Abstract
Although selection of chromosomally normal embryos has the potential to improve outcomes for patients undergoing IVF, the clinical impact of aneuploidy screening by fluorescence in situ hybridization (FISH) has been controversial. There are many putative explanations including sampling error due to mosaicism, negative impact of biopsy, a lack of comprehensive chromosome screening, the possibility of embryo self-correction and poor predictive value of the technology itself. Direct analysis of the negative predictive value of FISH-based aneuploidy screening for an embryo's reproductive potential has not been performed. Although previous studies have found that cleavage-stage FISH is poorly predictive of aneuploidy in morphologically normal blastocysts, putative explanations have not been investigated. The present study used a single nucleotide polymorphism (SNP) microarray-based 24 chromosome aneuploidy screening technology to re-evaluate morphologically normal blastocysts that were diagnosed as aneuploid by FISH at the cleavage stage. Mosaicism and preferential segregation of aneuploidy to the trophectoderm (TE) were evaluated by characterization of multiple sections of the blastocyst. SNP microarray technology also provided the first opportunity to evaluate self-correction mechanisms involving extrusion or duplication of aneuploid chromosomes resulting in uniparental disomy (UPD). Of all blastocysts evaluated (n = 50), 58% were euploid in all sections despite an aneuploid FISH result. Aneuploid blastocysts displayed no evidence of preferential segregation of abnormalities to the TE. In addition, extrusion or duplication of aneuploid chromosomes resulting in UPD did not occur. These findings support the conclusion that cleavage-stage FISH technology is poorly predictive of aneuploidy in morphologically normal blastocysts.
Keywords: FISH, PGD, microarray, mosaicism, uniparental isodisomy
Introduction
Development of new assisted reproductive technologies has improved the success of IVF. However, only ∼13% of embryos selected for transfer implant and develop into a healthy infant (Centers for Disease Control and Prevention, 2008). One potential factor accounting for the failure of the selected embryos to implant is aneuploidy. Fluorescence in situ hybridization (FISH) was among the first and has been the most widely used technology to screen for aneuploidy in human embryos. Initially FISH was employed because it can be rapidly performed on single cells at interphase. However, limitations exist, including the inability to comprehensively diagnose the abnormalities of all 24 chromosomes, and the lack of a clinically meaningful benefit in many randomized clinical trials (reviewed in Fritz, 2008). Poor clinical performance could be attributed to the embryonic chromosomal mosaicism that is present on Day 3 of development (when FISH is typically performed). Another significant limitation is the lack of a clinical trial to determine the predictive value of an abnormal FISH result. No study has ever documented that embryos designated as abnormal by FISH actually have little or no potential to become a healthy live born infant. This has left physicians and embryologists with a difficult dilemma: what should be done when an embryo blastulates normally, but has been designated as abnormal by cleavage-stage FISH?
Previous studies have determined the prevalence of euploidy in embryos designated as abnormal by cleavage-stage FISH and found that it varied from 10 to 71% (Li et al., 2005; Munne et al., 2005; Fragouli et al., 2008; Barbash-Hazan et al., 2009). Discrepant results have been attributed to mosaicism or embryo self-correction. However, previous studies have not systematically verified whether any self-correction actually occurs, and a recent study indicates that cleavage-stage mosaicism may be significantly overrepresented by FISH-based analyses (Treff et al., 2010a). New technologies, such as DNA microarray-based aneuploidy screening, that incorporate both chromosome copy number (CN) analysis and single nucleotide polymorphism (SNP) genotyping in parallel (Vanneste et al., 2009; Johnson et al., 2010; Treff et al., 2010b) may provide the first opportunity to investigate multiple explanations for the differences observed between cleavage-stage FISH and blastocyst reanalysis results.
Treff et al. (2010b) have demonstrated in a randomized and blinded fashion the ability to accurately identify aneuploidy of all 24 chromosomes using an SNP microarray-based method. Single cells from cell lines with known abnormalities were diagnosed with 98.6% accuracy and with no false positive aneuploidy diagnoses. Furthermore, SNP microarray-based 24 chromosome aneuploidy screening has been evaluated in a prospective non-selection randomized clinical trial (Scott et al., 2008), and demonstrated a high negative and positive predictive value for embryo reproductive potential. Therefore, SNP microarray-based 24 chromosome aneuploidy screening technology can be used to accurately predict the chromosomal complement of blastocysts reanalyzed after being designated as aneuploid by cleavage-stage FISH.
This study not only contributes additional reanalysis results using a clinically validated comprehensive aneuploidy screening technology, but also provides the first opportunity to investigate putative self-correction mechanisms including embryonic mosaicism, preferential segregation of chromosomal abnormalities to the trophectoderm (TE) and extrusion or duplication of aneuploid chromosomes resulting in uniparental disomy (UPD).
Materials and Methods
Experimental design
SNP microarray-based 24 chromosome aneuploidy screening technology was used to reassess embryos previously diagnosed as aneuploid and to evaluate mosaicism, confinement to the TE and extrusion or duplication of aneuploid chromosomes. Embryos that developed to morphologically normal blastocysts, and that were given an aneuploid FISH diagnosis from the cleavage-stage blastomere biopsy, were reanalyzed by SNP microarray-based 24 chromosome aneuploidy screening after preparing the embryo into three TE and one inner cell mass (ICM) sections. Mosaicism, preferential segregation to the TE layer and chromosome extrusion or duplication resulting in uniparental isodisomy (UPID), were all investigated as possible explanations for euploid observations. All materials were obtained and evaluated with informed patient consent and under Institutional Review Board approval.
Embryos
As a component of clinical care, a single blastomere was biopsied from embryos and underwent FISH-based preimplantation genetic diagnosis for aneuploidy screening (PGD-AS) due to repeated implantation failure, recurrent pregnancy loss or advanced maternal age. Embryos diagnosed as aneuploid by single blastomere FISH and that developed into morphologically normal blastocysts, as defined by Gardner and Schoolcraft, (1999), were cryopreserved and donated for research. A total of 50 embryos were evaluated from 24 patients. The mean maternal age was 35.1 ± 4.1 years.
Cell lines
Human B-lymphocytes from patients with known UPID were purchased from the Coriell Cell Repository (CCR, Camden, NJ) and cultured as recommended by CCR (repository numbers GM15603 and GM11496). Five cells (to model the minimum number of cells represented in blastocyst sections described below) were removed from media and picked up with a 275 µm micropipette under a dissecting microscope, and placed in a nuclease-free 0.2 ml polymerase chain reaction (PCR) tube in 1 µl of media for immediate lysis (as described below). Cell lysates underwent subsequent whole genome amplification (WGA) and 262 K microarray analysis as previously described (Treff et al., 2010b). In order to determine the ability of SNP microarray-based 24 chromosome aneuploidy screening to detect aneuploidy within a mosaic blastocyst biopsy, samples from a normal female and a normal male cell line were purchased from the CCR (Camden, NJ) and cultured as recommended by CCR (repository numbers GM00321 and GM00323). Samples were mixed at levels of 0, 25, 40, 60, 75 and 100% of one sample relative to the other. The ability to detect a monosomy X within a mixture of male (monosomy X) and female (disomy X) samples was evaluated as previously described (Treff et al., 2010b).
Fluorescence in situ hybridization
Blastomere biopsy was conducted on Day 3 of embryonic development as previously described (Csokmay et al., 2009). Biopsied embryos were rinsed repeatedly and placed individually in extended microdroplet culture for potential embryo transfer pending FISH analysis. The isolated blastomeres were individually placed on a glass slide, pre-incubated in a hypotonic solution and fixed using 3:1 methanol:acetic acid solution. Two rounds of FISH were performed on fixed blastomeres using probes specific for chromosomes 13, 15, 16, 17, 18, 21, 22, X and Y (Vysis, Downer's Grove, IL). The slides were analyzed using an automated Olympus BX61 fluorescence microscope (Center Valley, PA). The images were captured using Cytovision probe software (Applied Imaging Corp., San Jose, CA). Reconfirmation of inconclusive results was conducted as previously described (Colls et al., 2007).
SNP microarray-based 24 chromosome aneuploidy screening
Only embryos that had been previously diagnosed as abnormal by FISH and that fully blastulated were selected for reanalysis. Embryos chosen for reanalysis were slow-thawed and biopsied as previously described with slight modifications (Treff et al., 2010c). Instead of performing a single biopsy, four separate sections were processed (three individual TE samples and one ICM sample) to evaluate mosaicism and potential confinement to the TE. Briefly, TE biopsy was performed by opening a hole in the zona pellucida with a series of single, millisecond pulses by a 1–3 µm diode laser from an infrared 1.48 pulse duration at 100% power (Hamilton-Thorne Research, Beverly, MA). Sections of herniating TE cells were aspirated individually into a TE biopsy pipette (Humagen, Charlottesville, VA) and detached from the blastocyst by firing several pulses at the constricted area of cells at the end of the pipet. The biopsied pieces of TE tissue underwent three sequential washes in a hypotonic solution and were placed intact in nuclease-free 0.2 ml PCR tubes (Ambion, Austin, TX). A fourth sample consisting of cells comprising the ICM, was separated from the zona pellucida and placed into a PCR tube, as explained above. ICM was judged as free from TE cells by morphological assessment by an embryologist. In addition, each sample consisted of ∼5 cells, as judged by an embryologist. Immediately following biopsy, all cells were lysed, as previously described (Cui et al., 1989), and frozen at −20°C for SNP microarray-based 24 chromosome aneuploidy screening analysis. All sample tubes were coded by an embryologist and blinded for downstream microarray analysis.
All four sections taken from a blastocyst underwent SNP microarray-based 24 chromosome aneuploidy screening, as previously described (Treff et al., 2009). Briefly, WGA was conducted according to the recommended protocol beginning with library preparation (GenomePlex WGA4, Sigma Aldrich, St Louis, MO). WGA DNA was purified using the GenElute PCR Purification Kit as recommended (Sigma Aldrich). A WGA yield of 2.5 µg was considered successful amplification. Two-hundred and fifty nanograms of purified WGA DNA were processed through the NspI GeneChip Mapping 262 K microarray as recommended by the manufacturer (Affymetrix, Santa Clara, CA). Aneuploidy screening was performed by CN analysis of the microarray data using the Copy Number Analysis Tool (CNAT) version 4.0.1 (Affymetrix). The microarray data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE18932 (http://www.ncbi.nlm.nih.gov/geo).
Data analysis
Microarray reanalysis consistency with FISH
Embryos were selected for reanalysis based on one of three FISH diagnostic categories, (i) a single monosomy, (ii) a single trisomy or (iii) at least two or more abnormalities (complex aneuploid). This selection provided an opportunity to determine whether one type of FISH-based diagnosis was more prone to inconsistency with SNP microarray-based 24 chromosome aneuploidy screening reanalysis results. An ANOVA was performed to determine whether there were differences in consistencies between these three categories. Alpha was set at 0.05.
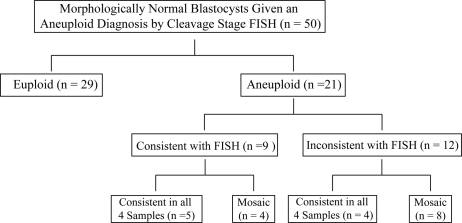
After each embryo section diagnosis was made by SNP microarray-based 24 chromosome aneuploidy screening, samples were decoded to reveal the relationship between each section. All evaluable sections from the same embryo were compared for similarity within the microarray data and against the original blastomere FISH result. Results were categorized into two main and four subset groups (Fig. 1). Results were first categorized as ‘euploid’ (all sections) or ‘aneuploid’ (at least one section). The aneuploid category was further separated into two subsets; either ‘consistent’ or ‘inconsistent’ with FISH when considering only the chromosomes evaluated by FISH. ‘Consistent’ aneuploid results were either consistent in all four sections or only consistent in some but not all of the sections (mosaic). Microarray ‘aneuploidy’ results that were ‘inconsistent’ with FISH had either the same diagnosis in all sections or had more than one diagnosis (mosaic).
Figure 1.
Diagram of categorical results of microarray reanalysis of morphologically normal blastocysts given a FISH-based aneuploidy diagnosis at the cleavage stage.
Mosaicism and TE confinement
SNP microarray-based 24 chromosome aneuploidy screening diagnosis was conducted on embryos that were separated into three sections containing TE cells and one section containing ICM cells. Evaluation of the rate of aneuploidy in TE sections was compared with the aneuploidy rate in ICM sections using a 2 × 2 contingency table of χ2 analysis. Alpha was set at 0.05. Mosaic blastocysts that included an aneuploidy consistent with the original FISH result were also specifically evaluated for preferential segregation of abnormalities to the TE.
Monosomy and trisomy rescue
Duplication (monosomy rescue) or extrusion (trisomy rescue) of an aneuploid chromosome represents an additional mechanism of self-correction and could be identified by the presence of UPD. All disomies derived from duplication of a monosomic chromosome would result in UPID, where both chromosomes have identical DNA sequences. In addition, a proportion of disomies derived from extrusion of a trisomic chromosome would result in UPID (Engel and Antonarkis, 2002). One hallmark of chromosomal UPID is a lack of any heterozygous SNPs (Suela et al., 2007). The absence or loss of heterozygosity (LOH) of SNPs in a chromosome was therefore used to indicate the presence of UPID. The chromosome-specific probability of LOH was calculated by averaging the probabilities assigned to each SNP for a given chromosome using CNAT 4.0 (Affymetrix Inc.). A DM algorithm setting of 0.01 was the only change to the default settings for analysis of LOH by CNAT 4.0.
In order to establish a mean LOH probability threshold for identifying the presence of UPID in only a few cells (i.e. an embryo TE or ICM section), four replicates of five cells each, from two different cell lines, with known UPID chromosomes (described above) were evaluated. LOH probability distributions of chromosomes known to be UPID in origin (positive controls), and of chromosomes known to be disomic with bi-parental inheritance (negative controls), were used to establish a UPID LOH probability threshold with 100% sensitivity and specificity. Chromosomes from embryo TE and ICM sections that reached the defined threshold were assigned an origin of UPID. UPID was evaluated for each of the chromosomes that were originally given a monosomy or a trisomy diagnosis by FISH but were shown to be disomic by microarray. In addition, the overall prevalence of UPID was determined by evaluating LOH probabilities in all copy neutral autosomes (i.e. CN of two) as assessed by SNP microarray-based 24 chromosome aneuploidy screening. The LOH probability of microarray copy neutral chromosomes was compared with UPID positive and negative control chromosomes using an ANOVA. Alpha was set at 0.05.
Results
Microarray reanalysis consistency with FISH
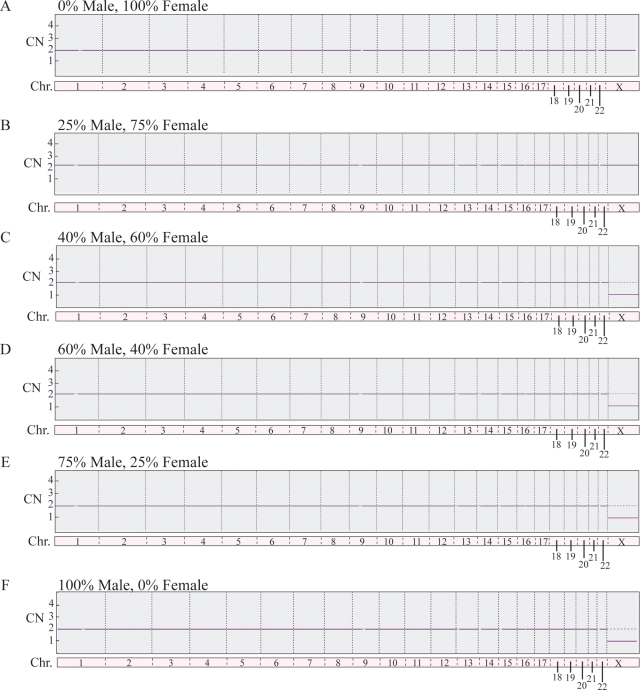
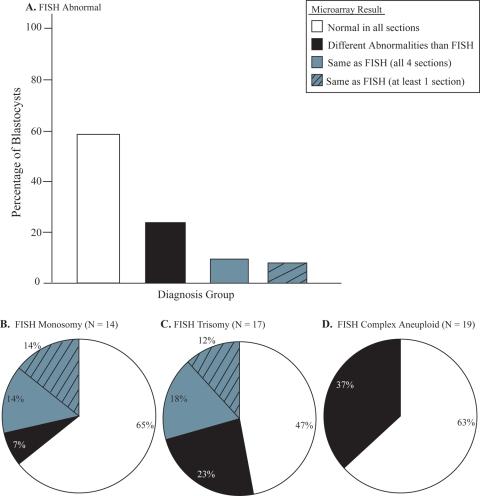
SNP microarray-based 24 chromosome aneuploidy screening demonstrated the ability to detect aneuploidy in samples that possess more than 25% aneuploidy (Fig. 2) indicating that the present technology is capable of detecting mosaicism when as few as 2 of the typical 5 cells within a blastocyst biopsy contain the same aneuploidy. Successful amplification and SNP microarray-based 24 chromosome aneuploidy screening were obtained for 192 of the 200 blastocyst sections processed (96%). After decoding identities for each section, data were assembled according to the embryo from which it came and compared with the original FISH result (Table I). Overall, reanalysis by microarray diagnosed 29 embryos as normal (58%) in all evaluable sections (Fig. 3A). Twenty-four percent of the embryos were aneuploid for chromosomes different from the original FISH result. Ten percent of the embryos were consistent with the original FISH result in all four sections evaluated by microarray. An additional 8% were consistent with the original FISH result in at least one section evaluated by microarray.
Figure 2.
Results of microarray-based aneuploidy screening of mixtures of normal female (46,XX) and a normal male (46,XY) samples indicating a sensitivity of aneuploidy detection (monosomy X) at >25%. As shown, 0–25% male samples (A, B) did not detect monosomy X whereas for 40, 60, 75 and 100% (C–F) monosomy X was detected (right hand column).
Table I.
Diagnosis of all cleavage-stage FISH abnormal embryos and reanalysis results of SNP microarray-based 24 chromosome aneuploidy screening at the blastocyst stage.
| Microarry blastocyst |
|||||
|---|---|---|---|---|---|
| Embryo | FISH aneuploidy | TE | TE | TE | ICM |
| Normal | |||||
| 1 | −13, −16, −17 | — | — | — | — |
| 2 | X, −17, −21, −22 | — | — | — | — |
| 3 | +21 | — | — | — | — |
| 4 | −16, −18 | — | — | — | — |
| 5 | −18 | — | — | — | — |
| 8 | −22 | — | — | — | — |
| 9 | +16 | — | — | — | — |
| 10 | +18 | — | — | — | — |
| 15 | −18 | — | — | — | — |
| 16 | +13, −17, −18 | — | — | — | — |
| 17 | +13, +18, +22 | — | — | — | — |
| 20 | −21 | — | — | — | — |
| 21 | −17, −18, −22 | — | — | — | — |
| 23 | −13 | — | — | — | — |
| 24 | −16 | — | — | — | — |
| 25 | −13, −15 | — | — | — | — |
| 26 | −17 | — | — | — | — |
| 27 | −21 | — | — | — | — |
| 28 | −17 | — | — | — | — |
| 29 | +13 | NA | — | NA | — |
| 30 | +15, +22 | — | — | — | — |
| 32 | +18 | — | — | — | — |
| 34 | +21 | — | — | — | — |
| 37 | +13 | — | — | — | — |
| 38 | +21 | — | — | — | — |
| 47 | −13, −16, −21, −22, +18 | — | — | — | — |
| 48 | −15, −17, +18 | — | — | — | — |
| 49 | −15, −17*, +18 | — | — | — | — |
| 50 | −16, +13, +22 | — | — | — | — |
| Different abnormality than FISH | |||||
| 18 | −15, −17 | −15 | −15 | −15 | −15 |
| 19 | X | NA | +11 | +11 | +11 |
| 31 | +13 | +2 | +2 | NA | +2 |
| 33 | +18 | −4 | — | −4 | −4 |
| 35 | +21 | — | — | −14, −18 | — |
| 36 | +13 | — | +16 | — | — |
| 39 | +13, +16 | −6 | −6, −16 | NA | NA |
| 42 | −16, −22, +13, +15 | −22 | −22 | −22 | −22 |
| 43 | −21, −22, +18 | +15, +17, +18 | +15, +17, +18 | +18 | +15, +17, +18 |
| 44 | −16, +22 | +22 | — | — | — |
| 45 | XYY, −16, +21 | −18, +21 | −18, +21 | −11, −18, +21 | −18, +21 |
| 46 | −21, +13, +22 | — | +22 | +22 | — |
| Same abnormality as FISH | |||||
| 6 | −22 | — | — | — | −22 |
| 7 | −16 | −16 | −16 | −16 | −16 |
| 11 | +21 | +21 | +21 | +21 | +21 |
| 12 | +16 | +16 | +16 | +16 | +16 |
| 13 | +15 | +15 | +15 | +15 | +15 |
| 14 | −22 | NA | −22 | −22 | −22 |
| 22 | −15 | −15 | −15 | — | −15 |
| 40 | +16 | +16 | +16 | +5, −9 | NA |
| 41 | +15 | +15 | — | +15 | +15 |
Legend: normal diagnosis (46,XX or XY) is represented by —. ‘Same as FISH’ indicates that at least one microarray result was consistent with the original FISH diagnoses. Abbreviations: TE, trophectoderm; ICM, inner cell mass; NA, no amplification; *, nullisomy.
Figure 3.
Results of microarray for (A) all blastocysts (n = 50), (B) blastocysts with a single monosomy diagnosis by FISH, (C) blastocysts with a single trisomy diagnosis by FISH or (D) blastocysts with two or more abnormalities diagnosed by FISH.
A subset analysis separated out the FISH results into three categories: monosomy, trisomy or complex aneuploid (Fig. 3B). Of those embryos diagnosed as monosomy by the original FISH analysis, 65% were euploid for all 24 chromosomes in all four sections evaluated by microarray. In another 7%, microarray showed a consistent abnormality in all evaluable sections. However, the abnormalities observed were different from the original FISH result. The remaining 28% of embryos showed a result consistent with FISH in one or more sections evaluated by microarray (Fig. 3B).
In those embryos called trisomy by FISH, 47% were euploid for all 24 chromosomes in all sections evaluated by microarray. In another 23%, microarray showed a consistent abnormality in all evaluable sections. However, the abnormalities observed were different from the original FISH result. The remaining 30% of embryos showed a consistent result with FISH in one or more sections evaluated by microarray (Fig. 3C).
Finally, of those embryos called complex aneuploid by FISH, 63% were euploid for all 24 chromosomes in all sections evaluated by microarray. In the remaining 37% of complex aneuploid FISH embryos, microarray showed a consistent abnormality in all evaluable sections. However, the abnormalities observed were different from the original FISH result. Remarkably, there were no complex aneuploid FISH embryos that were consistent with one or more sections evaluated by microarray (Fig. 3D). Embryos labeled by FISH as complex aneuploid were microarray normal more often than those that were trisomic or monosomic (P< 0.006).
Examination of self-correction mechanisms: mosaicism and TE confinement
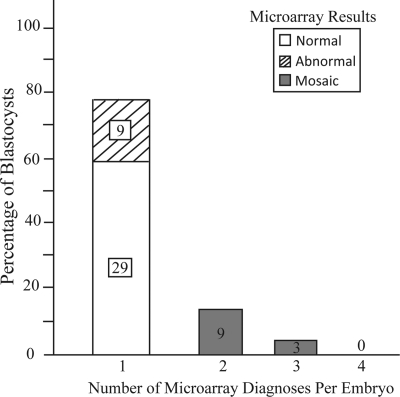
The rate of mosaicism observed by microarray reanalysis of multiple sections of 50 blastocysts was 24% (Fig. 4). The remaining 76% of blastocysts displayed the same euploid or aneuploid diagnosis in all evaluable sections (Fig. 4). Eight of the 12 mosaic embryos were entirely inconsistent with the original FISH result (Table I). Of the four mosaic embryos that had abnormalities possibly consistent with the original FISH result, only embryo number 6 (Table I) displayed preferential segregation of the abnormality. However, the abnormality was present in the ICM and not the TE. The ICM sample from one embryo (number 40, Table I) did not amplify. The other two embryos (numbers 22 and 41, Table I) showed no preferential segregation since abnormalities were found in both the TE and ICM. Aneuploidy was observed in 15 of 48 ICM samples (31%) and 46 of 144 TE samples (32%), showing no indication of preferential aneuploid cell migration to the TE layer (P = 0.9).
Figure 4.
The rate of mosaicsm in all embryos diagnosed as abnormal by FISH and reanalyzed in four separate blastocyst sections by microarray. Blastocysts are represented as having 1 diagnosis (no mosaicism) or having 2, 3 or 4 different diagnoses in the same embryo (mosaicism). Numbers inside each bar represents the number of embryos.
Monosomy and trisomy rescue
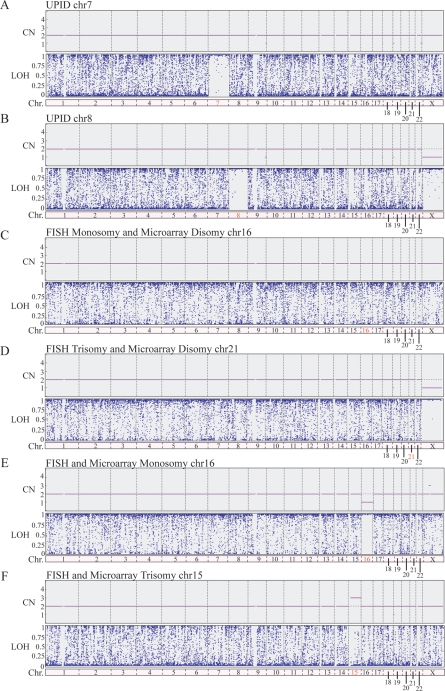
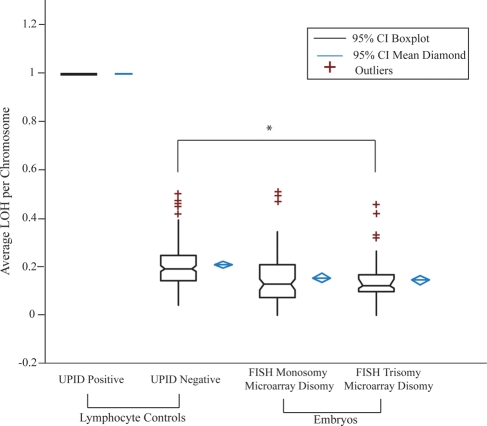
Monosomy and trisomy rescue have been proposed as a possible mechanism of embryo self-correction. This would result in UPID in all monosomy cases through a duplication event and in a proportion of trisomy rescue cases by extruding the extra chromosome. Cell lines with known chromosomal UPID were analyzed to establish a UPID-specific LOH probability threshold. Examples of LOH profiles are shown in Fig. 5, illustrating a marked reduction in the number of SNPs (blue dots) below the LOH probability of 1 for true UPID chromosomes (Fig. 5A and B, chromosomes 7 and 8, respectively). Chromosomes known to be UPID in origin (UPID positive controls, n = 8) displayed an average LOH probability of 0.998 ± 0.002 (Fig. 6). Copy neutral autosomes with bi-parental inheritance (UPID negative controls, n = 168) displayed an average LOH probability of 0.206 ± 0.090 (Fig. 6).
Figure 5.
Representative SNP microarray-based 24 chromosome aneuploidy screening images showing microarray results for CN and LOH probability. Microarray CN and LOH probability results for 5 cells from cell lines that possess UPID for either chromosome 7 (A) or 8 (B), where the CN equals 2 and the average LOH probability is greater than 0.996. Microarray results for a TE sample from embryo #24 (C) originally given a cleavage-stage FISH monosomy diagnosis for chromosome 16, and embryo #38 (D), originally given a cleavage-stage FISH diagnosis of trisomy 21. In both cases, the CN equals 2 and the average LOH probability equals less than 0.996 (∼0.2). Microarray results for a TE sample from embryo 7 (E), given a monsomy 16 diagnosis by FISH and microarray. The chromosome 16 CN equals 1 and the LOH probability is similar to UPID chromosomes. Microarray results for a TE sample from embryo 13 (F), given a trisomy 15 diagnosis by FISH and microarray. The chromosome 15 CN equals 3 and LOH equals less than 0.996. Red chromosome numbers indicated chromosomes of interest. Abbreviations: chr, chromosome.
Figure 6.
Defining UPID by LOH. Lymphocyte controls consist of 5 cells per sample. Box plots of distributions are shown for positive control chromosomes known to be UPID (n = 8), UPID negative control chromosomes from the same samples and known to be normal disomy (n = 168), and blastocyst microarray disomy chromosomes originally diagnosed as monosomy (n = 132), or trisomy (n = 72) by cleavage-stage FISH. *Indicates a significantly different average LOH probability of the UPID positive control chromosomes compared with either the UPID negative control chromosomes or the aneuploid FISH/disomy microarray chromosomes (P < 0.001).
As a result of these observations, a UPID-specific LOH probability threshold of 0.996 was used to systematically analyze chromosomes diagnosed as monosomy or trisomy by cleavage-stage FISH but subsequently shown at the blastocyst stage to be diploid by microarray. Examples of microarray results for embryos with either a FISH-based monosomy (Fig. 5C) or trisomy (Fig. 5D) illustrate a lack of differences in LOH probabilities. Indeed, all such chromosomes failed to exceed the UPID-specific LOH probability threshold of 0.996 (Fig. 6). Moreover, all of the UPID negative control chromosomes and the aneuploid FISH/disomy microarray chromosomes were significantly different from the UPID positive control chromosomes (P < 0.001). Examples of a microarray monosomy and trisomy are also presented. As expected, the monosomy chromosome (Fig. 5E) displayed a distinguishable LOH probability similar to known UPID chromosomes and consistent with previously published results on single cells (Treff et al., 2010b). In addition, a trisomy chromosome (Fig. 5F) displayed a distinguishable LOH probability consistent with previously published single cell results (Treff et al., 2010b). Not a single FISH aneuploid chromosome displayed UPID in any of the 192 samples from 50 blastocysts. Additional analysis of all microarray disomy autosomes indicated that none (0 of 4149) were UPID in origin.
Discussion
Several reports have commonly found euploidy in blastocysts that were previously diagnosed as aneuploid at the cleavage stage by FISH (Li et al., 2005; Munne et al., 2005; Fragouli et al., 2008; Barbash-Hazan et al., 2009) and in effect, this study reaffirms these observations. The authors of these studies elected to conclude that inconsistent results may be due to mosaicism, self-correction or technical error. Of those explanations, only one study explored the possibility of self-correction by examining preferential segregation of abnormalities to the TE in separate TE and ICM samples (Fragouli et al., 2008). However, that study found 100% consistency between the TE and ICM, and therefore provided no evidence to evaluate the etiology of the discrepancies through mosaicism or TE confinement. Indeed, none of the previous studies describing inconsistent results between the cleavage and blastocyst stages of development establish evidence for any of the proposed explanations.
The study presented here specifically evaluated the differences between the cleavage and blastocyst stages of development by investigating possible explanations of inconsistencies including mosaicism, TE confinement and self-correction through monosomy/trisomy rescue mechanisms. Preferential segregation or confinement of aneuploidy to the TE would result in a euploid fetus with placental mosaicism. Indeed, placental mosaicism has been well documented (Kalousek and Dill, 1983; Stetten et al., 2004) and is the primary experimental data used to support the concept of aneuploid confinement to the TE in human embryos. Interestingly, many cases of confined placental mosaicism have been identified as meiotic in origin (Robinson et al., 1997). Nonetheless, experimental evidence to support TE confinement of aneuploidy in the embryo is lacking (Evsikov and Verlinsky, 1998; Magli et al., 2000; Fragouli et al., 2008) and no preferential segregation of aneuploid cells to the TE was observed in our study.
Extrusion of an extra chromosome to rescue a trisomy or duplication of a chromosome to rescue a monosomy may also explain differences observed between the cleavage and blastocyst stages of development. By definition, all monosomy duplications would result in UPID with two copies of the same parental chromosome being present. A trisomy rescue event would lead to disomic chromosomes with bi-parental inheritance two-thirds of the time and UPD one-third of the time. Depending on the mechanism leading to the trisomy (i.e. non-disjunction during meiosis I or II, or premature separation of sister chromatids), a rescue event could either lead to uniparental heterodisomy or UPID (Engel and Antonarkis, 2002). Uniparental heterodisomy does not lead to long contiguous stretches of homozygosity and would require simultaneous analysis of parental SNPs in order to be identified. In the work reported here, the technical ability to accurately identify UPID in cells known to possess UPID chromosomes was established (Fig. 5). Subsequent use of the validated UPID detection technique on 4149 disomy autosomes from 192 sections of 50 human blastocysts demonstrated a 0% rate of UPID. Moreover, this rate of UPID was also observed after specific evaluation of chromosomes predicted to be a monosomy or trisomy by FISH at the cleavage stage and subsequent prediction of disomy in all sections of the blastocyst. This result indicates that monosomy/trisomy rescue resulting in UPID is not the explanation for euploid blastocyst development from a cleavage-stage embryo diagnosed as aneuploid by FISH.
Other explanations for the discrepancies observed between cleavage-stage FISH and blastocyst stage microarray analyses may include apoptotic elimination of aneuploid cells. Although this mechanism cannot be ruled out in the present study, its existence has yet to be experimentally demonstrated elsewhere. Another possible explanation may be that cells with reciprocal abnormalities from mitotic non-disjunction are present within the same blastocyst biopsy resulting in a euploid diagnosis by microarray. However, there are many reasons why this is highly unlikely. First, it is well established that human aneuploidy primarily originates from maternal meiosis (Hassold and Hunt, 2001), which has been supported by direct analysis of maternal meiosis in polar bodies (Kuliev et al., 2005). As a result, the majority of abnormalities observed by FISH at the cleavage stage should be present within the entire embryo (constitutive). Moreover, in some studies, mitotic non-disjunction is the least frequently observed form of mosaicism. For example, Delhanty and Handyside (1995) noted that, with respect to FISH-based analyses, embryos with predicted mitotic errors typically do not display complementary (reciprocal) abnormalities in other cells. Indeed, observation of reciprocal abnormalities is considered as evidence necessary to demonstrate the presence of mitotic non-disjunction. By this definition, comparative genomic hybridization-based methods (Voullaire et al., 2000; Wells and Delhanty, 2000) also found that mitotic non-disjunction was the least frequently observed form of mitotic mosaicism. Non-reciprocal abnormalities would indeed be detected by the methods used in the present study as long as more than 25% of the cells within the biopsy (i.e. two of five cells) possess the same aneuploidy (Fig. 2).
The argument can still be made that the discrepancies between cleavage-stage FISH results and blastocyst reanalysis results are largely due to true cleavage-stage mosaicism. This is supported by a high rate of mosaicism observed in many previous studies where multiple blastomeres from the same embryo have been studied. Although the present study cannot rule out this possibility, a recent study specifically evaluated the alternative interpretation of previous findings; that FISH may overestimate true cleavage-stage mosaicism (Treff et al., 2010a). In that study, blastomeres from the same embryos were blinded and randomized to analysis by two technologies; FISH and microarray-based aneuploidy screening. FISH estimated mosaicism to be present in 100% of the embryos evaluated. In contrast, microarray estimated, in the same embryos, mosaicism to be present in only 31% (P = 0.0005), despite evaluating more chromosomes per cell and more cells per embryo. In addition, the present study established the ability to detect mosaicism of above 25% within each blastocyst biopsy. Evaluation of four biopsies from each embryo also demonstrated that only 24% of blastocysts displayed mosaicism and that only 8% of embryos were mosaic for aneuploidy consistent with the original cleavage-stage FISH result. Together, these findings indicate that mosaicism is not the primary explanation for discrepancies between the cleavage-stage FISH result and the blastocyst stage reanalysis result.
There are now numerous studies indicating aneuploidy diagnosis in morphologically normal blastocysts is poorly predicted by cleavage-stage FISH. We have provided additional understanding to these findings by specifically evaluating mosaicism, confinement of aneuploidy to the TE and chromosome duplication or extrusion resulting in UPID as viable explanations. Given that many morphologically normal blastocysts diagnosed as aneuploid by cleavage-stage FISH are euploid, and that a blastocyst euploid diagnosis has positive predictive value for reproductive competence (Scott et al., 2008), it is possible that FISH-based aneuploidy screening on cleavage-stage embryos results in the erroneous disposal of reproductively competent blastocysts. At a minimum, retesting of morphologically normal blastocysts that develop despite an aneuploid cleavage-stage FISH diagnosis is strongly recommended.
Authors' roles
L.E.N., N.R.T. and R.T.S. designed the study. B.L. performed and supervised the FISH experiments and analyses. L.E.N. performed the microarray experiments and analyses. L.E.N. and N.R.T. wrote the manuscript. All authors discussed the results and implications and commented on the manuscript at all stages.
Acknowledgements
The authors would like to thank the anonymous reviewers for helpful comments to strengthen this work during the peer review process. We would also like to thank Xin Tao and Dr Deanne Taylor for assistance with microarray data analysis and the Embryology team at the Reproductive Medicine Associates of New Jersey for help with sample preparation.
References
- Barbash-Hazan S, Frumkin T, Malcov M, Yaron Y, Cohen T, Azem F, Amit A, Ben-Yosef D. Preimplantation aneuploid embryos undergo self-correction in correlation with their developmental potential. Fertil Steril. 2009;92:890–896. doi: 10.1016/j.fertnstert.2008.07.1761. doi:10.1016/j.fertnstert.2008.07.1761. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. 2006 Assisted reproductive technology success rates: preliminary data national summary and fertility clinic reports. 2008 [Google Scholar]
- Colls P, Escudero T, Cekleniak N, Sadowy S, Cohen J, Munne S. Increased efficiency of preimplantation genetic diagnosis for infertility using “no result rescue”. Fertil Steril. 2007;88:53–61. doi: 10.1016/j.fertnstert.2006.11.099. doi:10.1016/j.fertnstert.2006.11.099. [DOI] [PubMed] [Google Scholar]
- Csokmay J, Hill M, Cioppettini F, Miller KA, Scott RT, Jr, Fratterelli J. Live birth sex ratios are not influenced by blastocyst-stage embryo transfer. Fertil Steril. 2009;92:913–917. doi: 10.1016/j.fertnstert.2008.07.1741. doi:10.1016/j.fertnstert.2008.07.1741. [DOI] [PubMed] [Google Scholar]
- Cui XF, Li HH, Goradia TM, Lange K, Kasasian HH, Jr, Galas D, Arnheim N. Single-sperm typing: determination of genetic distance between the G gamma-globin and parathyroid hormone loci by using the polymerase chain reaction and allele-specific oligomers. Proc Natl Acad Sci USA. 1989;86:9389–9393. doi: 10.1073/pnas.86.23.9389. doi:10.1073/pnas.86.23.9389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhanty JD, Handyside AH. The origin of genetic defects in the human and their detection in the preimplantation embryo. Hum Reprod Update. 1995;1:201–215. doi: 10.1093/humupd/1.3.201. doi:10.1093/humupd/1.3.201. [DOI] [PubMed] [Google Scholar]
- Engel E, Antonarkis SE. Genomic Imprinting and Uniparental Disomy in Medicine: Clinical and Molecular Aspects. New York: John Wiley and Sons; 2002. [Google Scholar]
- Evsikov S, Verlinsky Y. Mosaicism in the inner cell mass of human blastocysts. Hum Reprod. 1998;13:3151–3155. doi: 10.1093/humrep/13.11.3151. doi:10.1093/humrep/13.11.3151. [DOI] [PubMed] [Google Scholar]
- Fragouli E, Lensi M, Ross R, Katz-Jaffe M, Schoolcraft WB, Wells D. Comprehensive molecular cytogenetic analysis of the human blastocyst stage. Hum Reprod. 2008;23:2596–2608. doi: 10.1093/humrep/den287. doi:10.1093/humrep/den287. [DOI] [PubMed] [Google Scholar]
- Fritz MA. Perspectives on the efficacy and indications for preimplantation genetic screening: where are we now? Hum Reprod. 2008;23:2617–2621. doi: 10.1093/humrep/den400. doi:10.1093/humrep/den400. [DOI] [PubMed] [Google Scholar]
- Gardner DK, Schoolcraft WB. In vitro culture of human blastocysts. In: Jansen R, Mortimer D, editors. Towards Reproductive Certainty: Infertility and Genetics Beyond. Carnforth: Parthenon Press; 1999. pp. 378–388. [Google Scholar]
- Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nature. 2001;2:280–291. doi: 10.1038/35066065. doi:10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- Johnson D, Gemelos G, Baner J, Ryan A, Cinnioglu C, Banjevic M, Ross R, Alper M, Barrett B, Frederick J, et al. Preclinical validation of a microarray method for full molecular karyotyping of blastomeres in a 24-h protocol. Hum Reprod. 2010 doi: 10.1093/humrep/dep452. advanced online access: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalousek DK, Dill FJ. Chromosomal mosaicism confined to the placenta in human conceptions. Science. 1983;221:665–667. doi: 10.1126/science.6867735. doi:10.1126/science.6867735. [DOI] [PubMed] [Google Scholar]
- Kuliev A, Cieslak J, Verlinsky Y. Frequency and distribution of chromosome abnormalities in human oocytes. Cytogenet Genome Res. 2005;111:193–198. doi: 10.1159/000086889. doi:10.1159/000086889. [DOI] [PubMed] [Google Scholar]
- Li M, DeUgarte CM, Surrey M, Danser H, DeCherney A, Hill DL. Fluorescence in situ hybridization reanalysis of day-6 human blastocysts diagnosed with aneuploidy on day 3. Fertil Steril. 2005;84:1395–1400. doi: 10.1016/j.fertnstert.2005.04.068. doi:10.1016/j.fertnstert.2005.04.068. [DOI] [PubMed] [Google Scholar]
- Magli MC, Jones GM, Gras L, Gianaroli L, Korman I, Trounson AO. Chromosome mosaicism in day 3 aneuploid embryos that develop to morphologically normal blastocysts in vitro. Hum Reprod. 2000;15:1781–1786. doi: 10.1093/humrep/15.8.1781. doi:10.1093/humrep/15.8.1781. [DOI] [PubMed] [Google Scholar]
- Munne S, Velilla E, Colls P, Garcia Bermudes M, Vemuri MC, Steuerwald N, Garrisi J, Cohen J. Self-correction of chromosomally abnormal embryos in culture and implications for stem cell production. Fertil Steril. 2005;84:1328–1334. doi: 10.1016/j.fertnstert.2005.06.025. doi:10.1016/j.fertnstert.2005.06.025. [DOI] [PubMed] [Google Scholar]
- Robinson WP, Barrett IJ, Bernard L, Telenius A, Bernasconi F, Wilson RD, Best RG, Howard-Peebles PN, Langlois S, Kalousek DK. Meiotic origin of trisomy in confined placental mosaicism is correlated with presence of fetal uniparental disomy, high levels of trisomy in trophoblast, and increased risk of fetal intrauterine growth restriction. Am J Hum Genet. 1997;60:917–927. [PMC free article] [PubMed] [Google Scholar]
- Scott RT, Jr, Miller K, Olivares R, Su J, Fratterelli J, Treff NR. Microarray based 24 chromosome preimplantation genetic diagnosis (mPGD) is highly predictive of the reproductive potential of human embryos: a prospective blinded non-selection trial. Fertil Steril. 2008;S90:22. [Google Scholar]
- Stetten G, Escallon CS, South ST, McMichael JL, Saul DO, Blakemore KJ. Reevaluating confined placental mosaicism. Am J Med Genet A. 2004;131:232–239. doi: 10.1002/ajmg.a.30363. [DOI] [PubMed] [Google Scholar]
- Suela J, Largo C, Ferreira B, Alvarez S, Robledo M, Gonzalez-Neira A, Calasanz MJ, Cigudosa JC. Neurofibromatosis 1, and Not TP53, seems to be the main target of chromosome 17 deletions in de novo acute myeloid leukemia. J Clin Oncol. 2007;25:1151–1152. doi: 10.1200/JCO.2006.09.3013. author reply 1152–1153. doi:10.1200/JCO.2006.09.3013. [DOI] [PubMed] [Google Scholar]
- Treff NR, Su J, Tao X, Miller KA, Levy B, Scott RT., Jr A novel single-cell DNA fingerprinting method successfully distinguishes sibling human embryos. Fertil Steril. 2009 doi: 10.1016/j.fertnstert.2009.03.067. April 24 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Treff NR, Levy B, Su J, Northrop L, Tao X, Scott RT., Jr SNP microarray based 24 chromosome aneuploidy screening is significantly more consistent than FISH. Mol Hum Reprod. 2010a doi: 10.1093/molehr/gaq039. May 19 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treff NR, Su J, Tao X, Levy B, Scott RT., Jr Accurate single cell 24 chromosome aneuploidy screening using whole genome amplification and single nucleotide polymorphism microarrays. Fertil Steril. 2010b doi: 10.1016/j.fertnstert.2010.01.052. February 24 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Treff NR, Su J, Kasabwala N, Miller KA, Levy B, Scott RT. Robust embryo identification using first polar body single nucleotide polymorphism (SNP) microarray-based DNA fingerprinting. Fertil Steril. 2010c;93:2453–2455. doi: 10.1016/j.fertnstert.2009.08.070. doi:10.1016/j.fertnstert.2009.08.070. [DOI] [PubMed] [Google Scholar]
- Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, Debrock S, Amyere M, Vikkula M, Schuit F, et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009;15:577–583. doi: 10.1038/nm.1924. doi:10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- Voullaire L, Slater H, Williamson R, Wilton L. Chromosome analysis of blastomeres from human embryos by using comparative genomic hybridization. Hum Genet. 2000;106:210–217. doi: 10.1007/s004390051030. doi:10.1007/s004390051030. [DOI] [PubMed] [Google Scholar]
- Wells D, Delhanty JD. Comprehensive chromosomal analysis of human preimplantation embryos using whole genome amplification and single cell comparative genomic hybridization. Mol Hum Reprod. 2000;6:1055–1062. doi: 10.1093/molehr/6.11.1055. doi:10.1093/molehr/6.11.1055. [DOI] [PubMed] [Google Scholar]