for genotyping was based largely on quality scores derived from the original sequence data and not on functional or site annotations. This ensured thorough coverage of the available sequence resource for loblolly pine (cf. http://dendrome.ucdavis.edu/adept2/). Genotyping of SNPs utilizing the Infinium platform (Illumina, San Diego) was carried out at the University of California Davis Genome Center. Arrays were imaged on a Bead Array reader (Illumina), and genotype calling was performed using BeadStudio v. 3.1.3.0 (Illumina). Information regarding the discovery and annotation, as well as PCR, genotyping, and DNA sequencing protocols for both marker types is available in the supporting information, File S1. The complete data are available in File S2 and File S3.
For each marker locus we calculated observed (HO) and expected (HE) heterozygosity as well as Wright's inbreeding coefficient (FIS = 1 − HO/HE). Loci with extreme values of FIS (|FIS| > 0.25) were removed prior to analysis. We used Fisher exact tests (Guo and Thompson 1992) with Bonferroni corrections to test for Hardy–Weinberg equilibrium (HWE) for each SNP and SSR marker, respectively. All analyses were preformed using the R environment (R Development Core Team 2007).
Environmental data:
Climate data were gathered from the WORLDCLIM 2.5-min geographical information system (GIS) layer using Diva-GIS version 5.4 (Hijimans et al. 2005; available at http://www.diva-gis.org/). Monthly minimum and maximum temperatures, monthly precipitation, and 19 bioclimatic variables were obtained from this layer (Table S1). The temperature and precipitation data were used to estimate potential evapotranspiration (PET) with the method of Thornthwaite (1948). An aridity index (AI) was defined as the ratio of precipitation to PET (File S1), with this ratio being defined quarterly. Annual quarters were defined starting with January 1 through March 31 as quarter one and are labeled as AI1 through AI4. We focus on aridity because it encapsulates water availability as a function of temperature and precipitation. Thus, the remainder of the article focuses solely on aridity.
Patterns of population structure:
Population genetic structure was analyzed by means of principal component analysis (PCA) on genotypes from individual trees. Briefly, PCA was performed on the normalized n × m matrix, M, of genotypes, where n is the number of trees and m is the number of loci. Similar analyses were conducted for SSR markers using a method that accounts for the dependence among alleles at a locus (van Heerwaarden et al. 2010). The eigenvalues corresponding to the principal components (PCs) were inspected to determine the number of major independent axes of genetic differentiation in the data. Following outlier removal and reanalysis of M, the significance of PCs was determined by comparing the value of each standardized eigenvalue to a Tracy–Widom distribution (Patterson et al. 2006). Outliers were defined as trees with PC scores >6 SDs away from the mean for any of the first 10 PCs. Trees were assigned to discrete genetic clusters on the basis of K − 1 significant PCs, where K is the number of clusters being considered (Paschou et al. 2007; van Heerwaarden et al. 2010). Specifically, Ward's hierarchical clustering algorithm was applied to the matrix of Euclidean distances, calculated from the significant PCs, and the resulting dendrogram was used to assign individuals to each cluster using the CUTREE function in R.
For comparison, we also used the program STRUCTURE version 2.2 to infer the number of genetic clusters and membership coefficients within those clusters using the 23 nuclear SSR markers (Pritchard et al. 2000; Falush et al. 2003). Analysis using the SNP data was avoided because of the lack of convergence among runs likely related to insufficient run times for the Markov chain Monte Carlo (MCMC) sampler (data not shown). We varied the number of genetic clusters (K) from 1 to 12, and for each value ran 50 independent MCMC simulations. Each run was carried out for 1.2 × 107 steps, with the first 2.0 × 106 steps being discarded as burn-in. We assumed further that allele frequencies were correlated among populations and that our data contained admixed trees. The optimal value of K was determined using the ΔK method (Evanno et al. 2005) and by inspection of the relationship between the log probability of the data and K. Average admixture coefficients in all cases were estimated for each value of K using the LargeKGreedy algorithm with 1000 random input orders as implemented in the program CLUMPP version 1.1 (Jakobsson and Rosenberg 2007). These values were visualized using bar plots constructed with DISTRUCT version 1.1 (Rosenberg 2004).
Associations between population structure, geography, and environment were studied by fitting a general linear model to each measure of structure (assignment probabilities, PCs) with per-county aridity indices and latitude and longitude as explanatory variables. The relation between the different environmental variables and genetic assignment was visualized by a biplot of the PCA on the environmental variables with plotted points colored according to their corresponding genetic cluster.
Environmental associations and outlier analysis:
Loci associated with environment were identified using a standard association mapping approach, substituting aridity for phenotype. Analysis was done separately for each variable and each SNP. Each tree was assigned a “phenotype” consisting of its corresponding county-level aridity index. Following Price et al. (2006), we used PCA analysis to correct for spurious associations due to confounding of ancestry and aridity. Briefly, PCA is performed as described above, but excluding the target SNP. Two vectors of ancestry-corrected residuals are obtained by multiple linear regression on environmental and genotypic values, using the k significant genetic PCs as independent variables. Association between genotype and aridity is described by the squared correlation r2 between the two vectors. For each SNP, scored in N individuals, the test statistic is calculated as (N − k − 1)r2, which is approximately χ2 distributed with one degree of freedom (Price et al. 2006). SNP loci showing the strongest association with different aridity indices were identified by Q–Q analysis of P-values. The magnitude of environmental differences among SNPs was evaluated using a general linear model with environment as a dependent variable and corrected genotypic values as explanatory variables. Differences of environment among SNP genotypes were evaluated using a general linear model with environment as a dependent variable and corrected genotypic values as explanatory variables. Multiple testing was accounted for using the false discovery rate method of Storey and Tibshirani (2003) with a significance threshold of Q = 0.05, although this method, or any multiple test correction method assuming multiple independent tests of the same null hypothesis, is conservative due to the correlations among environmental variables.
For comparison, we searched for patterns of adaptive differentiation among populations using fdist2 (Beaumont and Nichols 1996; Beaumont and Balding 2004); the populations were those identified using PCA or STRUCTURE. Outlier analyses were conducted only for the SNP data, as opposed to the SSR data, because these represent functional variation likely to be targets of natural selection. We used results from clustering of SSRs and SNPs to define populations because the range of loblolly pine is continuous, the number of discrete populations is relatively unknown, and studies employing FST outlier analysis in forest trees justify a priori population definitions with some form of structure analysis (Eveno et al. 2008; Namroud et al. 2008). We simulated 1.0 × 106 loci under two null models using the ms software (Hudson 2002): an island model and a Pleistocene refugial model (File S1, Figure S1). The latter model was used to account for the effects of historical demography on the null expectation of FST across loci (cf. Excoffier et al. 2009). An iterative approach was used to adjust the median FST (Weir and Cockerham 1984) and HE until they were within 5% of observed values. A modified form of the msstats software developed with the libsequence C++ library (Thornton 2003; available from: http://www.rilab.org/code/random_code.html) was used to estimate FST and HE, while the cplot and pvj programs, which are distributed as part of fdist2, were used to estimate 99% quantiles of the null distribution and P-values, respectively. Extreme values of FST conditional on expected heterozygosity were defined as those that lay above the 99% quantile of the null distribution.
RESULTS
Genotyping summary:
To our knowledge, this is the first successful application of the Illumina Infinium genotyping platform to a non-model plant species. Further details concerning conversion rates and quality scores are available in File S1. We selected one SNP that was typed successfully per EST locus for further analysis (n = 3082 SNPs). Forty-five of these SNPs deviated significantly from HWE proportions, with 23 also having FIS values >0.25. These 23 were removed prior to further analysis. An ascertainment bias was also apparent for the frequency distribution of the minor allele across all 3059 SNP loci (File S1, Figure S2). Call rates across the 23 SSR loci averaged 95%, with 18 of the 23 loci deviating significantly from HWE proportions. All 23 SSR loci had values of FIS <0.25 when only first-generation selections with known source localities were considered.
Integration of these data sets resulted in 622 trees sampled from 167 of the 188 county locations typed for 3059 SNPs and 23 SSRs. Summaries of these data are located in Table 1, with further descriptions of genotyping results presented in File S1 and Figure S3. These data were used to infer patterns of population structure and to search for multivariate and locus-specific environmental associations.
TABLE 1.
Diversity patterns across marker loci for the combined data set
| SSRs | SNPs | |
|---|---|---|
| No. of loci | 23 | 3059 |
| Missing data (%)a | 6.5 ± 5.7 | 2.2 ± 2.6 |
| Expected heterozygosity (HE)a | 0.72 ± 0.19 | 0.25 ± 0.16 |
| Fixation index (FIS)a | 0.11 ± 0.07 | 0.02 ± 0.06 |
| No. of loci in HWEb | 23 | 3037 |
n = 622 trees.
Numbers are averages (±1 SD).
Number of loci that do not deviate from HWE after correcting for multiple testing using a Bonferroni correction.
Patterns of environmental heterogeneity:
Strong correlations exist among environmental measures, as well as between those measures and geographical location (Figure S4). In general, sites located along the Gulf Coast Plain have the highest annual precipitation (1650 mm), whereas sites located in the northeast and southwest have the lowest (980 mm). The highest average annual temperatures are observed in the southeast (20.8°), while sites located in the northeast have the lowest (12.8°). These trends are apparent for measures of aridity both spatially and temporally (Figure 1). In general, all sites have water surpluses in the winter, with those located along the Gulf Coast Plain having the largest. Water deficits are apparent in the Northeast (spring) and Southwest (summer) as the year progresses. By fall, all sites have water surpluses, with those located in the extreme southeast being the driest.
Patterns of population structure:
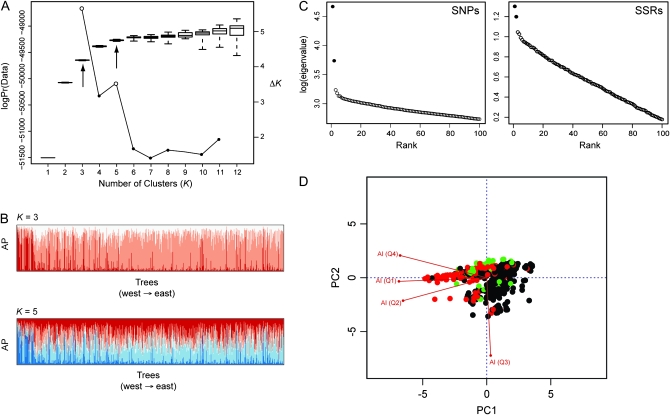
Patterns of population structure for loblolly pine are accounted for primarily by the Mississippi River discontinuity. This is apparent for results from PCA and STRUCTURE using both SSRs and SNPs. PCA analysis on the SNP data revealed the presence of seven significant PCs defining eight genetic clusters. Visual inspection of the eigenvalues, however, shows the presence of two major PCs explaining 2.4% of the total variation (56% of the significant variation), which indicates the presence of three clearly differentiated clusters (Figures 2 and 3). The remaining five significant clusters lack a strong geographical basis. These three clusters are largely divided along the Mississippi River Valley, with a further division of the eastern cluster into Gulf and Atlantic Coast clusters. Similar results were observed for the SSR markers using PCA, with two of the three significant PCs clustered across the Mississippi River discontinuity (Figures 2 and 3).
Figure 2.—
Patterns of population structure for loblolly pine are correlated with geography and aridity. (A) The relationship between the log probability of the data and the number of clusters (K) using the SSR data. Arrows denote values of K assumed during further analysis. (B) Assignment probabilities (AP) for trees arranged from west to east assuming K = 3 or K = 5. (C) Screen plots for PCA analysis using SNPs (left) and SSR markers (right). Major eigenvalues are denoted by black circles. (D) A biplot indicating correlations among aridity variables, as well as with population structure. The PCs were derived by PCA on the four aridity indices. Colors denote cluster memberships as determined using PCA on the SNP data.
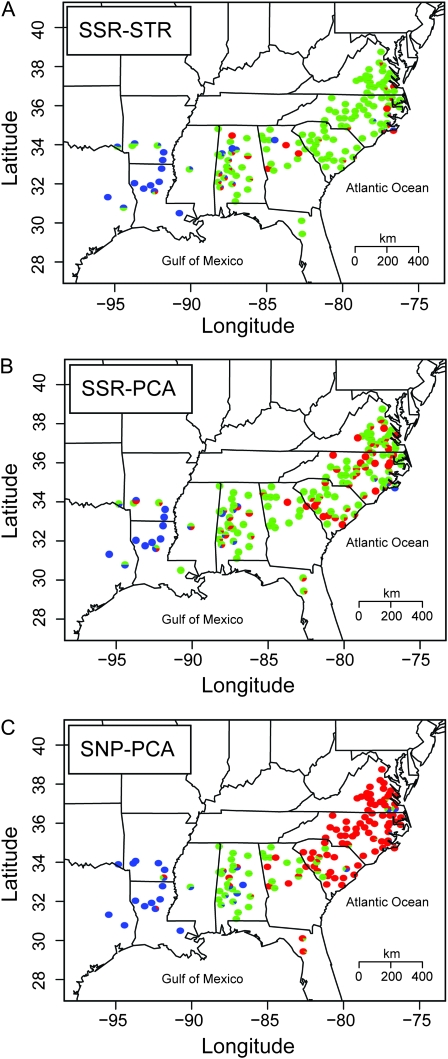
Figure 3.—
Cluster assignments are not dramatically affected by choice of clustering method (STRUCTURE and PCA) and marker type (SSRs and SNPs). Illustrated are assignments summarized by county for each method-marker pair used to assess patterns of population structure for K = 3. Each point denotes the proportion of trees in that county belonging to a specific genetic cluster.
The optimal value of K was 2 as determined by the ΔK statistic using STRUCTURE with the SSRs. Inspection of bar plots for the admixture coefficient when K = 2, averaged across the 50 replicated MCMC runs, indicated that each cluster is geographically based, with one cluster corresponding to trees that are located primarily west of the Mississippi River and the other to those located to the east (Figure S5). The use of ΔK to choose an optimal value of K = 2, however, is difficult, because ΔK in this case compares the lack of structure (K = 1) to some structure (K = 2 and 3). We chose to use values of 3 and 5 for further analysis, because K = 3 had the second largest value of ΔK, and K = 5 is where the median value of the log probability of the data leveled off (Figure 2). Geographical trends in cluster assignments for these values of K reflected the west–east division, as well as further divisions of the eastern cluster along a southwest to northeast axis (Figure 3).
The method of clustering did not dramatically affect the assignment of trees to clusters or the geographical basis of those clusters (Figure 3). The use of PCA and STRUCTURE on the SSR data yielded similar patterns for K = 3, with values of FST calculated for each assignment scheme being significantly correlated (r2 = 0.50, P < 2.2 × 10−16). Some discrepancy between the methods was apparent. For example, PCA placed the trees located in Livingston County, Louisiana, into the eastern population, while STRUCTURE associated them with the western population, which has been noted previously (Schmidtling 2003).
Multilocus population structure was strongly confounded with aridity. Geography was correlated with patterns of population structure and aridity for all clustering methods and choices of K (Table 2, Table S2). The first two PCs from PCA using the SNP data were correlated with geography and aridity. The four aridity indices, longitude, and latitude together explained 61% of the variance in PC1 and 39% of the variance in PC2, thus illustrating that the genetic clusters identified previously differ with respect to aridity (Figure S6). All aridity indices also showed significant correlations with either longitude or latitude (Figure S4). This was reflected by the fact that aridity and geography by themselves explained much of the variance in PC1 (r2 = 0.46 for all four aridity indices, P < 2.2 × 10−16; r2 = 0.51 for latitude and longitude, P < 2.2 × 10−16) and PC2 (r2 = 0.25, P < 2.2 × 10−16 for all four aridity indices; r2 = 0.16 for latitude and longitude, P < 2.2 × 10−16). Three of the four aridity indices (AI1, AI2, and AI4) were also significantly correlated (minimum r2 = 0.53, P < 2.2 × 10−16). A visual representation of the correlations between these variables and genetic structure is given in Figure 2. Similar yet weaker correlations were apparent for PCs and STRUCTURE assignment probabilities using the SSR data (Table 2).
TABLE 2.
Population structure, geography, and environmental heterogeneity are correlated (Pearson's r) across the range of loblolly pine
| SSR-STRa |
SSR-PCAb |
SNP-PCAb |
|||||
|---|---|---|---|---|---|---|---|
| AP1 | AP2 | AP3 | PC1 | PC2 | PC1 | PC2 | |
| Latitude | −0.21*** | 0.20*** | 0.02 | 0.22*** | −0.04 | −0.39*** | −0.39*** |
| Longitude | −0.37*** | 0.34*** | 0.04 | 0.39*** | −0.03 | −0.71*** | −0.34*** |
| AI1 | 0.05 | 0.13** | 0.13** | −0.13** | 0.04 | 0.08 | 0.49*** |
| AI2 | 0.23*** | −0.25*** | 0.03 | −0.27*** | 0.04 | 0.34*** | 0.38*** |
| AI3 | −0.12** | 0.12** | 0.01 | 0.16*** | 0.06 | −0.33*** | −0.08* |
| AI4 | 0.21*** | −0.25*** | 0.07 | −0.26*** | 0.04 | 0.36*** | 0.41*** |
K = 3 for population structure. *P < 0.05, **P < 0.005, ***P < 0.0005.
Correlations are with the assignment probabilities (AP) for each cluster derived using STRUCTURE.
Correlations are with the first two principal components (PCs), which define three clusters.
Environmental associations and outlier analysis:
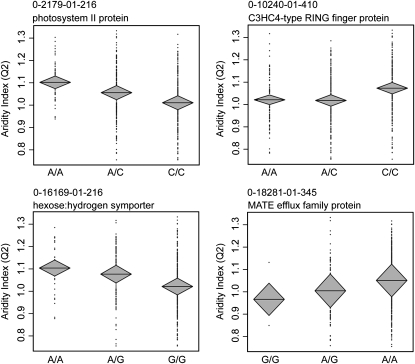
Association analysis resulted in the identification of five loci with significant correlations to aridity (Table 3, Figure S7). The strongest associations were between four loci and aridity during the second quarter (AI2), with subsets of these loci also associating with aridity during the first (AI1) and fourth quarters (AI4). Only a single locus was found to associate significantly with aridity during the third quarter (AI3). The four significant loci associated with aridity during quarters 1, 2, and 4 together explained 9.2% of the variance in AI2 (Figure 4), 5.6% of the variance in AI1, and 4.7% of the variance in AI4 (15.2%, 9.1%, and 8.1% of the ancestry-corrected environment). For comparison, the average SNP explained 0.1% ± 0.2% of the variance in AI2 (0.2% ± 0.3% for ancestry-corrected environment). The single locus associated with AI3 explained 2.6% of the variation (3.0% of variation in ancestry-corrected environment). All five significant SNPs were located in loci with high sequence similarity to coding sequences in Arabidopsis that primarily affect abiotic and pathogenic stress responses (see discussion; Table 3). Two of these five SNPs are mapped to linkage group 3. Three of the five SNPs are located in synonymous positions, while the remaining two are located in an intron and a 3′ UTR.
TABLE 3.
Significant associations between genotypes and aridity gradients
| SNP locusa | Gene product | At locusb | E-valuec | AId | r2e | Pf | Q |
|---|---|---|---|---|---|---|---|
| 0-16169-01-216syn | Hexose:hydrogen symporter | At5g26340 | 3E-118 | AI1 | 0.027 | 4.79E-08 | 1.84E-04 |
| 3E-118 | AI2 | 0.040 | 1.79E-10 | 1.03E-06 | |||
| 3E-118 | AI4 | 0.015 | 2.74E-05 | 3.49E-02 | |||
| 0-2179-01-216nc(utr) | Photosystem II protein | At2g06520 | 2E-24 | AI1 | 0.034 | 6.31E-07 | 1.81E-03 |
| 2E-24 | AI2 | 0.051 | 4.48E-11 | 5.14E-07 | |||
| 2E-24 | AI4 | 0.029 | 1.30E-06 | 2.50E-03 | |||
| 0-10240-01-410syn | C3HC4-type RING finger | At4g26400 | 1E-05 | AI2 | 0.039 | 9.63E-07 | 2.21E-03 |
| 0-18281-01-345nc(intron) | MATE efflux family protein | At4g25640 | 9E-72 | AI2 | 0.022 | 8.23E-06 | 1.35E-02 |
| UMN-6195-01-397syn | UDP-galactose transporter | At3g59360 | 2E-88 | AI3 | 0.029 | 2.50E-05 | 3.49E-02 |
False discovery rate Q < 0.05.
nc, noncoding; ns, nonsynonymous; syn, synonymous; utr, untranslated region.
“At locus” refers to the locus tag for Arabidopsis thaliana.
E-values from tBLASTx analysis of the loblolly pine EST contigs against the NCBI refseq RNA database for Arabidopsis.
AI, aridity index.
r2 is that from a general linear model with environment as the dependent variable and corrected genotypic values as explanatory variables.
P-values listed in boldface type are also significant using a Bonferroni correction. Significance refers to the (N − k − 1)r2 statistic of Price et al. (2006).
Figure 4.—
Genotypic associations for four loci with the aridity index during the second quarter (AI2) illustrate small yet significant correlations. Horizontal lines within diamonds denote the genotypic mean, with diamonds representing the upper and lower bounds of the 95% confidence interval for the mean. Ancestry-corrected environmental indices are plotted on the y-axis. For clarity, the SNP genotypes are plotted on the x-axis, although ancestry-corrected SNP genotypes are used in the association analysis.
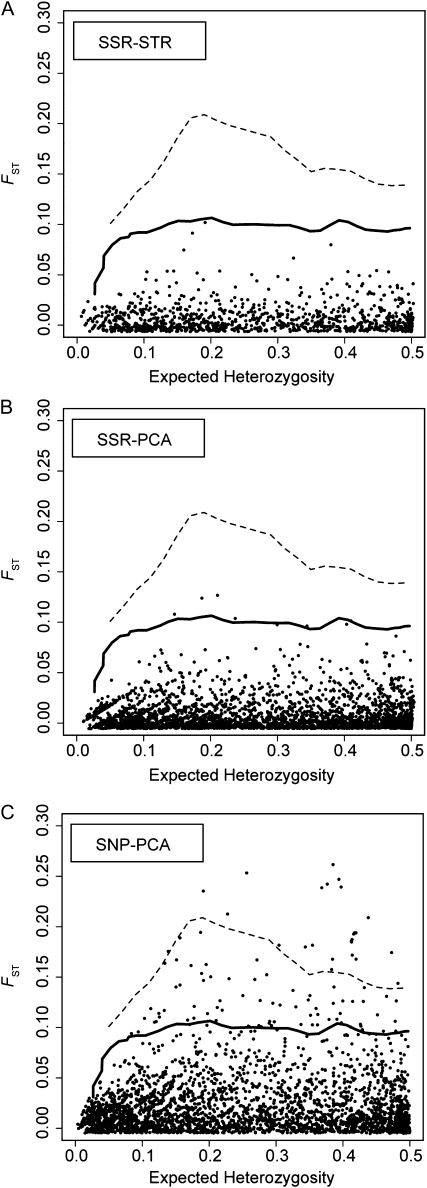
Clusters defined using the SSR markers resulted in few outlier SNPs, patterns indicative of residual within cluster substructure and multilocus values of FST <1% (Figure 5). Most of these outliers can be accounted for by including a dual Pleistocene refugia model as the null model. This model fit the frequency distribution of the minor allele as well as, if not better than, that of drift alone or an island model (File S1, Figure S3). Three loci had significantly elevated FST estimates, however, when K = 5.
Figure 5.—
The quantity and identity of FST outliers under island and dual Pleistocene refugia models are affected by cluster assignments. Each point represents a SNP locus (n = 3059), with lines denoting the upper 99th percentile of the null distribution (solid: island model; dashed: two-refugia model). Each plot represents a different set of cluster assignments based on differing marker sets for K = 3. (A) Cluster assignments from STRUCTURE based on SSR markers. (B) Cluster assignments from PCA using SSR markers. (C) Cluster assignments from PCA using SNP markers.
A different pattern emerges when the cluster memberships are based on SNP loci. In this case, 24 and 15 loci are significant outliers after accounting for demography when K = 3 or K = 5, respectively. In both cases, the multilocus values of FST (0.022 for K = 3, 0.016 for K = 5) are more similar to those reported previously (Schmidtling 2003). Only 7 loci are shared between lists of outlier loci for each value of K; however, the outlier loci unique to a particular value of K were always located in the upper tail of the distribution for FST of the alternative value of K. We focus on the results when K = 3; results for K = 5 are shown in Table S3, Figure S8 and Figure S9. All outlier loci have values of FST that are ∼6- to 12-fold larger than the average FST across all SNPs (Table 4).
TABLE 4.
Significant FST outliers based on K = 3 using a two-refugia model have values of FST 6- to 12-fold greater than the average across all loci
| SNP locusa | Gene productb | At locusc | E-valued | FST |
|---|---|---|---|---|
| 0-1126-02-419ns | O-methyltransferase | At5g54160 | 1E-17 | 0.182 |
| 0-11531-01-379ns | Ovate family protein | At2g18500 | 2E-23 | 0.185 |
| 0-12076-01-310ns | Hypothetical protein | At1g01500 | 7E-05 | 0.157 |
| 0-13230-02-146ns | Eukaryotic translation initiation factor | At3g60240 | 6E-50 | 0.144 |
| 0-14415-01-190ns | Unknown | — | — | 0.209 |
| 0-15241-02-133ns | Hypothetical protein | At2g01300 | 5E-16 | 0.262 |
| 0-17238-01-290nc(intron) | MtN21 family protein | At3g28050 | 3E-20 | 0.213 |
| 0-2784-01-297ns | Nucleoporin family protein | At1g59660 | 2E-10 | 0.247 |
| 0-6427-02-341nc(utr) | Unknown | — | — | 0.186 |
| 0-7745-01-176syn | Unknown | — | — | 0.239 |
| 0-8922-01-645nc(intron) | TIFY domain containing protein | At4g32570 | 1E-11 | 0.194 |
| 1-3327-01-113ns | Unknown | — | — | 0.242 |
| 2-1087-01-86nc(utr) | Ubiquitin-specific protease | At1g04860 | 2E-15 | 0.235 |
| 2-1205-02-71nc(utr) | 3-ketoacyl-CoA synthase | At1g68530 | 2E-54 | 0.180 |
| 2-2953-01-168nc(utr) | Unknown | — | — | 0.176 |
| 2-3591-03-186nc(utr) | Zinc finger-homeodomain protein 2 | At4g24660 | 1E-53 | 0.174 |
| 2-7619-01-191syn | VHS/GAT domain-containing protein | At5g16880 | 8E-29 | 0.168 |
| 2-9190-01-446syn | Auxin responsive protein | At4g30080 | 2E-26 | 0.182 |
| CL1569Contig1-03-45nc(utr) | Hypothetical protein | At3g16070 | 2E-08 | 0.193 |
| CL1799Contig1-04-224nc(intron) | Chlorophyll a/b binding protein | At3g54890 | 1E-112 | 0.168 |
| CL872Contig1-03-287ns | Unknown | — | — | 0.239 |
| UMN-1592-02-53ns | SWIB/MDM2 domain-containing protein | At4g34290 | 2E-30 | 0.172 |
| UMN-897-01-82nc(utr) | Unknown | — | — | 0.253 |
| UMN-CL194Contig1-04-130syn | Unknown | — | — | 0.187 |
P < 0.01 for significant FST outliers.
nc, noncoding; ns, nonsynonymous; syn, synonymous, utr, untranslated region.
All loci listed as unknown have putative homologs in Sitka spruce (P. sitchensis) EST libraries.
Locus tag from Arabidopsis thaliana (At).
E-values from tBLASTx analysis of the loblolly pine EST contigs against the NCBI refseq RNA database for Arabidopsis.
Nineteen of the 24 loci identified as outliers had significant tBLASTx hits to annotated loci in Arabidopsis (Table 4). The remaining 5 loci had little sequence similarity to known protein sequence in plants, although high nucleotide sequence similarity was detected in Sitka spruce [Picea sitchensis (Bong.) Carr.] EST libraries for 4 of those 5 using blastn. Putative functions of gene products in Arabidopsis range from growth regulators to pathogenic stress response. Approximately half of the SNPs located in outlier loci are nonsynonymous point mutations, with the remainder located largely in synonymous positions. Eleven of the 24 outliers are mapped and span five different linkage groups. Six of those 11 are located in close proximity (<2 cM) on linkage group 8.
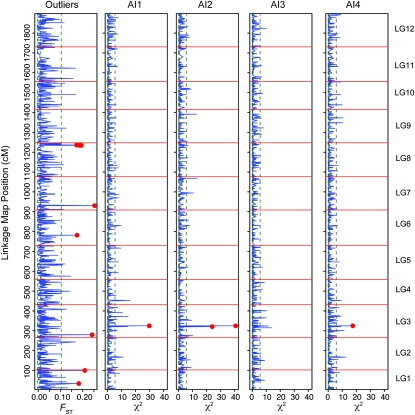
There was no overlap between the loci associated with aridity gradients and those identified as outliers. Loci associated with aridity gradients had values of FST within the range of the mean value across loci (FST = 0–0.035), while FST outliers showed no meaningful correlation with aridity gradients (maximum r2 = 0.008). Correlations between FST and χ2 statistics for aridity during each quarter, moreover, were nonsignificant and the percentage variance explained was <0.5% in all cases. The genomic location of both sets of loci was different, with two of the five loci associated with aridity located ∼4 cM apart on linkage group 3 and the FST outliers spread across linkage groups 1, 3, 6, 7, and 8 (Figure 6).
Figure 6.—
Loci associated with aridity and those identified as FST outliers are distributed across the linkage map of loblolly pine. Plotted are trend lines across the 1898-cM linkage map for FST and χ2 statistics for four aridity indices (AI1, AI2, AI3, and AI4). Red circles denote significant outliers (n = 11) or associated SNPs (n = 2), while dashed green lines denote the 2.5%, 50%, and 97.5% quantiles of the genomic distribution for each statistic. Three of the five SNPs associated significantly to aridity were not segregating within the cross that was used to construct the linkage map and are thus unmapped.
DISCUSSION
Population structure in loblolly pine:
This study presents the first genome-wide analysis for population structure of functional genetic variation among natural populations of loblolly pine. Our results are broadly consistent with previous conclusions regarding patterns of population structure for loblolly pine and conifers in general (Ledig 1998; Scmidtling et al. 1999; Al-Rabab'ah and Williams 2002, 2004; Schmidtling 2003; González-Martínez et al. 2006, 2007; Xu et al. 2008): genetic structure is weak, primarily accounted for by the Mississippi River discontinuity, and is consistent with a dual Pleistocene refugial model. Our results also lay the foundation for performing population structure corrections during association mapping.
We detected further substructure across the range of loblolly pine that was apparent only in the SNP data set. While this result could possibly be a function of the ascertainment bias for the SNP data, such biases are not expected to affect the relative placement of trees in PC space (McVean 2009). However, the same three genetic clusters accounted for most of the geographical patterns observed for levels of population structure across values of K ranging from 3 to 8 (Figure S10), thus justifying our focus on K = 3. This is also reflected in the correlations of genetic structure with aridity (Table 2) because two to three clusters accounted for most of the significant correlations. Geographical trends in cluster assignments show clearly that trees in Florida, a putative refugium, are strongly clustered with those along the Atlantic rather than Gulf Coastal Plain and that trees located to the west of the Mississippi River are genetically distinct. This pattern supports expansion from dual refugia, one located in southern Florida and one in southern Texas or Mexico (Schmidtling 2003).
Novel to the analyses presented here is the placement of the Gulf Coast trees in a unique genetic cluster (Figure 3C). This result is consistent with a scenario of expansion with differentiation or admixture. These trees were intermediate to the eastern and western clusters along the first PC, which is consistent with admixture. Testing of these hypotheses, however, is complicated by introgression between loblolly pine and closely related sympatric pines (Chen et al. 2004), shared ancestral polymorphism (Syring et al. 2007), and sample size differences among the populations considered (McVean 2009).
Environmental associations and FST outliers:
Environmental association analysis identified five genes associated with aridity gradients. The primary functions of gene products encoded by these loci were abiotic and biotic stress responses. All five loci have putative orthologs in Arabidopsis that are responsive to abscisic (ABA) or jasmonic (JA) acid, two plant hormones with well-documented correlations to abiotic stress responses (Glazebrook et al. 2003; Wong et al. 2006; Wasternack 2007; Fabro et al. 2008; Mizuno and Yamashino 2008). In addition, gene expression for four of the five loci in Arabidopsis or close relatives was repressed at low temperatures (Kreps et al. 2002; Vergnolle et al. 2005; Wong et al. 2006), with one locus significantly upregulated in the presence of chitin (Libault et al. 2007) and another upregulated during infection by biotrophic fungi (Fabro et al. 2008). The protein products encoded by these genes also likely affect general osmotic stress responses because drought cold-tolerance responses are often correlated (Beck et al. 2007). These five genes did not have extreme values of FST for any of the six clustering assignments, with values similar to the mean across all loci in each case.
FST outliers encoded proteins involved in a wide range of plant processes including response to viral infection (0–13230; Nicaise et al. 2007), cuticular wax biosynthesis (2–1205; Costaglioli et al. 2005), floral and gametophytic development (2–3591; Tan and Irish 2006), and structural components of photosystem complexes (CL1799Contig1; Jansson 1994). Additional loci have been previously associated with growth (0–14415), leaf nitrogen content (2–1087), and pitch canker resistance (1–3324) for loblolly pine (P. Cumbie, A. Eckert, J. Wegrzyn, R. Whetten, D. Neale and B. Goldfarb, unpublished results; T. Quesada, V. Gopal, W. P. Cumbie, A. J. Eckert, J. L. Wegrzyn, D. Neale, B. Goldfarb, D. Huber, G. Casella and J. Davis, unpublished results). None of these 24 loci, nor the additional 8 FST outlier loci for K = 5, showed strong correlations with aridity.
Many of the outlier loci showed striking geographical trends along the major axis of differentiation (southwest to northeast), with some approaching fixation of different alleles at opposite ends of this axis (Figure S11). One of these was locus CL3949Contig1, which encodes a peptidyl-prolyl cis-trans isomerase affecting protein folding and signal transduction in Arabidopsis (Chou and Gasser 1997). A SNP in this locus was an FST outlier when K = 5 (Table S3) and was in the upper tail of the distribution of FST for K = 3. A putative ortholog of this locus in coastal Douglas fir [Pseudotsuga menziesii (Mirb.) Franco var. menziesii] was also identified as an FST outlier during a scan across candidate genes for associations with cold-hardiness phenotypes (Eckert et al. 2009). Trees in the northeast part of the range for loblolly pine were almost fixed for the A allele at this SNP, whereas the C allele was at high frequency in the southwest. Genome-wide data sets thus open the door to comparative association analysis across natural populations of conifers, where replication is across evolutionary lineages sampled across similar environmental gradients (cf. Turner et al. 2008).
Comparison between environmental association and FST outlier approaches:
A multitude of correlations between environmental variables and genetic variation has been noted across diverse taxa (Mitton 1997). To our knowledge, this is the first study to use the analytical machinery of association analysis to discover environmental associations between functional genetic markers and environmental gradients. This approach identified an independent set of genes relative to the FST outlier approach, which highlights the fundamental differences between these methods. The association approach used here assesses the effect of natural selection along specific environmental gradients, while FST outlier methods aim to identify loci influenced by natural selection driving allele-frequency differences among populations (i.e., ancestral groups) and are thus agnostic about the environmental gradients driving the extreme values of FST. We highlight the advantages and appropriate uses of each method by comparing and contrasting the interpretations attributed to significant results for these two approaches (see also Latta 1998; Barton 1999; Le Corre and Kremer 2003).
Environmental gradients are defined a priori in the association approach, and correlation with them is tested after corrections for population structure. Significant associations with an environmental gradient, therefore, likely represent those polymorphisms underlying functional responses to that gradient. In contrast, post hoc interpretations of environmental differences are attributed to the cause of FST outliers. Geography, however, can create genetic structure that is correlated to environment solely through neutral processes such as barriers to gene flow, distance effects, and historical population size and range changes (Manel et al. 2003; Storfer et al. 2007). As seen here, aridity does differ significantly among the identified genetic clusters, and a tempting conclusion to draw about FST outliers is one related to WDS. This implies that caution should be used when interpreting the biological significance of outlier loci, especially if overall correlations of structure to geography and environmental heterogeneity have not been assessed.
The association approach does not require identification of discrete genetic clusters. As illustrated here, the process of defining populations affects the identification of FST outliers. Although geographical patterns of cluster membership did not change dramatically between methods, they did become clearer in the full SNP data set. Differing definitions of populations may, therefore, lead to spurious results even if the island model is a good descriptor of the underlying genetic structure, unless the “islands” are known without error. A better approach in this case would be to define populations on the basis of environment, while accounting for within-population substructure, rather than neutral marker-based estimates. This is often done for species with subdivided ranges by organizing population samples along environmental or geographical gradients (cf. Eveno et al. 2008).
The identification of genetic clusters may be difficult or inappropriate for species such as loblolly pine that are distributed continuously across large geographical expanses and for which paleobotanical (cf. references in Schmidtling 2003) and population genetic evidence suggests historical fluctuations in population size (Waples and Gaggiotti 2006; Guillot 2009). Invocation of the island model in these cases to derive the null distribution of test statistics will result in undesirable statistical behaviors. Populations of most North American conifers are likely far from demographic equilibrium, and acknowledging this may lead to better inferences of selection (Westfall and Millar 2004). As illustrated here, incorporation of demographic models into FST approaches dramatically reduces the number of outliers detected (Figure 5). Even under equilibrium, however, misspecification of the hierarchical nature of population structure may increase the false-positive rate by nearly an order of magnitude (Excoffier et al. 2009). Results from analyses of FST outlier analyses that do not consider these scenarios (cf. Namroud et al. 2008) should be viewed with some skepticism.
In contrast with association methods, the FST outlier approach identifies loci independently of environment. Genome-wide scans for outliers thus enable identification of loci underlying phenotypic responses to diverse environmental gradients correlated to ancestry (but see McKay and Latta 2002; Le Corre and Kremer 2003). In contrast, the association approach has low power when there is complete confounding between environment gradients and axes of ancestry. Thus, lists of outlier loci are useful because they may allow a reverse engineering of the relevant phenotypes and possibly the environmental gradients on which the phenotypes are selected through comparative genomics, functional experimentation, and GIS analysis. This would in principle help clarify the relationship between genotype, adaptive phenotypes, and fitness (Luikart et al. 2003; Storz 2005; Ross-Ibarra et al. 2007).
We are not the first to note differences between results obtained using association vs. FST outlier approaches. In an analysis of SNPs associated with human diseases, there was no significant difference in FST across associated vs. randomly chosen genes (Lohmueller et al. 2009). This suggests that environment, as opposed to ancestry, plays a significant role in disease risk for many causative polymorphisms in humans. Analogously, natural selection may not be driving large-scale adaptive differences among lineages of loblolly pine, but may instead be selecting genotypes along environmental gradients regardless of ancestry. Selection may thus be operating across different spatial and evolutionary scales in forest trees, and our ability to detect it will depend upon the scale of sampling and the method employed.
Alternatively, quantitative genetic theory predicts that clinal variation for phenotypic traits is derived from small frequency differences at many loci (Barton 1999), with the among-population component of linkage disequilibrium across loci being largely responsible for changes in phenotypic means (Le Corre and Kremer 2003). In this case, loci underlying a quantitative trait are expected to illustrate only small allele-frequency differences among populations, but manifest large phenotypic effects (Latta 1998; McKay and Latta 2002; Le Corre and Kremer 2003). An observation consistent with this expectation was made previously in European aspen (Populus tremula L.) for two SNPs associated with bud phenology (Ingvarsson et al. 2008). This is consistent with the low FST for SNPs associated with aridity and suggests that scans for FST outliers may miss many loci underlying adaptive phenotypes.
While our approach to identifying ecologically relevant genetic variants has numerous advantages, it is not without its limitations. Within-county sampling may be more effective for association analyses than the county-level approach taken here because fine-scale environmental variation may be of considerable biological relevance (Mitton et al. 1998; Parisod and Christin 2008). Alternatively, allele-frequency approaches utilizing local populations have also been shown to be powerful in identifying loci affected by diversifying selection (Hancock et al. 2008). Our SNP data, moreover, represent only a fraction of the coding portion of the loblolly pine genome, and further SNP discovery and analysis would enable truly genomic approaches to ecologically relevant genetic variation. Our FST outlier approach is limited by the assumption that our demographic model reflects the true phylogeographical history of loblolly pine. While model misspecification can lead to erroneous results, we note that our model is likely more biologically plausible than the standard model used in FST outlier analyses and in fact explains the observed data as well as, if not better than, drift or the island model alone. Finally, future approaches using sets of targeted candidate genes, as opposed to genome scans, may be more fruitful in identifying loci correlated with specific environmental gradients.
Conclusions:
We identified general patterns of population structure, environmental correlates to those patterns, and 29 SNP loci that were associated with aridity (n = 5) or had extreme values of FST (n = 24) for loblolly pine. The 5 SNP loci associated with aridity were located in genes encoding proteins primarily involved with biotic and abiotic stress responses, while the 24 outlier loci were located in genes encoding proteins involved with a diverse set of physiological functions. Increased awareness of the strengths, weaknesses, and complementarity of the methods employed during scans for ecologically important genetic variation is needed as truly population genomic data sets emerge for non-model species.
Acknowledgments
We thank Sedley Josserand, Dennis Deemer, Craig Echt, Valerie Hipkins, Vanessa K. Rashbrook, Charles M. Nicolet, John D. Liechty, Benjamin N. Figueroa, and Gabriel G. Rosa for laboratory and bioinformatics support. We also thank Andrew Bower for advice on GIS-derived climate data, W. Patrick Cumbie and Barry Goldfarb for providing geographic information for sampled trees, and Aslam Mohamed for creating the linkage map. The manuscript was also much improved by critical and insightful comments made by two anonymous reviewers. This work was supported by a National Science Foundation (grant no. DBI-0501763).
Supporting information is available at http://www.genetics.org/cgi/content/full/genetics.110.115543/DC1.
References
- Aitken, S. N., K. L. Kavanagh and B. J. Yoder, 1995. Genetic variation in seedling water-use efficiency as estimated by carbon isotope ratios and its relationship to sapling growth in Douglas-fir. For. Genet. 2 199–206. [Google Scholar]
- Allard, R. W., G. R. Babbel, M. T. Clegg and A. L. Kahler, 1972. Evidence for coadaptation in Avena barbata. Proc. Natl. Acad. Sci. USA 69 3043–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Rabab'ah, M. A., and C. G. Williams, 2002. Population dynamics of Pinus taeda L. based on nuclear microsatellites. For. Ecol. Manage. 163 263–271. [Google Scholar]
- Al-Rabab'ah, M. A., and C. G. Williams, 2004. An ancient bottleneck in the Lost Pines of central Texas. Mol. Ecol. 13 1075–1084. [DOI] [PubMed] [Google Scholar]
- Antonovics, J., 1971. The effects of a heterogeneous environment on the genetics of natural populations. Am. Sci. 59 593–599. [PubMed] [Google Scholar]
- Antonovics, J., and A. D. Bradshaw, 1970. Evolution in closely adjacent plant populations. 8. Clinal patterns at a mine boundary. Heredity 25 349–362. [DOI] [PubMed] [Google Scholar]
- Aukland, L., T. Bui, Y. Zhou, M. Shepherd and C. G. Williams, 2002. Conifer Microsatellite Handbook. Texas A&M University, College Station, TX. 57 pp.
- Baltunis, B. S., T. A. Martin, D. A. Huber and J. M. Davis, 2008. Inheritance of foliar stable carbon isotope discrimination and third-year height in Pinus taeda clones on contrasting sites in Florida and Georgia. Tree Genet. Genomes 4 797–807. [Google Scholar]
- Barton, N. H., 1999. Clines in polygenic traits. Genet. Res. 74 223–236. [DOI] [PubMed] [Google Scholar]
- Beaumont, M. A., and R. A. Nichols, 1996. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. Lond. B 263 1619–1626. [Google Scholar]
- Beaumont, M. A., and D. J. Balding, 2004. Identifying adaptive genetic divergence among populations from genome scans. Mol. Ecol. 13 969–980. [DOI] [PubMed] [Google Scholar]
- Beck, E. H., S. Fettig, C. Knake, K. Hartig and T. Bhattarai, 2007. Specific and unspecific responses of plants to cold and drought stress. J. Biosci. 32 501–510. [DOI] [PubMed] [Google Scholar]
- Bray, E. A., 2004. Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. J. Exp. Bot. 55 2331–2341. [DOI] [PubMed] [Google Scholar]
- Brendel, O., D. Pot, C. Plomion, P. Rozenberg and J. M. Guehl, 2002. Genetic parameters and QTL analysis of δ13C and ring width in maritime pine. Plant Cell Environ. 25 945–953. [Google Scholar]
- Chen, J., C. G. Tauer, G. Bai, Y. Huang, M. E. Payton et al., 2004. Bidirectional introgression between Pinus taeda and Pinus echinata: evidence from morphological and molecular data. Can. J. For. Res. 34 2508–2516. [Google Scholar]
- Chou, I. T., and C. S. Gasser, 1997. Characterization of the cyclophilin gene family of Arabidopsis thaliana and phylogenetic analysis of known cyclophilin proteins. Plant Mol. Biol. 35 873–892. [DOI] [PubMed] [Google Scholar]
- Costaglioli, P., J. Joubès, C. Garcia, M. Stef, B. Arveiler et al., 2005. Profiling candidate genes involved in wax biosynthesis in Arabidopsis thaliana by microarray analysis. Biochim. Biophys. Acta 1734 247–258. [DOI] [PubMed] [Google Scholar]
- Dubos, C., and C. Plomion, 2003. Identification of water-deficit responsive genes in maritime pine (Pinus pinaster Ait.) roots. Plant Mol. Biol. 51 249–262. [DOI] [PubMed] [Google Scholar]
- Dudley, S. A., 1996. a Differing selection on plant physiological traits in response to environmental water availability: a test of adaptive hypotheses. Evolution 50 92–102. [DOI] [PubMed] [Google Scholar]
- Dudley, S. A., 1996. b The response to differing selection on plant physiological traits: evidence for local adaptation. Evolution 50 103–110. [DOI] [PubMed] [Google Scholar]
- Eckert, A. J., A. D. Bower, J. L. Wegrzyn, B. Pande, K. D. Jermstad et al., 2009. Association genetics of coastal Douglas-fir (Pseudotsuga menziesii var. menziesii, Pinaceae). I. Cold-hardiness related traits. Genetics 182 1289–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno, G., S. Regnaut and J. Goudet, 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14 2611–2620. [DOI] [PubMed] [Google Scholar]
- Eveno, E., C. Collada, M. A. Guevara, V. Leger, A. Soto et al., 2008. Contrasting patterns of selection at Pinus pinaster Ait. drought stress candidate genes as revealed by genetic differentiation analyses. Mol. Biol. Evol. 25 417–437. [DOI] [PubMed] [Google Scholar]
- Excoffier, L., T. Hofer and M. Foll, 2009. Detecting loci under selection in a hierarchically structured population. Heredity 103 285–298. [DOI] [PubMed] [Google Scholar]
- Fabro, G., J. A. Di Rienzo, C. A. Voigt, T. Savchenko, K. Dehesh et al., 2008. Genome-wide expression profiling Arabidopsis at the stage of Golovinomyces cichoracearum haustorium formation. Plant Physiol. 146 1421–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush, D., M. Stephens and J. K. Pritchard, 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J., W. Chen, B. Estes, H. S. Chang, C. Nawrath et al., 2003. Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 34 217–228. [DOI] [PubMed] [Google Scholar]
- González-Martínez, S. C., E. Ersoz, G. R. Brown, N. C. Wheeler and D. B. Neale, 2006. DNA sequence variation and selection of tag SNPs at candidate genes for drought-stress response in Pinus taeda L. Genetics 172 1915–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González–Martínez, S.C., N. C. Wheeler, E. Ersoz, C. D. Nelson and D. B. Neale, 2007. Association genetics in Pinus taeda L. I. Wood property traits. Genetics 175 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González–Martínez, S. C., D. A. Huber, E. Ersoz, J. M. Davis and D. B. Neale, 2008. Association genetics in Pinus taeda L. II. Carbon isotope discrimination. Heredity 101 19–26. [DOI] [PubMed] [Google Scholar]
- Gram, W. K., and V. L. Sork, 2001. Association between environmental and genetic heterogeneity in forest tree populations. Ecology 82 2012–2021. [Google Scholar]
- Guillot, G., 2009. On the inference of spatial genetic structure from population genetics data. Bioinformatics 25 1796–1801. [DOI] [PubMed] [Google Scholar]
- Guo, S. W., and E. A. Thompson, 1992. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics 48 361–372. [PubMed] [Google Scholar]
- Hamrick, J. L., and R. W. Allard, 1972. Microgeographical variation in allozyme frequencies in Avena barbata. Proc. Natl. Acad. Sci. USA 69 2100–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock, A. M., D. B. Witonsky, A. S. Gordon, G. Eshel, J. K. Pritchard et al., 2008. Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genet. 4 e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijimans, R. J., S. E. Cameron, J. L. Parra, P. G. Jones, and A. Jarvis, 2005. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25 1965–1978. [Google Scholar]
- Hudson, R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18 337–338. [DOI] [PubMed] [Google Scholar]
- Ingram, J., and D. Bartels, 1996. The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 377–403. [DOI] [PubMed] [Google Scholar]
- Ingvarsson, P. K., M. V. Garcia, V. Luquez, D. Hall and S. Jansson, 2008. Nucleotide polymorphism and phenotypic associations within and around the phytochrome B2 locus in European aspen (Populus tremula, Salicaceae). Genetics 178 2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson, M., and N. A. Rosenberg, 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23 1801–1806. [DOI] [PubMed] [Google Scholar]
- Jansson, S., 1994. The light-harvesting chlorophyll a/b-binding proteins. Biochim. Biophys. Acta 1184 1–19. [DOI] [PubMed] [Google Scholar]
- Johnsen, K. H., L. B. Flanagan, D. A. Huber and J. E. Major, 1999. Genetic variation in growth, carbon isotope discrimination, and foliar N concentration in Picea mariana: analyses from a half-diallel mating design using field grown trees. Can. J. For. Res. 29 1727–1735. [Google Scholar]
- Joost, S., A. Bonin, M. W. Bruford L. Després, C. Conord et al., 2007. A spatial analysis method (SAM) to detect candidate loci for selection: towards a landscape genomics approach to adaptation. Mol. Ecol. 16 3955–3969. [DOI] [PubMed] [Google Scholar]
- Kreps, J. A., Y. Wu, H.-S. Chang, T. Zhu, X. Wang et al., 2002. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 130 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlet, O., 1971. Two hundred years of genecology. Taxon 20 653–722. [Google Scholar]
- Latta, R. G., 1998. Differentiation of allelic frequencies at quantitative trait loci affecting locally adaptive traits. Am. Nat. 151 283–292. [DOI] [PubMed] [Google Scholar]
- Le Corre, V., and A. Kremer, 2003. Genetic variability at neutral markers, quantitative trait loci and trait in a subdivided population under selection. Genetics 164 1205–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledig, T. F., 1998. Genetic variation in Pinus, pp. 251–280 in Ecology and Biogeography of Pinus, edited by D. M. Richardson. Cambridge University Press, Cambridge, UK.
- Libault, M., J. Wan, T. Czechowski, M. Udvardi and G. Stacey, 2007. Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol. Plant-Microbe Interact. 8 900–911. [DOI] [PubMed] [Google Scholar]
- Linhart, Y. B., and M. C. Grant, 1996. Evolutionary significance of local genetic differentiation in plants. Annu. Rev. Ecol. Syst. 27 237–277. [Google Scholar]
- Lohmueller, K. E., M. M. Mauney, D. Reich and J. M. Braverman, 2009. Variants associated with common disease are not unusually differentiated in frequency across populations. Am. J. Hum. Genet. 78 130–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart, G., P. R. England, D. Tallmn, S. Jordan and P. Taberlet, 2003. The power and promise of population genomics: from genotyping to genome typing. Nat. Rev. Genet. 4 981–994. [DOI] [PubMed] [Google Scholar]
- Manel, S., M. K. Schwartz, G. Luikart and P. Taberlet, 2003. Landscape genetics: combining landscape ecology and population genetics. Trends Ecol. Evol. 18 189–197. [Google Scholar]
- McKay, J. K., and R. G. Latta, 2002. Adaptive population divergence: markers, QTL and traits. Trends Ecol. Evol. 17 285–291. [Google Scholar]
- McVean, G., 2009. A genealogical interpretation of principal components analysis. PLoS Genet. 5 e1000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitton, J. B., 1997. Selection in Natural Populations. Oxford University Press, Oxford.
- Mitton, J. B., M. C. Grant and A. M. Yoshino, 1998. Variation in allozymes and stomatal size in pinyon (Pinus edulis, Pinaceae), associated with soil moisture. Am. J. Bot. 85 1262–1265. [PubMed] [Google Scholar]
- Mizuno, T., and T. Yamashino, 2008. Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol. 49 481–487. [DOI] [PubMed] [Google Scholar]
- Morgenstern, E. K., 1996. Geographic Variation in Forest Trees. UBC Press, Vancouver, BC, Canada.
- Namkoong, G., 1979. Introduction to quantitative genetics in forestry. USDA Forest Service Tech. Bull. no. 1588.
- Namroud, M-C., J. Beaulieu, N. Juge, J. Laroche and J. Bousquet, 2008. Scanning the genome for gene single nucleotide polymorphisms involved in adaptive population differentiation in white spruce. Mol. Ecol. 17 3599–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, R. J., E. A. Funkhouser, F. Fong and C. G. Tauer, 1991. Molecular and physiological genetics of drought tolerance in forest species. For. Ecol. Manage. 43 225–250. [Google Scholar]
- Nicaise, V., J-L. Gallois, F. Chafiai, L. M. Allen, V. Schurdi–Levraud et al., 2007. Coordinated and selective recruitment of eIF4E and eIF4G factors for potyvirus infection in Arabidopsis thaliana. FEBS Lett. 581 1041–1046. [DOI] [PubMed] [Google Scholar]
- Olivas-Garcia, J. M., B. M. Cregg and T. C. Hennessey, 2000. Genotypic variation in carbon isotope discrimination and gas exchange of Ponderosa pine seedlings under two levels of water stress. Can. J. For. Res. 30 1581–1590. [Google Scholar]
- Parisod, C., and P-A. Christin, 2008. Genome-wide association to fine-scale ecological heterogeneity within a continuous population of Biscutella laevigata (Brassicaceae). New Phytol. 178 436–447. [DOI] [PubMed] [Google Scholar]
- Paschou, P., P. Drineas, J. Lewis, C. M. Nievergelt, D. A. Nickerson et al., 2007. Tracing sub-structure in the European American population with PCA-informative markers. PLoS Genet. 4 e1000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, N., A. L. Price and D. Reich, 2006. Population structure and eigenanalysis. PLoS Genet. 2 e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, A. L., N. J. Patterson, R. M. Plenge, M. E. Weinblatt, N. A. Shadick et al., 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38 904–909. [DOI] [PubMed] [Google Scholar]
- Pritchard, J. K., M. Stephens and P. Donnelly, 2000. Inference of population structure using multilocus genotype data. Genetics 155 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, 2007. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna http://www.R-project.org.
- Ross-Ibarra, J., P. L. Morrell, and B. S. Gaut, 2007. Plant domestication, a unique opportunity to identify the genetic basis of adaptation. Proc. Natl. Acad. Sci. USA 104 8641–8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, N. A, 2004. Distruct: a program for the graphical display of population structure. Mol. Ecol. Notes 4 137–138. [Google Scholar]
- Savolainen, O., T. Pyhäjärvi and T. Knürr, 2007. Gene flow and local adaptation in forest trees. Annu. Rev. Ecol. Evol. Syst. 38 595–619. [Google Scholar]
- Schmidtling, R. C., 2003. The southern pines during the Pleistocene. ISHS Acta Horticulturae 615 203–209. [Google Scholar]
- Schmidtling, R.C., E. Carroll and T. LaFarge, 1999. Allozyme diversity of selected and natural loblolly pine populations. Silvae Genet. 48 35–45. [Google Scholar]
- Shinozaki, K., and K. Yamaguchi-Shinozaki, 2000. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signalling pathways. Curr. Opin. Plant Biol. 3 217–223. [PubMed] [Google Scholar]
- Shinozaki, K., and K. Yamaguchi-Shinozaki, 2007. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58 221–227. [DOI] [PubMed] [Google Scholar]
- Soltis, D. E., A. B. Morris, J. S. McLachlan, P. S. Manos and P. S. Soltis, 2006. Comparative phylogeography of unglaciated eastern North America. Mol. Ecol. 15 4261–4293. [DOI] [PubMed] [Google Scholar]
- Squillace, A. E., and O. O. Wells, 1981. Geographic variation of monoterpenes in cortical oleoresin of loblolly pine. Silvae Genet. 30 127–135. [Google Scholar]
- Storey, J. D., and R. Tibshirani, 2003. Statistical significance for genome-wide studies. Proc. Natl. Acad. Sci. USA 100 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storfer, A., M. A. Murphy, J. S. Evans, C. S. Goldberg, S. Robinson et al., 2007. Putting the ‘landscape’ in landscape genetics. Heredity 98 128–142. [DOI] [PubMed] [Google Scholar]
- Storz, J. F., 2005. Using genome scans of DNA variability to infer the genetic basis of adaptive population divergence. Mol. Ecol. 14 671–688. [DOI] [PubMed] [Google Scholar]
- Syring, J., K. Farrell, A. Liston and R. Cronn, 2007. Widespread genealogical nonmonophyly in species from Pinus subgenus Strobus (Pinaceae). Syst. Biol. 56 163–181. [DOI] [PubMed] [Google Scholar]
- Tan, Q. K-G., and V. F. Irish, 2006. The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development. Plant Physiol. 140 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornthwaite, C. W., 1948. An approach toward a rational classification of climate. Geogr. Rev. 38 55–94. [Google Scholar]
- Thornton, K., 2003. libsequence: a C++ class library for evolutionary genetic analysis. Bioinformatics 19 2325–2327. [DOI] [PubMed] [Google Scholar]
- Turner, T. L., M. T. Levine, M. L. Eckert and D. J. Begun, 2008. Genomic analysis of adaptive differentiation in Drosophila melanogaster. Genetics 179 455–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heerwaarden, J., J. Ross-Ibarra, J. Doebley, J. C. Glaubitz, J. J. Sánchez-González et al., 2010. Fine scale genetic structure in the wild ancestor of maize (Zea mays spp. parviglumis). Mol. Ecol. 19 1162–1173. [DOI] [PubMed] [Google Scholar]
- Vasemägi, A., and C. R. Primmer, 2005. Challenges for identifying functionally important genetic variation: the promise of combining complementary research strategies. Mol. Ecol. 14 3623–3642. [DOI] [PubMed] [Google Scholar]
- Vergnolle, C., M-N. Vaultier, L. Taconnat, J-P. Renou, J-C. Kader, et al., 2005. The cold-induced early activation of phospholipase C and D pathways determines the response of two distinct clusters of genes in Arabidopsis cell suspensions. Plant Physiol. 139 1217–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waples, R. S., and O. Gaggiotti, 2006. What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol. Ecol. 15 1419–1439. [DOI] [PubMed] [Google Scholar]
- Warren, C. R., J. F. McGrath and M. A. Adams, 2001. Water availability and carbon isotope discrimination in conifers. Oecologia 127 476–486. [DOI] [PubMed] [Google Scholar]
- Wasternack, C., 2007. Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 100 681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkinson, J. I., A. A. Siosan, C. Vasquez-Robinet, M. Shukla, D. Kumar et al., 2003. Photosynthetic acclimation is reflected in specific patterns of drought-stressed loblolly pine. Plant Physiol. 133 1702–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir, B. S., and C. C. Cockerham, 1984. Estimating F-statistics for the analysis of population structure. Evolution 38 1358–1370. [DOI] [PubMed] [Google Scholar]
- Wells, O. O., and P. C. Wakeley, 1966. Geographic variation in survival, growth and fusiform infection of planted loblolly pine. For. Sci. Monogr. 11 1–40. [Google Scholar]
- Wells, O. O., G. L. Switzer and R. C. Schmidtling, 1991. Geographic variation in Mississippi loblolly pine and sweetgum. Silvae Genet. 40 105–118. [Google Scholar]
- Westfall, R. D., and M. T. Conkle, 1992. Allozyme markers in breeding zone designation. New Forests 6 279–309. [Google Scholar]
- Westfall, R. D., and C. I. Millar, 2004. Genetic consequences of forest population dynamics influenced by historic climatic variability in the western USA. For. Ecol. Manage. 197 159–170. [Google Scholar]
- Wong, C. E., Y. Li, A. Labbe, D. Guevara, P. Nuin et al., 2006. Transcriptional profiling implicates novel interactions between abiotic stress and hormonal responses in Thellungiella, a close relative of Arabidopsis. Plant Physiol. 140 1437–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S., C. G. Tauer and C. D. Nelson, 2008. Genetic diversity within and among populations of shortleaf pine (Pinus echinata Mill.) and loblolly pine (Pinus taeda L.). Tree Genet. Genomes 4 859–868. [Google Scholar]
- Yang, S-H., and C. A. Loopstra, 2005. Seasonal variation in gene expression for loblolly pines (Pinus taeda) from different geographical regions. Tree Physiol. 25 1063–1073. [DOI] [PubMed] [Google Scholar]
- Yu, J., G. Pressoir, W. H. Briggs, I. V. Bi, M. Yamasaki et al., 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38 203–208. [DOI] [PubMed] [Google Scholar]
- Zhang, J., and J. D. Marshall, 1994. Population differences in water-use efficiency of well-watered and water-stressed western larch seedlings. Can. J. For. Res. 24 92–99. [Google Scholar]