Abstract
Many studies estimate that chromosomal mosaicism within the cleavage-stage human embryo is high. However, comparison of two unique methods of aneuploidy screening of blastomeres within the same embryo has not been conducted and may indicate whether mosaicism has been overestimated due to technical inconsistency rather than the biological phenomena. The present study investigates the prevalence of chromosomal abnormality and mosaicism found with two different single cell aneuploidy screening techniques. Thirteen arrested cleavage-stage embryos were studied. Each was biopsied into individual cells (n = 160). The cells from each embryo were randomized into two groups. Those destined for FISH-based aneuploidy screening (n = 75) were fixed, one cell per slide. Cells for SNP microarray-based aneuploidy screening (n = 85) were put into individual tubes. Microarray was significantly more reliable (96%) than FISH (83%) for providing an interpretable result (P = 0.004). Markedly different results were obtained when comparing microarray and FISH results from individual embryos. Mosaicism was significantly less commonly observed by microarray (31%) than by FISH (100%) (P = 0.0005). Although FISH evaluated fewer chromosomes per cell and fewer cells per embryo, FISH still displayed significantly more unique genetic diagnoses per embryo (3.2 ± 0.2) than microarray (1.3 ± 0.2) (P < 0.0001). This is the first prospective, randomized, blinded and paired comparison between microarray and FISH-based aneuploidy screening. SNP microarray-based 24 chromosome aneuploidy screening provides more complete and consistent results than FISH. These results also suggest that FISH technology may overestimate the contribution of mitotic error to the origin of aneuploidy at the cleavage stage of human embryogenesis.
Keywords: aneuploidy screening, SNP microarray, FISH, randomized blastomere comparison, chromosomal mosaicism
Introduction
The concept of aneuploidy screening of human embryos to enhance IVF outcomes is based on sound principles. Multiple technologies have demonstrated that aneuploidy is common in preimplantation embryos and involves monosomies and trisomies of all 22 autosomes as well as the sex chromosomes (Fragouli et al., 2009; Vanneste et al., 2009; Johnson et al., 2010; Treff et al., 2010). The prevalence of abnormalities at the time of antenatal screening and in live born infants is dramatically lower than that found in embryos (reviewed in Hassold and Hunt, 2001). The difference in these rates is reflective of the fact that aneuploid embryos either fail to implant or arrest in their development during the early phases of gestation in most cases. A small residual clinical risk remains for ongoing aneuploid gestations (typically involving trisomies or monosomy X) which may remain viable and are ultimately live born. If embryos could be accurately screened prior to transfer, those which are aneuploid could be eliminated. By transferring only euploid embryos, the diminution in reproductive efficiency attributable to aneuploidy might be reduced or eliminated. Clinical benefits should include higher implantation rates, lower pregnancy loss rates and a reduction in the risk for delivery of an anomalous infant.
Unfortunately, clinical results with preimplantation genetic screening (PGS) for aneuploidy have not demonstrated the theoretical improvements which were anticipated (Staessen et al., 2004, 2008; Mastenbroek et al., 2007; Hardarson et al., 2008; Schoolcraft et al., 2009). In fact, every randomized clinical trial performed to date has failed to demonstrate improved outcomes on an intent-to-treat basis. Putative explanations for this clinical failure have included mosaicism, self-correction of aneuploidy within the embryo, evaluation of a very limited number of chromosomes with the most commonly used technology—fluorescence in situ hybridization (FISH), an adverse impact of embryo biopsy which overwhelms any positive influence gained by aneuploidy screening and technical limits of the screening technology itself (reviewed in Practice Committee of the Society for Assisted Reproductive Technology and Practice Committee of the American Society for Reproductive Medicine, 2007; Anderson and Pickering, 2008; American Congress of Obstetricians and Gynecologists Committee Opinion, 2009).
While all of these factors may have contributed to the lack of success of FISH-based aneuploidy screening, the most concerning are the issues relating to technical accuracy. Evaluation of a single blastomere is challenging. Unfortunately, direct evaluation of the reliability of FISH on human embryonic blastomeres has not been done. Scientists have made great efforts to optimize FISH in this setting and have provided estimates of error rates (e.g. Colls et al., 2007). These calculations require a variety of assumptions. When multiple cells are evaluated from a single embryo and differing results are attained, it is difficult to know whether those differences should be attributed to genuine mosaicism, or if they reflect a failure of the technique to provide reproducible results.
Some investigators have addressed this question by taking embryos which were diagnosed as aneuploid following biopsy and FISH evaluation of a blastomere on Day 3 and then re-evaluating the embryo at the blastocyst stage. In fact, non-concurrence rates following an abnormal Day 3 FISH result and re-analysis at the blastocyst stage on Day 5 of development are substantial (Magli et al., 2000; Li et al., 2005; Munne et al., 2005; Fragouli et al., 2008; Barbash-Hazan et al., 2009). Euploidy rates in these embryos previously designated as abnormal may be as high as 71% (Munne et al., 2005).
Interpreting the data from re-evaluation studies is further complicated by the use of differing definitions for ‘concurrence’. If the blastocyst is aneuploid but has a different chromosomal abnormality than that identified on the original Day 3 biopsy, should the original diagnosis be considered concurrent or non-concurrent? If the standard is purely clinical, then the detection of aneuploidy in both samples would lead to the same clinical designation (non-viable) and it is possible to consider that the original clinical diagnosis was assigned correctly even though the technical results clearly differed. This is an extremely low standard and would not be suitable for evaluating the reliability of a technique.
One option for evaluating the precision of aneuploidy screening would be to analyze multiple cells from the same embryos with two different technologies. For example, a cleavage-stage embryo which has arrested and been discarded for research could be dispersed into individual cells and then randomly assigned to analysis by either FISH or another method such as copy number analysis by SNP microarray-based 24 chromosome aneuploidy screening (Treff et al., 2010). While some mosaicism is almost inevitable, the paired nature of the comparison would control for its impact on the results obtained by each technique. In other words, mosaicism should be equally common with both techniques. Any consistent disparity in results with one relative to the other would strongly suggest that the techniques are not providing equivalent results. This study is designed to evaluate the consistency in aneuploidy assessment using two aneuploidy screening techniques.
Materials and Methods
Population
Supernumerary embryos were donated to research by infertile patients participating in the IVF program. Embryos which arrested in extended culture were considered for the study. Embryos on Day 5 of in vitro development which had failed to progress through compaction to form either a morulae or blastocyst were selected for the study. As such, these embryos were not considered viable or suitable for cryopreservation for future clinical use. All patients gave informed consent to have their non-viable embryos donated to the research program by following an IRB approved protocol.
Experimental design
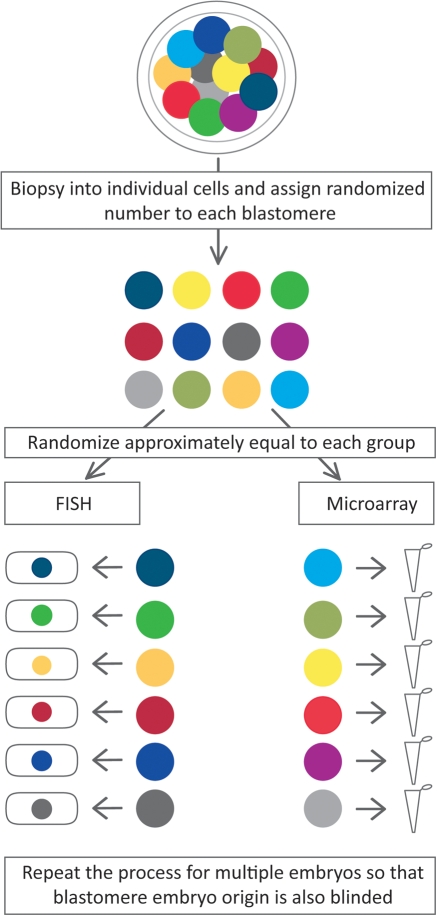
The goal of this research was to determine the consistency in diagnostic results obtained with FISH and microarray-based aneuploidy screening. To that end, it was necessary to evaluate and compare multiple cells from the same embryo. The experimental design is outlined in Fig. 1. Embryos which had failed to compact but which had eight or more cells on Day 5 were specifically selected for study. The cells all needed to be intact, with a visible nucleus, and no evidence of multinucleation in any cell at any point during embryonic development.
Figure 1.
Blastomeres from the same cleavage-stage embryos were biopsied into individual cells and randomized to analysis by either FISH or microarray. Blastomeres from multiple embryos were randomized together so that the embryo specific origin of each blastomere was blinded. The paired randomized design controls for true mosaicism and the blinded analysis avoids bias in interpretation from knowledge of each blastomere's embryo of origin.
The selected embryos were biopsied into individual cells. It was then necessary to randomly assign the cells from that individual embryo to either FISH or microarray-based analysis. As such, each embryo had its own unique randomization table. Using that table, the blastomeres from each embryo were randomly assigned to FISH or microarray analysis. Cells which were randomized to FISH-based analysis were fixed one cell per slide to avoid bias when interpreting the FISH results. Blastomeres randomized to microarray-based analysis were placed into individual PCR tubes for cell lysis.
While there was a unique randomization table for each individual embryo to make certain that the cells were distributed approximately equally between the two study groups, the numbers used to label the glass slides and lysis buffer tubes were assigned from a separate single large randomization table. This was essential to assure blinding of the laboratory team. It was not possible for the laboratory scientists to know which slides or tubes came from the same embryos. They were also unaware of how many cells came from each embryo or even how many embryos were being evaluated in the study.
Assays
The isolated blastomeres which were randomized to undergo FISH analysis were fixed, one cell to a glass slide. Each cell had been pre-incubated in a hypotonic solution and fixed using 3:1 methanol:acetic acid solution. Two rounds of FISH were performed on fixed blastomeres using probes specific for chromosomes 13, 15, 16, 17, 18, 21, 22, X and Y (Vysis, Downer's Grove, IL, USA). The slides were analyzed using an automated Olympus BX61 fluorescence microscope (Center Valley, PA, USA). The images were captured using Cytovision probe software (Applied Imaging Corp, San Jose, CA, USA). Reconfirmation of inconclusive results was conducted as previously described (Colls et al., 2007).
Blastomeres which were randomized to undergo microarray-based analysis were placed into individual reaction tubes containing lysis buffer. Each then underwent microarray-based copy number analysis for whole chromosome aneuploidy as previously described (Treff et al., 2010). This microarray technology previously demonstrated 98.6% accuracy in a randomized blinded analysis of single cells from cell lines with whole chromosome aneuploidies previously identified by conventional cytogenetics (Treff et al., 2010). Accuracy per chromosome was greater than 99%. However, we have not validated this single cell microarray technology for identification of segmental aneuploidy and therefore it could not be used for this purpose in the present study. Briefly, whole genomic amplification was conducted according to the recommended protocol beginning with library preparation (GenomePlex WGA4, Sigma Aldrich, St. Louis, Missouri). Whole genome amplified DNA was then purified using the GenElute PCR Purification Kit as recommended (Sigma Aldrich). Two-hundred and fifty nanograms of purified DNA were then processed through the NspI GeneChip Mapping 262 K microarray as recommended by the manufacturer (Affymetrix, Santa Clara, CA, USA). Aneuploidy screening was performed by copy number analysis of the microarray data using the Copy Number Analysis Tool (CNAT) version 4.0.1 (Affymetrix). The microarray data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE20 975 (http://www.ncbi.nlm.nih.gov/geo).
Statistics
Samples were analyzed and a genetic result assigned. The fact that the samples undergoing FISH were evaluated for nine chromosomes while those undergoing microarray analysis were evaluated for 24 chromosomes (all twenty two autosomes, and X and Y) provides an unequal opportunity to identify aneuploidy and thus to have discrepant results. In particular, when comparing the consistency of genetic diagnoses, the difference in the number of chromosomes being evaluated would provide an advantage for the FISH group where there would be only nine chances to obtain a different result.
The results obtained from each technology for each individual cell were then collated by embryo for comparison. Since both techniques will occasionally fail to provide an interpretable result, the no-result rates were compared using a Pearson's chi square test for concurrence. Following this comparison, those cells which failed to provide a genetic diagnosis were excluded from further consideration.
The genetic diagnoses for each technology were reviewed for each individual embryo. If all the genetic diagnoses were the same with a particular technology then there would be perfect concurrence of results. In contrast, if any of the diagnoses using a particular technology were different, then that embryo was designated as being mosaic according to that technology. The number of unique genetic diagnoses made by FISH was compared with the number made by microarray using a paired Student's t-test. The prevalence of mosaicism identified by each method was compared using a Fisher exact test.
Results
One hundred and sixty cells were evaluated from thirteen arrested cleavage-stage embryos (median of 13 cells per embryo; range 9–16). No cells were lost during the biopsy process. Seventy five cells were randomized to FISH-based analysis with the remaining 85 cells designated to undergo evaluation by microarray. The number of interpretable results obtained for each technique for each embryo is presented in Table I. Microarray analyses produced interpretable results in 82 of 85 cells (96%). In contrast, FISH analyses produced interpretable results in only 62 of 75 cells (83%). This difference was statistically significant (P = 0.004).
Table I.
Blastomeres that were randomized from 13 cleavage-stage embryos and produced interpretable results by either FISH or microarray.
| Embryo No. | Total No. of Randomized Cells | FISH |
Microarray |
||
|---|---|---|---|---|---|
| No. of Cells Randomized | No. of Interpretable Results | No. of Cells Randomized | No. of Interpretable Results | ||
| 1 | 14 | 7 | 5 | 7 | 6 |
| 2 | 14 | 7 | 7 | 7 | 6 |
| 3 | 10 | 4 | 3 | 6 | 6 |
| 4 | 15 | 7 | 5 | 8 | 8 |
| 5 | 11 | 5 | 5 | 6 | 5 |
| 6 | 9 | 4 | 4 | 5 | 5 |
| 7 | 16 | 8 | 6 | 8 | 8 |
| 8 | 10 | 4 | 2 | 6 | 6 |
| 9 | 13 | 6 | 5 | 7 | 7 |
| 10 | 15 | 7 | 5 | 8 | 8 |
| 11 | 13 | 6 | 6 | 7 | 7 |
| 12 | 9 | 4 | 3 | 5 | 5 |
| 13 | 11 | 6 | 6 | 5 | 5 |
| Total No. of Reliability | 160 | 75 | 62 (83%) | 85 | 82 (96%a) |
aIndicates microarray interpretation of the results is significantly more reliable than FISH (P = 0.004).
The specific genetic diagnoses obtained with each technique for each cell are presented in Table II. Eight of the 13 embryos (62%) were diagnosed as uniformly diploid (all cells) by microarray. The remaining 5 embryos displayed at least one aneuploid cell by microarray; 2 embryos were mosaic aneuploid, 2 were mosaic diploid/aneuploid, and 1 was a uniformly aneuploid embryo. In contrast, 10 of the 13 embryos were diagnosed as mosaic diploid/aneuploid by FISH including the same 8 embryos that were diagnosed as uniformly diploid by microarray. The remaining 3 embryos were diagnosed as mosaic aneuploid by FISH. The prevalence of mosaicism was therefore 100% in the FISH group. In contrast, the prevalence of mosaicism was 31% in the microarray group, and was significantly lower than the FISH group (P = 0.0005).
Table II.
Number of unique diagnoses made by FISH and microarray from multiple blastomeres randomized, blinded and paired after biopsy from the same embryos.
| Embryo No. | FISH Result |
Microarray Result |
||||
|---|---|---|---|---|---|---|
| Results (no. of cells) | No. Unique Results | Interpretation | Results (no. of cells) | No. Unique Results | Interpretation | |
| 1 | Dip female(3) | 2 | Mosaic Diploid/Aneuploid | 46,XX(6) | 1 | Uniformly Diploid |
| +18 female(2) | ||||||
| 2 | Dip male(5) | 3 | Mosaic Diploid/Aneuploid | 46,XY(6) | 1 | Uniformly Diploid |
| −18 male(1) | ||||||
| −15,−17,−18,−21 male(1) | ||||||
| 3 | Dip male(1) | 3 | Mosaic Diploid/Aneuploid | 46,XY(6) | 1 | Uniformly Diploid |
| −13 male(1) | ||||||
| +13,−16,−21 male(1) | ||||||
| 4 | Dip male(3) | 3 | Mosaic Diploid/Aneuploid | 46,XY(8) | 1 | Uniformly Diploid |
| −16 male(1) | ||||||
| −13 male(1) | ||||||
| 5 | Dip male(2) | 4 | Mosaic Diploid/Aneuploid | 46,XY(5) | 1 | Uniformly Diploid |
| tet male(1) | ||||||
| XXY(1) | ||||||
| +13,+18 male(1) | ||||||
| 6 | +22 female(2) | 3 | Mosaic Aneuploid | 46,XX,−19,+22(4) | 2 | Mosaic Aneuploid |
| complex aneuploid(1) | 45,XX,−19(1) | |||||
| tet female(1) | ||||||
| 7 | Dip female(2) | 5 | Mosaic Diploid/Aneuploid | 46,XX(8) | 1 | Uniformly Diploid |
| −13,−15 female(1) | ||||||
| −22,+18 female(1) | ||||||
| −13,−16,−22 female(1) | ||||||
| −18,XXX(1) | ||||||
| 8 | −22 female(1) | 2 | Mosaic Aneuploid | 45,XX,−22(6) | 1 | Uniformly Aneuploid |
| −22,XXX(1) | ||||||
| 9 | Dip female(3) | 3 | Mosaic Diploid/Aneuploid | 46,XX(7) | 1 | Uniformly Diploid |
| −16 female(1) | ||||||
| complex aneuploid(1) | ||||||
| 10 | Dip female(2) | 4 | Mosaic Diploid/Aneuploid | 46,XX(7) | 2 | Mosaic Diploid/Aneuploid |
| −13,−17 female(1) | 47,XX,+18(1) | |||||
| −13 female(1) | ||||||
| −13,−16,−18 female(1) | ||||||
| 11 | Dip female(3) | 4 | Mosaic Diploid/Aneuploid | 46,XX(2) | 3 | Mosaic Diploid/Aneuploid |
| −18,−22 female(1) | 45,XX,−17(4) | |||||
| −13,−18,−21 female(1) | 47,XX,+19(1) | |||||
| complex aneuploid(1) | ||||||
| 12 | complex aneuploid(1) | 3 | Mosaic Aneuploid | Complex aneuploid(5) | 1 | Mosaic Aneuploid |
| XYYY(1) | ||||||
| −16,−17,−18 male(1) | ||||||
| 13 | Dip female(4) | 3 | Mosaic Diploid/Aneuploid | 46,XX(5) | 1 | Uniformly Diploid |
| +13 female(1) | ||||||
| −15,−17,XXY(1) | ||||||
| Mean | Mean | |||||
| 3.2 ± 0.2 | 1.3 ± 0.2a | |||||
aIndicates significantly fewer unique diagnoses by microarray compared with FISH for the same embryos (P < 0.0001). dip, diploid; tet, tetraploid.
Perhaps more important than the overall prevalence of mosaicism is the number of unique genetic diagnoses which would have been assigned to each embryo. In the FISH group, there were almost as many different genetic diagnoses as there were cells analyzed. For example, embryo number seven had 6 cells evaluable and received five different genetic diagnoses. The mean number of unique genetic diagnoses assigned to each individual embryo was 3.2 ± 0.2 by FISH and 1.3 ± 0.2 by microarray. Microarray displayed significantly fewer unique genetic diagnoses (P < 0.0001) despite evaluating more chromosomes per cell (24 by microarray compared with 9 by FISH), and evaluating more cells per embryo.
Discussion
The phenomenon of chromosomal mosaicism in the developing human embryo has complicated the interpretation of inconsistent aneuploidy diagnoses of multiple samples from the same embryo. Indeed, there are at least two interpretations of inconsistencies observed after analysis of multiple blastomeres from within the same embryo. One interpretation is that true mosaicism, which likely occurs as a result of errors in chromosome segregation during mitotic cell division, was present in the embryo. Indeed, mosaicism represents the primary source of variation that has been used to explain the inconsistencies observed by FISH-based aneuploidy screening. However, a largely overlooked interpretation is that the observed inconsistencies are due to the FISH technique itself. Although high rates of mosaicism have been found by other methods, including conventional comparative genomic hybridization (CGH) (Voullaire et al., 2000; Wells and Delhanty, 2000), and array CGH combined with a multiple displacement amplification based SNP microarray analysis (Vanneste et al., 2009), no previous studies have been performed using the present study design (where blastomeres from the same embryos were randomized to blinded analysis by two independent technologies). Until such a study design has been implemented to evaluate other technologies, it will remain unclear as to whether previous estimates of the prevalence of mitotic aneuploidy made by these other technologies are based on true mosaicism or technical limitations.
Randomization of cells within the same embryo and blinded analysis by two different methods provided a unique opportunity to isolate the putative individual contributions made by true mosaicism and technical limitations to the inconsistency in aneuploidy diagnoses. Blinded analysis is a critical component since knowledge of whether specific blastomeres belong to the same embryo can significantly bias interpretation of the results. For example, interpretation of a blastomere that would otherwise be difficult to make might be influenced by a diagnosis made for another blastomere from the same embryo. Although previous reports describing FISH-based diagnosis of aneuploidy have elected to attribute inconsistencies primarily to mosaicism, the present study indicates that the most likely explanation is inconsistency of the technique itself.
First, the microarray process was significantly more reliable for producing an interpretable result (P = 0.004). This could be due to the specific method of sample processing for microarray-based analyses which is independent of cell spreading or fixation known to be an unreliable process that is required for FISH. It could also be related to the nature of the blastomeres in this study, in that they were derived from arrested embryos rather than developmentally normal embryos. The present study found that 13 blastomeres (17%) failed to provide an interpretable FISH result; 8 were anucleated, 2 were lost during fixation, and 3 were not analyzable (covered in debris). Although this rate of failure is considerably higher than the approximately 6% failure rate recently reported by the ESHRE PGD consortium for blastomeres derived from developmentally normal cleavage-stage embryos (Goossens et al., 2009), it is below that of a previously reported FISH analysis of 719 arrested cleavage-stage embryos, where an overall failure rate of 23% was found; 106 cells were anucleated, 17 were broken during biopsy, 25 were lost during fixation, and 17 were not analyzable (Munne et al., 1994).
Microarray-based aneuploidy screening also displayed significantly higher consistency in diagnosis for blastomeres randomized from the same embryos that underwent FISH-based analysis (P < 0.0001). FISH predicted a rate of mosaicism of 100%, while microarray predicted a significantly lower rate of 31% (P = 0.0005). As a result of the randomized and paired design, it was not possible that cells with true mosaicism were assigned to the FISH analysis group only. Moreover, since microarray analyses were more reliable for providing an interpretable result, there were more blastomeres analyzed per embryo by microarray than by FISH, thus providing a larger opportunity for true mosaicism to impact observations made by microarray. Microarray analysis also included diagnosis of all 24 chromosomes, while FISH analysis only included 9 chromosomes, again providing a larger opportunity for true mosaicism to impact the observations made by microarray-based analysis.
Together with previous studies which indicate poor predictive value of cleavage-stage FISH for aneuploidy in morphologically normal blastocysts (Li et al., 2005; Munne et al., 2005; Fragouli et al., 2008; Barbash-Hazan et al., 2009), and the absence of clinical benefit observed in many randomized controlled trials, it is becoming clear that FISH-based technology is inadequate for the diagnosis of aneuploidy in human embryos. One important implication of the current study is that previous interpretation of FISH-based analysis of the origin of human aneuploidy may be incorrect. For example, in this study, a significantly larger proportion of embryos would have been assigned as having mitotic error when estimated by FISH-based analysis (100%) compared with microarray-based analysis (31%). Therefore, aneuploidy screening technologies with better consistency may provide better estimates of the origin of aneuploidy in human embryos that are more in line with the primarily observed maternal meiotic origins of aneuploidy in products of conception (reviewed in Hassold et al., 2007).
Despite the growing evidence that FISH-based aneuploidy screening does not work, the concept of improving outcomes by aneuploidy screening should not be considered invalid. Indeed, new comprehensive technologies such as SNP microarray-based 24 chromosome aneuploidy screening may result in the realization of the expected clinical benefit. However, development and validation of these new technologies should be held to a higher standard than FISH. These standards should include demonstration of preclinical accuracy, consistency, and reliability. In addition, completion of randomized trials with class I strength of evidence for clinical benefit, and experimentally demonstrating acceptable risk of clinical implementation (i.e. a negligible impact of biopsy), should be required before any new aneuploidy screening technologies are offered as a clinical routine (reviewed in Scott and Treff, 2010).
Authors' roles
R.T.S. and N.R.T. designed the study. J.S., X.T. and L.E.N. performed the microarray analyses. B.L. performed the FISH analyses. N.R.T., R.T.S. and B.L. wrote the manuscript.
References
- American Congress of Obstetricians and Gynecologists Committee Opinion. Preimplantation Genetic Screening for Aneuploidy. Obstet Gynecol. 2009;113:766–767. doi: 10.1097/AOG.0b013e31819e9f05. [DOI] [PubMed] [Google Scholar]
- Anderson RA, Pickering S. The current status of preimplantation genetic screening: British Fertility Society Policy and Practice Guidelines. Hum Fertil (Camb) 2008;11:71–75. doi: 10.1080/14647270802041607. doi:10.1080/14647270802041607. [DOI] [PubMed] [Google Scholar]
- Barbash-Hazan S, Frumkin T, Malcov M, Yaron Y, Cohen T, Azem F, Amit A, Ben-Yosef D. Preimplantation aneuploid embryos undergo self-correction in correlation with their developmental potential. Fertil Steril. 2009;92:890–896. doi: 10.1016/j.fertnstert.2008.07.1761. doi:10.1016/j.fertnstert.2008.07.1761. [DOI] [PubMed] [Google Scholar]
- Colls P, Escudero T, Cekleniak N, Sadowy S, Cohen J, Munne S. Increased efficiency of preimplantation genetic diagnosis for infertility using “no result rescue”. Fertil Steril. 2007;88:53–61. doi: 10.1016/j.fertnstert.2006.11.099. doi:10.1016/j.fertnstert.2006.11.099. [DOI] [PubMed] [Google Scholar]
- Fragouli E, Lensi M, Ross R, Katz-Jaffe M, Schoolcraft WB, Wells D. Comprehensive molecular cytogenetic analysis of the human blastocyst stage. Hum Reprod. 2008;23:2596–2608. doi: 10.1093/humrep/den287. doi:10.1093/humrep/den287. [DOI] [PubMed] [Google Scholar]
- Fragouli E, Jaffe-Katz M, Alfarawati M, Stevens J, Colls P, Goodall N, Tormasi S, Mateo-Gutierres C, Prates R, Schoolcraft WB, et al. Comprehensive chromosome screening of polar bodies and blastocysts from couples experiencing repeated implantation failure. Hum Reprod. 2009 doi: 10.1016/j.fertnstert.2009.04.053. June 21 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Goossens V, Harton G, Moutou C, Traeger-Synodinos J, Van Rij M, Harper JC. ESHRE PGD Consortium data collection IX: cycles from January to December 2006 with pregnancy follow-up to October 2007. Hum Reprod. 2009;24:1786–1810. doi: 10.1093/humrep/dep059. doi:10.1093/humrep/dep059. [DOI] [PubMed] [Google Scholar]
- Hardarson T, Hanson C, Lundin K, Hillensjo T, Nilsson L, Stevic J, Reismer E, Borg K, Wikland M, Bergh C. Preimplantation genetic screening in women of advanced maternal age caused a decrease in clinical pregnancy rate: a randomised controlled trial. Hum Reprod. 2008;23:2806–2812. doi: 10.1093/humrep/den217. doi:10.1093/humrep/den217. [DOI] [PubMed] [Google Scholar]
- Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. doi: 10.1038/35066065. doi:10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- Hassold T, Hall H, Hunt P. The origin of human aneuploidy: where we have been, where we are going. Hum Mol Genet. 2007;16:R203–R208. doi: 10.1093/hmg/ddm243. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Gemelos G, Baner J, Ryan A, Cinnioglu C, Banjevic M, Ross R, Alper M, Barrett B, Frederick J, et al. Preclinical validation of a microarray method for full molecular karyotyping of blastomeres in a 24-h protocol. Hum Reprod. 2010;25:1066–1075. doi: 10.1093/humrep/dep452. doi:10.1093/humrep/dep452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, DeUgarte CM, Surrey M, Danser H, DeCherney A, Hill DL. Fluorescence in situ hybridisation reanalysis of day-6 human blastocysts diagnosed with aneuploidy on day 3. Fertil Steril. 2005;84:1395–1400. doi: 10.1016/j.fertnstert.2005.04.068. doi:10.1016/j.fertnstert.2005.04.068. [DOI] [PubMed] [Google Scholar]
- Magli MC, Jones GM, Gras L, Gianaroli L, Korman I, Trounson AO. Chromosome mosaicism in day 3 aneuploid embryos that develop to morphologically normal blastocysts in vitro. Hum Reprod. 2000;15:1781–1786. doi: 10.1093/humrep/15.8.1781. doi:10.1093/humrep/15.8.1781. [DOI] [PubMed] [Google Scholar]
- Mastenbroek S, Twisk M, van Echten-Arends J, Sikkema-Raddatz B, Korevaar JC, Verhoeve HR, Vogel NE, Arts EG, de Vries JW, Bossuyt PM, et al. In vitro fertilization with preimplantation genetic screening. N Engl J Med. 2007;357:9–17. doi: 10.1056/NEJMoa067744. doi:10.1056/NEJMoa067744. [DOI] [PubMed] [Google Scholar]
- Munne S, Grifo J, Cohen J, Weier HU. Chromosome abnormalities in human arrested preimplantation embryos: a multiple-probe FISH study. Am J Hum Genet. 1994;55:150–159. [PMC free article] [PubMed] [Google Scholar]
- Munne S, Velilla E, Colls P, Garcia Bermudes M, Vemuri MC, Steuerwald N, Garrisi J, Cohen J. Self-correction of chromosomally abnormal embryos in culture and implications for stem cell production. Fertil Steril. 2005;84:1328–1334. doi: 10.1016/j.fertnstert.2005.06.025. doi:10.1016/j.fertnstert.2005.06.025. [DOI] [PubMed] [Google Scholar]
- Practice Committee of the Society for Assisted Reproductive Technology and Practice Committee of the American Society for Reproductive Medicine. Preimplantation genetic testing: a Practice Committee opinion. Fertil Steril. 2007;88:1497–1504. doi: 10.1016/j.fertnstert.2007.10.010. doi:10.1016/j.fertnstert.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Schoolcraft WB, Katz-Jaffe MG, Stevens J, Rawlins M, Munne S. Preimplantation aneuploidy testing for infertile patients of advanced maternal age: a randomized prospective trial. Fertil Steril. 2009;92:157–162. doi: 10.1016/j.fertnstert.2008.05.029. doi:10.1016/j.fertnstert.2008.05.029. [DOI] [PubMed] [Google Scholar]
- Scott RT, Jr, Treff NR. Assessing the reproductive competence of individual embryos: a proposal for the validation of new “-omics” technologies. Fertil Steril. 2010 doi: 10.1016/j.fertnstert.2010.03.041. Epub ahead of print May 4. [DOI] [PubMed] [Google Scholar]
- Staessen C, Platteau P, Van Assche E, Michiels A, Tournaye H, Camus M, Devroey P, Liebaers I, Van Steirteghem A. Comparison of blastocyst transfer with or without preimplantation genetic diagnosis for aneuploidy screening in couples with advanced maternal age: a prospective randomised controlled trial. Hum Reprod. 2004;19:2849–2858. doi: 10.1093/humrep/deh536. doi:10.1093/humrep/deh536. [DOI] [PubMed] [Google Scholar]
- Staessen C, Verpoest W, Donoso P, Haentjens P, Van der Elst J, Liebaers I, Devroey P. Preimplantation genetic screening does not improve delivery rate in women under the age of 36 following single-embryo transfer. Hum Reprod. 2008;23:2818–2825. doi: 10.1093/humrep/den367. doi:10.1093/humrep/den367. [DOI] [PubMed] [Google Scholar]
- Treff NR, Su J, Tao X, Levy B, Scott RT., Jr Accurate single cell 24 chromosome aneuploidy screening using whole genome amplification and single nucleotide polymorphism microarrays. Fertil Steril. 2010 doi: 10.1016/j.fertnstert.2010.01.052. [Epub ahead of print, February 24] [DOI] [PubMed] [Google Scholar]
- Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, Debrock S, Amyere M, Vikkula M, Schuit F, et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009;15:577–583. doi: 10.1038/nm.1924. doi:10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- Voullaire L, Slater H, Williamson R, Wilton L. Chromosome analysis of blastomeres from human embryos by using comparative genomic hybridization. Hum Genet. 2000;106:210–217. doi: 10.1007/s004390051030. doi:10.1007/s004390051030. [DOI] [PubMed] [Google Scholar]
- Wells D, Delhanty JD. Comprehensive chromosomal analysis of human preimplantation embryos using whole genome amplification and single cell comparative genomic hybridization. Mol Hum Reprod. 2000;6:1055–1062. doi: 10.1093/molehr/6.11.1055. doi:10.1093/molehr/6.11.1055. [DOI] [PubMed] [Google Scholar]