Abstract
Knowing mutation rates and the molecular spectrum of spontaneous mutations is important to understanding how the genetic composition of viral populations evolves. Previous studies have shown that the rate of spontaneous mutations for RNA viruses widely varies between 0.01 and 2 mutations per genome and generation, with plant RNA viruses always occupying the lower side of this range. However, this peculiarity of plant RNA viruses is based on a very limited number of studies. Here we analyze the spontaneous mutational spectrum and the mutation rate of Tobacco etch potyvirus, a model system of positive sense RNA viruses. Our experimental setup minimizes the action of purifying selection on the mutational spectrum, thus giving a picture of what types of mutations are produced by the viral replicase. As expected for a neutral target, we found that transitions and nonsynonymous (including a few stop codons and small deletions) mutations were the most abundant type. This spectrum was notably different from the one previously described for another plant virus. We have estimated that the spontaneous mutation rate for this virus was in the range 10−6−10−5 mutations per site and generation. Our estimates are in the same biological ballpark that previous values reported for plant RNA viruses. This finding gives further support to the idea that plant RNA viruses may have lower mutation rates than their animal counterparts.
THE rate of spontaneous mutation is a key parameter to understanding the genetic structure of populations over time. Mutation represents the primary source of genetic variation on which natural selection and genetic drift operate. Although the exact value of the mutation rate is important for several evolutionary theories, accurate estimates are available only for a handful of organisms. RNA viruses show mutation rates that are orders of magnitude higher than those of their DNA-based hosts and in the range of 0.03–2 per genome and replication round (Drake et al. 1998; Drake and Holland 1999; Chao et al. 2002). This difference results from the lack of proofreading activity of the virus-encoded RNA-dependent RNA polymerases (Steinhauer et al. 1992). The evolutionary causes of such elevated mutation rates remain unknown and it is commonly accepted that they may be beneficial as a mechanism to escape from the strong selective pressures imposed by the host's defense mechanisms, although not necessarily evolved in response to natural selection (Elena and Sanjuán 2005; Clune et al. 2008). Indeed, in the short term, a too high mutation rate has pernicious effects on viral fitness since most of the mutations produced are deleterious (Bonhoeffer et al. 2004; Sanjuán et al. 2004).
In the case of plant RNA viruses, it has been repeatedly reported that their populations are highly genetically stable (Rodríguez-Cerezo et al. 1991; Fraile et al. 1997; Marco and Aranda 2005; Herránz et al. 2008) in comparison with their animal counterparts, although reports of higher substitution rates also exist (Fargette et al. 2008; Gibbs et al. 2008). This peculiar behavior might be due in part to stronger stabilizing selection, weaker immune-mediated positive selection (García-Arenal et al. 2001), the existence of strong bottlenecks during cell-to-cell movement and systemic colonization of distal tissues (Hall et al. 2001; Sacristán et al. 2003; Li and Roossinck 2004), severe bottlenecks during vector-mediated transmission (Ali et al. 2006; Moury et al. 2007; Betancourt et al. 2008), or differences in the replication mode compared to lytic animal viruses (French and Stenger 2003; Sardanyés et al. 2009). Another more obvious possibility is that, indeed, plant viruses have lower mutation rates than other RNA viruses. Indeed the only two available direct estimates of mutation rates for plant viruses are both in the lower side of the range usually accepted for animal riboviruses: 0.10–0.13 per genome and generation for Tobacco mosaic virus (TMV) (Malpica et al. 2002) and 0.28 for Tobacco etch virus (TEV) (Sanjuán et al. 2009). However, none of these estimates is perfect. Although in the TMV experiments particular care was taken to measure mutation rate in a long target protected from the action of purifying selection (hence deleterious mutations remain in the population), uncertainties related to the number of infection cycles elapsed during the mutation–accumulation phase and the fraction of mutations that produced a selectable phenotype exist. In the case of TEV, the estimate should be taken as an upper limit because selection was operating during the mutation–accumulation phase. Furthermore, the estimate is of the same order of magnitude as the methodological error.
To further evaluate whether plant RNA viruses show unusually low mutation rates, we have developed a new empirical method that allows estimating the mutation rate and the spectrum of spontaneous mutations produced during an in vivo infectious process. The viral model system chosen for this experiment has been TEV (family Potyviridae, genus Potyvirus), a prototypical example of positive sense RNA virus that has also become a model for virus experimental evolution. The method is based on the analysis of the temporal accumulation of mutations in a 1536-nt-long neutral viral target. TEV genome size is 9539 nt long (GeneBank DQ986288) and encodes a large polyprotein of 346 kDa that self-processes into at least nine mature proteins. One of these proteins, the nuclear inclusion protein b (NIb) has RNA-dependent RNA-polymerase activity (Urcuqui-Inchima et al. 2001). This protein forms inclusions in the nucleus of infected plants and is required in the cytoplasm for replication complexes during viral RNA synthesis. NIb is the only protein that can be provided functionally in trans (Li and Carrington 1995). Taking advantage of this property, we infected Nicotiana tabacum transgenic plants expressing TEV NIb and followed the accumulation of mutations in the viral copy of NIb. This experimental system minimizes the effect of purifying selection on the virus-encoded NIb due to complementation by the transgene.
MATERIALS AND METHODS
Virus and plants:
The pTEV7DA infectious clone (Dolja et al. 1992) was used as source for TEV. A TEV genotype that lacked the full replicase gene (ΔNIb) was produced by inverse PCR using PfuTurbo DNA polymerase (Stratagene) and primers conserving the proteolytic NIa–NIb and NIb–CP sites (5′-TTGCGAGTACACCAATTCACTCATGAGTTGAGTCGCTTCCTT-3′ and 5′-AGTGGCACTGTGGGTGCTGGTGTTGACGCTGGTAAGAAGAAA-3′, respectively). The resulting clone was named pTEV7DA–ΔNIb.
Two different genotypes of N. tabacum L. were used in these experiments, the wild-type tobacco var. Xanthi and the transgenic Nt∷NIb line derived from var. Samsun by Li and Carrington (1995). These transgenic plants express TEV NIb protein in a stable and functional manner. Prior to starting our experiments, the presence of the transgene was confirmed by PCR using Taq polymerase (Roche) and the primers F90–95 (5′-GCTGTATTGAAAGTGCGAC-3′ identical to bases 7767–7786 of TEV NIb) and R86–91 (5′-AGGCCCAACTCTCCGAAAG-3′ complementary to bases 8084–8102 of TEV NIb). The expression of the gene also was confirmed by RT–PCR. Moloney murine leukemia virus reverse transcriptase (MMLV RT) (Fermentas) was used to obtain cDNA from plants RNA extracts using primer R92–96 (5′-GCAAACTGCTCATGTGTGG-3′ complementary to bases 8761–8779 of CP gene). Then this cDNA was amplified using Taq and primers F90–95 and R86–91. Finally, the biological activity of the NIb protein encoded by the transgene was confirmed by inoculating batches of Nt∷NIb plants with infectious RNAs from both viruses. All Nt∷NIb plants inoculated with either TEV (n = 20) or TEV–ΔNIb (n = 10) developed a systemic infection after 6–7 days postinoculation (dpi). By contrast, none of the wild-type plants inoculated with TEV–ΔNIb (n = 5) became infected, while all plants inoculated with TEV (n = 5) were so. Furthermore, these results confirm that the presence of any putative RNA secondary folding structure within the NIb coding sequence is necessary for completing the infectious cycle of the virus.
Experimental procedure:
Infectious plasmid pTEV7DA was linearized with BglII (Takara) and transcribed into 5′-capped RNAs using the SP6 mmESSAGE mmACHINE kit (Ambion Inc). Transcripts were precipitated (1.5 vol of DEPC-treated water, 1.5 vol of 7.5 m LiCl, 50 mm EDTA), collected, and resuspended in DEPC-treated water (Carrasco et al. 2007). RNA integrity was assessed by gel electrophoresis and its concentration spectrophotometrically determined using a Biophotometer (Eppendorf). Twenty 4-week-old Nt∷NIb plants were inoculated mechanically on the third true leaf with TEV transcripts (4–7 μg) and 10% of inoculation buffer (100 mg/ml carborundum, 0.5 m K2HPO4, 3% PEG8000, pH 7). In all cases, first symptoms appeared 6–7 dpi.
Total RNA was extracted using RNeasy Plant Mini Kit (Quiagen) from symptomatic leaves of 3 Nt∷NIb plants at 5, 10, 15, 20, 25, and 60 dpi. One of the plants at 20 dpi was not sampled because it dried out. The full NIb gene was reverse transcribed using MMLV RT and primer R92–96 and PCR amplified using the high-fidelity PrimeSTAR HS DNA polymerase (Takara Bio Inc.) and primers F73–80 (5′-TCATTACAAACAAGCACTTG-3′ identical to bases 6377–6396 of TEV NIa gene) and R92–96. By using this pair of primers we ensure that the mRNA from the transgene is not amplified and only NIb sequences from viral genomes will be so. PCR products of 2403 pb were gel purified with Zymoclean (Zymo Research), cloned into the plasmid pUC19/SmaI (Fermentas), and used to transform Escherichia coli DH5α. At least 25 clones per plant were purified and sent out for sequencing by GenoScreen (http://www.genoscreen.fr) using BIGDYE 3.1 and a 96-capillars ABI3730XL sequencing system (Applied Biosystems). The following five internal primers were used for fully sequencing NIb with overlapping readouts: F1 5′-GCAAACCTGAAGAGCCTTTTCAG-3′; F2 5′-GCATGCTCATCACAAAGCTCAAG-3′; F3 5′-GTGGATGATTTCAACAATCAATTTTATGAT-3′; F4 5′-ACCAGCGTCAACACCAGCAC-3′; F5 5′-GATCTGTCCCATTCCAAAATAGAAAC-3′. Contigs were assembled using GENEIOUS version 4.7 (http://www.geneious.com). The number of clones that rendered useful sequences was 472 (instead of the 500 submitted for sequencing). The number of sequenced clones per plant ranged between 12 and 34, with a median value of 24.
Mutation rate estimations:
Two different approaches have been used to estimate TEV mutation rate. In the first approach, we proceeded as follows. For a given plant the number of clones sequenced that contained zero, one, two, … , k mutations was fitted to a Poisson distribution with parameter λ = μlT, where λ is the expected number of mutations per clone, T the number of generations of viral replication, l = 1536 the length of the amplicon, and μ the mutation rate per base and per generation (m/b/g). Defining generations in vivo for a plant virus is troublesome, given that a viral population colonizing a plant is not replicating synchronously but with overlapping generations. A good approximation is to define viral generations as the number of cycles of cell infections (Malpica et al. 2002). For this definition to be operative, it is necessary first to have an estimate of the average number of viruses produced per infected cell. By performing one-step accumulation curves in tobacco protoplasts, F. Martínez, S. F. Elena and J. A. Daròs (unpublished results) have estimated that, on average, an infected cell yields 1555 genomes (quantified by real-time quantitative RT–PCR). To estimate the number of generations experienced by TEV at the time points where the samples were taken, we revisited previously published data on the kinetics of TEV accumulation (Carrasco et al. 2007). Reanalyzing these data, we found that the model that better describes TEV accumulation within an infected plant was a four-parameters Gompertz growth equation (R2 = 0.975) (Campbell and Madden 1990). From the parameters of the model and using the above estimate of virus yield per cell, it is possible to calculate that during the exponential growth phase, the viral population experienced 3.2 generations per day, but this number reduces as the growth rate flats off and the carrying capacity of the system is reached. After estimating the number of generations corresponding to each sampling day, it is then possible to transform the above per-clone mutation rate values into the biologically meaningful scale of mutations per base and per generation using the simple expression μ = λ/lT. Each plant has been treated as an independent replicate, rendering 19 estimates of μ.
For the second approach, we focused only on putatively lethal mutations, that is, mutations generating frameships or stop codons. Readers need to recall that the only ORF encoded by TEV genome is translated into a single polyprotein. Our method is based on the fact that amino acid substitutions affecting NIb would in turn be neutral because the trans complementation provided by the host (and the best evidence of such active trans complementation is the ability of TEV–ΔNIb to infect Nt∷NIb plants). However, frameship mutations and stop codons affecting the NIb sequence would be lethal because they will produce a virus deficient not only in NIb but also in CP, the gene downstream from NIb, which is not complemented by the host. In haploid populations at the mutation–selection balance, the frequency of deleterious mutations, p, is given by p = μ/s, where s is the selection coefficient. For lethal mutations, however, s = 1, then μL = p and the equilibrium is reached instantaneously because all lethal mutations have been generated in the previous generation (Crow and Kimura 1970). In other words, this method provides an estimate of mutation rate per replication event (m/b/r) rather than by generation, as in the first method. Following Cuevas et al. (2009), it is possible to calculate a mutation rate for the ith amplicon using the expression
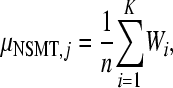 |
(1) |
where n is the total number of nonsense mutational targets (NSMT, i.e., sites that can generate a stop codon after a single nucleotide substitution) in an amplicon, Wi is a weighting factor for the two types of nonsense mutations (Wi = 3 if only one of the three possible mutations in a NSMT produces a stop codon and Wi = 1.5 if two out of three possible produce a stop codon), and K is the total number of observed nonsense mutations in the amplicon. According to the standard genetic code, there are 18 NSMT-containing codons and 19 different NSMTs (the UGG codon contains two). In our experiments, we have 472 independent estimates of μNSMT. If the frequency of insertions and deletions is μindel (it can be computed using the Poisson distribution, as described above), then μL can be estimated as
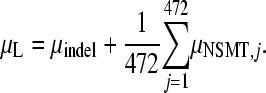 |
(2) |
where μNSMT,j is estimated using Equation 1.
Hereafter, we use the notation μL when referring to the estimates based on the frequency of lethals (units of m/b/r) and reserve the notation μ for the Poisson estimate (units of m/b/g).
Statistical analyses:
All statistical tests have been performed using SPSS version 16. All molecular evolutionary analyses were done using MEGA4 (Tamura et al. 2007).
RESULTS AND DISCUSSION
Characterization of the mutant spectrum:
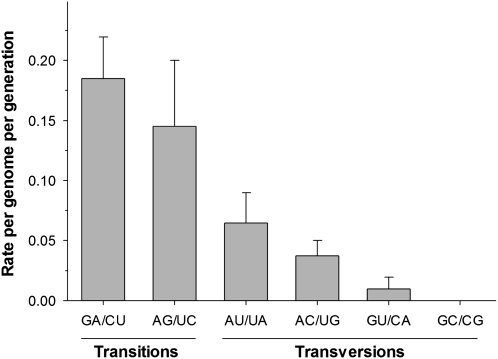
Table 1 summarizes the spectrum of mutations characterized for the 472 clones sequenced (724992 nt). Fifty-two mutations have been identified, 46 of which were nucleotide substitutions and six deletions. Four hundred twenty-six amplicons had no mutation, 41 carried a single mutation, and 5 had two mutations. This distribution does not depart from the Poisson expectation (Kolmogorov–Smirnov test, P = 1). Among base substitutions, 33 were transitions and 13 transversions. Consistent with the principle that transitions are biochemically more likely than transversions, the maximum composite likelihood estimate of the overall transitions to transversions rates ratio was 2.161. This excess also occurs when purines (4.262) or pyrimidines (6.681) are considered separately. Indeed, the observed frequencies of transitions among purines and among pyrimidines are similar (Figure 1) and by far the most frequent type of mutation (Table 1 and Figure 1). Therefore, we can conclude that TEV polymerase produces, on average, 2/3 transitions and 1/3 transversions. If purifying selection would not be canceled out by NIb trans complementation, the ratio would be more biased toward transitions because they are more often silent.
TABLE 1.
Numbers of mutations by type and observed substitution matrix
| Type of mutation | Number | Substitution matrix | ||||
|---|---|---|---|---|---|---|
| Total | 51 | A | U | G | C | |
| Base substitutions | 46 | A | — | 3 | 4 | 7 |
| Transitions | 33 | U | 3 | — | 7 | 0 |
| Transversions | 13 | G | 0 | 9 | — | 0 |
| Synonymous | 16 | C | 10 | 2 | 1 | — |
| Nonsynonymous | 30 (2 stops) | |||||
| Deletions | 6 | |||||
| 1 nt | 3 | |||||
| 3 nt | 1 | |||||
Figure 1.—
Observed frequencies for the different types of nucleotides substitutions. Each column groups mutations rendering complementary pairs and, thus, can occur during the synthesis of the genomic or antigenomic strains. The LaPlace estimator of the frequency has been used to minimize the bias due to small sample size (Agresti and Coull 1998). Error bars represent the 95% confidence interval for the estimator.
Under the observed mutational spectrum, the equilibrium base composition achieved by mutation in the absence of selection would be 31.1% A, 25.4% U, 17.8% C, and 25.7% G. This distribution significantly deviates from what is expected just by sheer chance (χ2 = 55.505, 3 d.f., P < 0.001). The deviation is mainly driven by the unbalanced composition in purines, with a large excess of A (24.5%) that compensates for the large defect in G (−28.9%).
We have observed that 16 mutations were synonymous and 30 were nonsynonymous (including two stop codons). At least eight of the nonsynonymous substitutions could induce a major deformation on NIb folding by replacing polar or charged side chains by apolar ones (E20G, Q462P, H355L, and E507A) or apolar side chains by polar ones (F106S, G200S, and W417R). Three substitutions (L143P, D146H, and D276Y) lead to a strong change in the side chain length. Nine substitutions (D248N, A270V, D276Y, R283Q, I302L, D348N, H355L, T381I, and Q387Stop) may be affecting the putative NIb active site (PFAM00680). Among deletions, three involved a single nucleotide; one involved three contiguous nucleotides. To evaluate whether this pattern of synonymous and nonsynonymous changes is compatible with a model of neutral evolution, we have estimated the difference between substitution rates per nonsynonymous (dN) and synonymous (dS) sites (using Nei–Gojobori's modified method and bootstrap SEM). The observed value of dN − dS = (5.537 ± 4.133) × 10−4 is not significantly different from zero (z = 1.340, P = 0.090), failing to reject the null hypothesis of neutral evolution and validating our methodology for protecting a viral sequence from purifying selection.
Comparison of TEV mutant spectrum with that observed for other plant viruses:
TEV spontaneous mutational spectrum differs in several aspects from the other only one reported for plant viruses, TMV (Malpica et al. 2002). First, TMV mutational spectrum is dominated by insertions and deletions (69% of all mutations belong to these categories). Deletions were both short (five cases with 1–3 nt deleted) and long [seven cases with up to 100 nt deleted). Insertions were also short (1 nt) and large (four cases with poly(A) insertions]. In sharp contrast, only 9.8% of mutations in TEV mutational spectrum were short deletions, and not a single insertion has been observed. This significant difference (Fisher's exact test, P < 0.001) suggests either that TEV NIb is more processive in vivo than TMV replicase or that the difference is due to the experimental setup. In this regard, Malpica et al. (2002) used the MP protein expressed in trans as a target for measuring mutation rate on the viral copy of MP. However, MP has a positive regulatory effect on the formation of TMV replication complex (Beachy and Heinlein 2000) and may favor template switching and a higher rate of deletions and insertions. By contrast, in our experiments NIb is expressed in excess concentration from the transgene and, thus, replicases may remain attached to the RNA molecules reducing the likelihood of template switching.
The second noticeable difference between both mutational spectra refers to the ratio of synonymous to nonsynonymous substitutions. TMV ratio is 1:10, whereas for TEV it is about five times larger (16:30). This difference may reflect that the method employed by Malpica et al. (2002) was less efficient than our method to protect deleterious point mutations from purifying selection, although this explanation is unsatisfactory given the large amount of deletions maintained in TMV populations. Nonetheless, this fivefold difference was not statistically significant (Fisher's exact test, P = 0.146).
A third difference is that the ratio of transitions to transversions was roughly 1.0 for TMV whereas it was > 2.0 for TEV. Given that it is biochemically easier to produce transitions than transversions, the deficit of the former type observed for TMV may reflect a preference of its replicase for transversions or imperfect sampling (Malpica et al. 2002).
Finally, Malpica et al. (2002) found striking the frequency of mutant genotypes carrying multiple mutations. The distribution of mutations per mutant TMV amplicon had a median of one and a range of 1–3. In our case, the distribution had also a median of one and a range of 1–2. From a statistical point of view, both distributions are undistinguishable in shape (Kolmogorov–Smirnov test, P = 0.199) and location (Mann–Whitney test, P = 0.929). Therefore, we would not consider it striking to find a minor proportion of amplicons carrying more than one mutation: it is just what is expected from the Poisson model.
Estimates of the mutation rate:
Applying the first method described in material and methods, we have obtained 19 independent estimates of the spontaneous mutation rate. The estimates ranged from 0 ≤ μ ≤ 1.340 × 10−5 m/b/g. The distribution of estimates was Gaussian (Kolmogorov–Smirnov test, P = 0.944) with mean μ̄ = 4.754 × 10−6 m/b/g and standard deviation sμ = 3.540 × 10−6 m/b/g. As a way to evaluate the statistical power associated with this estimate, we constructed the 95% confidence interval around the mean as 3.048 × 10−6 ≤ μ̄ ≤ 6.460 × 10−6 m/b/g, which excludes the zero. Therefore, according to these values, we conclude that the genomic mutation rate of TEV is 0.045 ± 0.008 (SEM) per generation.
Next, we applied the lethal alleles method. To compute the first term in Equation 2, we proceeded as above and fitted the observed number of deletions per amplicon per plant to a Poisson distribution, obtaining 19 independent estimates of μindel. The average rate of deletion mutations was μindel = (3.787 ± 1.558) × 10−7 deletions/b/r. Next we focused on the computation of the second term in Equation 2. Only two of the ∼725-kb sequences were stop codons (hence K = 2 in Equation 2). As a consequence of codon usage bias, the actual number of NSMT in our sample is 7.46% instead of the expected 10.34%. Taking this source of bias into consideration and after correcting for the three possible nucleotide substitutions per site, the second term in Equation 2 results in (6.295 ± 0.556) × 10−5 m/NSMT/r. Therefore, the estimate of the spontaneous lethal mutation rate is μL = (6.299 ± 0.558) × 10−5 m/b/r or, expressed on the per genome scale, 0.601 ± 0.053 per replication event.
This μL value is 13.4 times higher than the μ estimate obtained using the first method, the difference being highly significant (two-samples t-test, t36 = 10.328, P < 0.001). What may produce this discrepancy? The lethality method has the advantage of being independent from generation time. However, it is strongly dependent on whether the mutations considered are truly lethal. Deviations from this assumption imply that the estimate immediately becomes an upper limit of the true value. In infected cells wherein multiple genomes may coexist, genomes carrying deletions or NSMTs can still be replicated by the pool of polymerases, encapsidated into wild-type capsides, and moved cell-to-cell and even systemically. In other words, complementation with functional proteins makes lethal alleles behave as effectively neutral. An alternative consideration is that, as defined above, one generation involves many replication rounds. Assuming that μL has not been biased by complementation, the 13.4-fold difference between estimates can be interpreted as the number of replication events within an infected cell. Nonetheless, we can conservatively conclude that the above μL estimate must be taken as an upper-limit estimate of the true mutation rate.
Comparison of TEV mutation rate with those obtained for other RNA viruses:
The only previous direct estimate of mutation rate for another plant virus, TMV, was in the range 1.452 × 10−5−2.060 × 10−5 m/b/g (Malpica et al. 2002), values lying well within our two estimates. In a recent study, Sanjuán et al. (2009) estimated TEV upper-limit mutation rate as (2.96 ± 0.32) × 10−5 m/b/g, a value also within both our estimates. In the same study, these authors performed a literature survey for upper-limit estimates of per-site mutation rates for four plant viruses. All the compiled studies were methodologically similar and relied on characterizing the mutant spectrum from individual plants inoculated with a viral clone (i.e., close to zero starting genetic diversity). In neither of these studies was genetic variation protected from purifying selection (Sanjuán et al. 2009). The median upper-limit mutation rate estimated was 7.74 × 10−4 m/b/g, which was in the range of values estimated for animal RNA viruses and some bacteriophages (Drake and Holland 1999) but still 12.3-fold larger than our upper-limit estimate.
Our data allow us to conclude that the mutation rate of TEV is slightly lower than previously estimated by Sanjuán et al. (2009) and very similar to the only other direct estimation available for another RNA plant virus, TMV (Malpica et al. 2002). All these estimates are within a narrow range of values in the lower side of estimates reported for RNA animal viruses and bacteriophages. This agreement suggests that plant RNA viruses have lower mutation rates than those of their animal counterparts. Indeed, this difference in mutation rates may help to partially explain why the rates of molecular evolution of most RNA plant viruses are apparently lower than those observed for RNA animal viruses (Rodríguez-Cerezo et al. 1991; Fraile et al. 1997; Marco and Aranda 2005; Herránz et al. 2008). This difference between animal and plant RNA viruses raises an intriguing question: Given that plant and animal RNA viruses do not form separated phylogenetic groups and that they are replicated by similar polymerases, why do plant RNA viruses show lower mutation rates? We can imagine several scenarios. First, this may not be the rule and just by chance TEV and TMV turn out to have polymerases of particularly good fidelity. A second obvious possibility is that most values for animal RNA viruses are upper-limit estimations. In this sense, it has been reported that Yellow fever virus polymerase has an error rate as low as 1.9 × 10−7 m/b/g (Pugachev et al. 2004). Third, the difference is real and results from differences in the selective pressures that modulated the evolution of mutation rates in both types of hosts; this implies that the mutation rate is higher for animal RNA viruses because animals represent a more stressful environment, perhaps in the form of more diverse cell types or stronger antiviral responses (e.g., the adaptive immune system; Kamp et al. 2003). However, whether virus mutation rates have been optimized by natural selection or are by-products of a parasitic fast lifestyle needs to be confirmed (Elena and Sanjuán 2005).
Potential pitfalls and considerations:
In this study we have used a high-fidelity DNA polymerase to minimize the probability that observed mutations may be due to PCR errors. PrimeSTAR HS DNA polymerase is about two times more accurate than Pfu due to its improved and robust 3′ → 5′ exonuclease activity and its error rate has been estimated to be 1.60 × 10−6 m/b/PCR cycle (http://catalog.takara-bio.com/research.html). Since we run PCRs for 30 cycles, we expect an error rate per amplicon of 4.8 × 10−5 m/b. Henceforth, we may expect ∼34 mutations in our sample to be due to errors during PCR. Unfortunately, this is not the only source of error; the error rate of MMLV RT is around 3.3 × 10−5 m/b/r (Arezi and Hogrefe 2007), which means that we may expect as well ∼24 mutations to be produced during retrotranscription. Since we have obtained 52 mutations, someone may argue that all of them must result from errors during either retrotranscription or PCR amplification. This being the case, the mutation rate of TEV would be <10−9 m/b/g, a value that is, by all means, absurdly low. Furthermore, the estimate of the error rate of PrimeSTAR HS polymerase should be taken with strong precaution. It is surprising that the manufacturer claims that the enzyme has improved fidelity compared with Pfu but the estimate they provide is indistinguishable from values reported for Pfu, 1.3 × 10−6 m/b/PCR cycle (Cline et al. 1996; Bracho et al. 1998). Therefore, we can conclude that even if unwanted mutations are produced during the RT–PCR amplification, the estimated mutation rates are still on the low side of previous reports.
Acknowledgments
We thank J. C. Carrington (Oregon State University) for kindly providing the N. tabacum NIb transgenic plants and the pTEV7DA clone, J. A. Daròs, R. Sanjuán, and our labmates for advice and discussion, and F. de la Iglesia for excellent technical assistance. This study was supported by grants BFU2006-14819-C02-01/BMC and BFU2009-06993 from the Spanish Ministerio de Ciencia e Innovación (MICINN). N.T. is supported by a Formación del personal Investigador fellowship from MICINN.
References
- Agresti, A., and B. A. Coull, 1998. Approximate is better than “exact” for interval estimation of binomial proportions. Am. Stat. 52 119–126. [Google Scholar]
- Ali, A., H. Li, W. L. Schneider, D. J. Sherman, S. Gray et al., 2006. Analysis of genetic bottlenecks during horizontal transmission of Cucumber mosaic virus. J. Virol. 80 8345–8350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arezi, B., and H. H. Hogrefe, 2007. Escherichia coli DNA polymerase III epsilon subunit increases Moloney murine leukemia virus reverse transcriptase fidelity and accuracy of RT-PCR procedures. Anal. Biochem. 360 84–91. [DOI] [PubMed] [Google Scholar]
- Beachy, R. N., and M. Heinlein, 2000. Role of P30 in replication and spread of TMV. Traffic 1 540–544. [DOI] [PubMed] [Google Scholar]
- Betancourt, M., A. Fereres, A. Fraile and F. García-Arenal, 2008. Estimation of the effective number of founders that initiate an infection after aphid transmission of a multipartite plant virus. J. Virol. 82 12416–12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhoeffer, S., C. Chappey, N. T. Parkin, J. M. Whitcomb and C. J. Petropoulos, 2004. Evidence for positive epistasis in HIV-1. Science 306 1547–1550. [DOI] [PubMed] [Google Scholar]
- Bracho, M. A., A. Moya and E. Barrio, 1998. Contribution of Taq polymerase-induced errors to the estimation of RNA virus diversity. J. Gen. Virol. 79 2921–2928. [DOI] [PubMed] [Google Scholar]
- Campbell, C. L., and L. V. Madden, 1990. Introduction to Plant Disease Epidemiology. John Wiley & Sons, New York.
- Carrasco, P., J. A. Daròs, P. Agudelo-Romero and S. F. Elena, 2007. A real-time RT-PCR assay for quantifying the fitness of Tobacco etch virus in competition experiments. J. Virol. Meth. 139 181–188. [DOI] [PubMed] [Google Scholar]
- Chao, L., C. U. Rang and L. E. Wong, 2002. Distribution of spontaneous mutants and inferences about the replication mode of the RNA bacteriophage φ6. J. Virol. 76 3276–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, J., J. C. Braman and H. H. Hogrefe, 1996. PCR fidelity of Pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 24 3546–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clune, J., D. Misevic, C. Ofria, R. E. Lenski, S. F. Elena et al., 2008. Natural selection fails to optimize mutation rates for long-term adaptation on rugged fitness landscapes. PLoS Comp. Biol. 4 e1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow, J. F., and M. Kimura, 1970. An Introduction to Population Genetics Theory. Harper & Row, New York.
- Cuevas, J. M., F. González-Candelas, A. Moya and R. Sanjuán, 2009. Effect of ribavirin on the mutation rate and spectrum of Hepatitis C virus in vivo. J. Virol. 83 5760–5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolja, V. V., H. J. Mcbride, J. C. Carrington, 1992. Tagging of plant potyvirus replication and movement by insertion of beta-glucuronidase into the viral polyprotein. Proc. Natl. Acad. Sci. USA 89 10208–10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake, J. W., and J. J. Holland, 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 96 13910–13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake, J. W., B. Charlesworth, D. Charlesworth and J. F. Crow, 1998. Rates of spontaneous mutation. Genetics 148 1667–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena, S. F., and R. Sanjuán, 2005. Adaptive value of high mutation rates of RNA viruses: separating causes from consequences. J. Virol. 79 11555–11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fargette, D., A. Pinel, M. Rakotomalala, E. Sangu, O. Traoré et al., 2008. Rice yellow mottle virus, an RNA plant virus, evolves as rapidly as most RNA animal viruses. J. Virol. 82 3584–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraile, A., F. Escriu, M. A. Aranda, J. M. Malpica, A. J. Gibbs et al., 1997. A century of tobamovirus evolution in an Australian population of Nicotiana glauca. J. Virol. 71 8316–8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French, R., and D. C. Stenger, 2003. Evolution of Wheat streak mosaic virus: dynamics of population growth within plants may explain limited variation. Annu. Rev. Phytopathol. 41 199–214. [DOI] [PubMed] [Google Scholar]
- García-Arenal, F., A. Fraile, and J. M. Malpica, 2001. Variability and genetic structure of plant virus populations. Annu. Rev. Phytopathol. 39 157–186. [DOI] [PubMed] [Google Scholar]
- Gibbs, A. J., K. Ohshima, M. J. Phillips and M. J. Gibbs, 2008. The prehistory of potyviruses: their initial radiation was during the dawn of agriculture. PLoS One 3 e2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, J. S., R. French, G. L. Hein, J. Morris and D. C. Stenger, 2001. Three distinct mechanisms facilitate genetic isolation of sympatric Wheat streak mosaic virus lineages. Virology 282 230–236. [DOI] [PubMed] [Google Scholar]
- Herránz, M. C., M. Al Rwahnih, J. A. Sánchez-Navarro, S. F. Elena, E. Choueiri et al., 2008. Low genetic variability in the coat and movement proteins of American plum line pattern virus isolates from different geographic origins. Arch. Virol. 153 367–373. [DOI] [PubMed] [Google Scholar]
- Kamp, C., C. O. Wilke, C. Adami and S. Bornholdt, 2003. Viral evolution under the pressure of an adaptive immune system: optimal mutation rates for viral escape. Complexity 8 28–33. [Google Scholar]
- Li, H., and M. J. Roossinck, 2004. Genetic bottlenecks reduce population variation in an experimental RNA virus population. J. Virol. 78 10582–10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. H., and J. C. Carrington, 1995. Complementation of Tobacco etch potyvirus mutants by active RNA polymerase expressed in transgenic cells. Proc. Natl. Acad. Sci. USA 92 457–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpica, J. M., A. Fraile, A., I. Moreno, C. I. Obies, J. W. Drake, et al., 2002. The rate and character of spontaneous mutation in an RNA virus. Genetics 162 1505–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco, C. F., and M. A. Aranda, 2005. Genetic diversity of a natural population of Cucurbit yellow stunting disorder virus. J. Gen. Virol. 86 815–822. [DOI] [PubMed] [Google Scholar]
- Moury, B., F. Fabre and R. Senoussi, 2007. Estimation of the number of virus particles transmitted by an insect vector. Proc. Natl. Acad. Sci. USA 104 17891–17896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugachev, K. V., F. Guirakhoo, S. W. Ocran, F. Mitchell, M. Parsons et al., 2004. High fidelity of Yellow fever virus RNA polymerase. J. Virol. 78 1032–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Cerezo, E., S. F. Elena, A. Moya and F. García-Arenal, 1991. High genetic stability in natural populations of the plant RNA virus Tobacco mild green mosaic virus. J. Mol. Evol. 32 328–332. [Google Scholar]
- Sacristán, S., J. M. Malpica, A. Fraile and F. García-Arenal, 2003. Estimation of population bottleneck during systemic movement of Tobacco mosaic virus in tobacco plants. J. Virol. 77 9906–9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán, R., A. Moya and S. F. Elena, 2004. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc. Natl. Acad. Sci. USA 101 8396–8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán, R., P. Agudelo-Romero and S. F. Elena, 2009. Upper limit mutation rate estimation for a plant RNA virus. Biol. Lett. 5 394–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardanyés, J., R. V. Solé and S. F. Elena, 2009. Replication mode and landscape topology differentially affect RNA virus mutational load and robustness. J. Virol. 83 12579–12589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer, D. A., E. Domingo and J. J. Holland, 1992. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene 122 281–288. [DOI] [PubMed] [Google Scholar]
- Tamura, K., J. Dudley, M. Nei and S. Kumar, 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. [DOI] [PubMed] [Google Scholar]
- Urcuqui-Inchima, S., A. L. Haenni and F. Bernardi, 2001. Potyvirus proteins: a wealth of functions. Virus Res. 74 157–175. [DOI] [PubMed] [Google Scholar]