Abstract
The protein-synthesizing ribosome undergoes large motions to effect the translocation of tRNAs and mRNA; here the domain motions of this system are explored with a coarse-grained elastic network model using normal mode analysis. Crystal structures are used to construct various model systems of the 70S complex with/without tRNA, elongation factor Tu and the ribosomal proteins. Computed motions reveal the well-known ratchet-like rotational motion of the large subunits, as well as the head rotation of the small subunit and the high flexibility of the L1 and L7/L12 stalks, even in the absence of ribosomal proteins. This result indicates that these experimentally observed motions during translocation are inherently controlled by the ribosomal shape and only partially dependent upon GTP hydrolysis. Normal mode analysis further reveals the mobility of A- and P-tRNAs to increase in the absence of the E-tRNA. In addition, the dynamics of the E-tRNA is affected by the absence of the ribosomal protein L1. The mRNA in the entrance tunnel interacts directly with helicase proteins S3 and S4, which constrain the mRNA in a clamp-like fashion, as well as with protein S5, which likely orients the mRNA to ensure correct translation. The ribosomal proteins S7, S11 and S18 may also be involved in assuring translation fidelity by constraining the mRNA at the exit site of the channel. The mRNA also interacts with the 16S 3’ end forming the Shine-Dalgarno complex at the initiation step; the 3’ end may act as a ‘hook’ to reel in the mRNA to facilitate its exit.
PACS: 87.10.Pq; 87.15.bk; 87.15.kj; 87.16.dj; 87.16.dr
Keywords: ribosome processing, functional motions, elastic network models, structural control of motions, mRNA tunnel
1. Introduction
The ribosome is a large protein-RNA complex, which is a molecular machine essential for most cellular functions; it synthesizes proteins based on the genetic information encoded in the mRNA. In bacteria, the 70S ribosomal complex contains two subunits: 1) the smaller 30S subunit, which has the large 16S rRNA and 21 ribosomal proteins and 2) the larger 50S subunit containing the 23S rRNA, the smaller 5S rRNA, and more than 30 ribosomal proteins. Figure 1 displays the 30S and 50S subunits labeled for its functional regions and the proteins discussed in this study. Each subunit plays a particular role in the protein synthesis. The mRNA carrying the genetic code wraps around the neck of the 30S subunit, which forms part of the three tRNA binding sites, designated A- (aminoacyl), P- (peptidyl) and E- (exit).1,2 Assisted by the elongation factor (EF) Tu, each tRNA-amino acid enters the ribosome, with the cognate tRNA moving into the A-site. During elongation, the peptidyl transferase center residing in the 50S subunit catalyzes the peptide bond formation between the two amino acids attached to the A- and P-tRNAs, and the nascent polypeptide grows by one amino acid pushing into an exit tunnel formed mainly of rRNAs.3
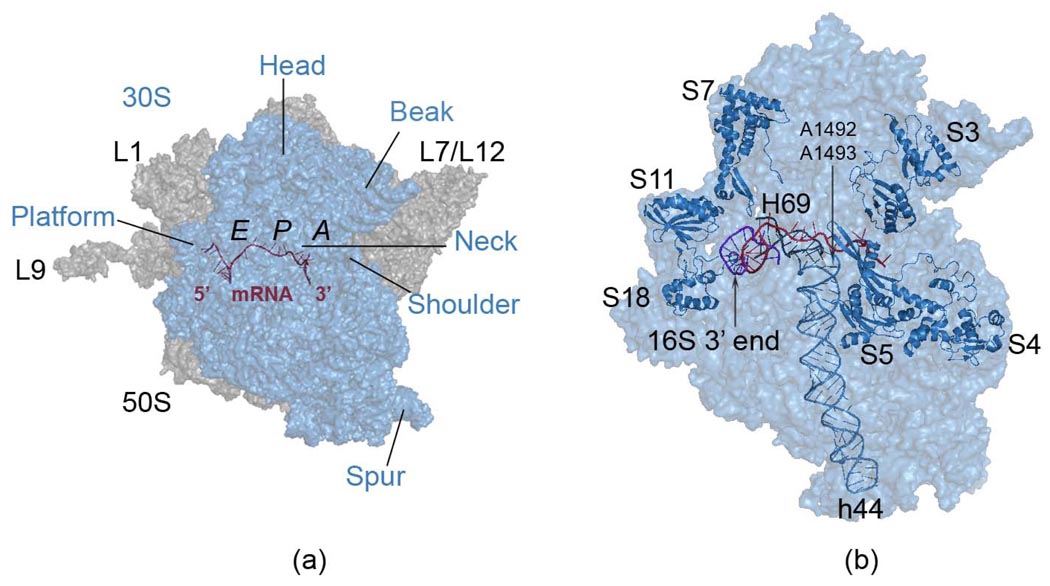
Figure 1.
Functionally important regions of the 70S ribosome complex (panel a). The small 30S subunit shown in transparent blue (panel a and b) contains the mRNA binding region, as well as its entrance and exit channels, together with the decoding center residues A1492 and A1493 residing on helix 44, at the interface of two subunits. The tRNA binding sites A-, P- and E- are indicated (panel a).The large 50S subunit in gray (panel a) has two prominent mobile stalks L1 and L7/L12, involved in controlling the tRNA exit and tRNA entrance, respectively. Extended ribosomal protein L9 of 50S is removed in some simulations to prevent its large dominant motions in normal mode analysis. The 30S subunit head has been reported to rotate during subunit association,10 tRNA selection9 and translocation,5 and this same type of rotation was also found in empty 70S E. coli ribosome crystal structure.11 The ribosomal proteins studied and the3’ end of 16S rRNA of the small subunit are shown in cartoon representation (panel b). All molecular graphics were prepared using PyMol.89
After the peptide bond formation is completed, the deacylated tRNA remains bound at the P-site with the polypeptide remaining attached to the A-tRNA. In order to vacate the A-site for an incoming new aminoacylated-tRNA, translocation of the tRNAs is required. Translocation is catalyzed by the EF-G.GTP binding to the L7/L12 stalk base, accompanied by large conformational rearrangements in the ribosome complex,4–6 but the exact mechanism is not known. According to the hybrid-states model,7 first the regions of the A- and P-tRNAs interacting with the large subunit move to the P- and E-sites while their anticodons remain bound to the mRNA in the 30S subunit. This transition state is called A/P and P/E for the two tRNAs. Then, subsequently the anticodon stems in the 30S subunit move to the P- and E-sites, catalyzed by EF-G.GTP. At the end of these translocation steps the deacylated tRNA is located at the E-site and the polypeptide carrying the tRNA at the P-site.
During protein synthesis, some regions of the ribosome undergo conformational changes, such as those regions important for the translocation of the tRNAs (Fig 1), which must assure maintenance of the correct frame reading. Cryo-electron microscopy (cryo-EM) studies have shown that upon binding of EF-G, the ribosomal subunits undergo a ratchet-like rotation.8 Other experimental studies also showed that small subunit head rotates during tRNA selection9, translocation,5 and subunit association,10 even in the absence of ligands.11 Such experimental evidence indicates that the 30S head rotation possesses inherent motions essential for protein synthesis. Another functionally important region is Helix 44 on the 30S subunit that contains the universally conserved decoding center A1492 and A1493 forming the major part of the inter-subunit bridges.12 Experimental evidence suggests that a shuttling motion of top region of Helix 44 is essential for tRNA and mRNA translocation.13 Another significant rearrangement during protein synthesis is observed at L1 and L7/L12 stalks of the large subunit:14–16 The L1 stalk plays an important role in the translocation process helping E-tRNA to leave the ribosome14 and was also confirmed in our previous simulations.17 On the other hand, the L7/L12 stalk, where EF-G or EF-Tu binds at its base to catalyze the translocation or to deposit a tRNA respectively, also undergoes the largest conformational change in the large subunit.5,15 This stalk that comprises the ribosomal protein L10 and multiple copies of protein L7/L12,18 is one of the most mobile parts of the large subunit.5,19–22 The other mobile components include the base of the L7/L12 stalk, the L1 stalk,14,21,23,24 the GTPase-associated centre25,26 and the C-terminal domain of protein L9.27,28
The correct positioning of the mRNA within the ribosome is critical for correct frame reading during protein synthesis. We have previously shown in our simulations how it is confined so strictly that it moves as a fully rigid body.17 However, little is known about the details of the mechanism of mRNA translocation during protein synthesis. A recent study based on a comparison of ribosome conformations from different crystal structures has elucidated the formation of the initiation complex where the anti-Shine-Dalgarno (S-D) sequence (AGGAGG) (i.e. the 3’-end of 16S shown in Fig 1 (b)), interacts with the 5’-region of the mRNA –together called S-D duplex- and is tightly constrained between the 30S platform and neck.29–31 It was also suggested that after a clock-wise rotation of the small subunit head, the mRNA undergoes a relaxation between initiation and post-initiation steps causing the S-D duplex to lengthen and to rotate about 70°, providing the correct positioning of the start codon at the P-site.30,31 After the first peptide bond is added, the S-D interactions are destabilized, and the 3’ end of 16S rRNA allows mRNA to advance along the exit channel,32 accompanied by a screw-like motion of the S-D duplex.33
Various structural features and regions are also essential for the ribosome to function properly. Among those, the secondary structure of the mRNA plays an important role in correct frame reading. Recent mutation studies on ribosomal proteins S3 and S434 revealed a previously unknown helicase processivity of the ribosome, functionally similar to the sliding clamp that holds DNA polymerase on DNA during replication.35 The ribosomal protein S5 is also important for translation fidelity, where the highly conserved residue Glycine 28 on loop 2 plays a significant role.36 A recent experimental study has revealed that the translocation is a non-continuous process divided by intrinsic short pauses due to the energy barriers originating in the secondary structure of the mRNA, and that the RNA unwinding and translocation processes are coupled.37 Symmetrical to this site, ribosomal proteins S7, S11 and S18 are located around the mRNA channel exit (Fig 1 (b)). S7 is one of the ribosomal proteins starting the assembly process of the small subunit.38,39 It is located near the decoding site and the tRNAs and its β-hairpin is positioned near the Watson-Crick face of the E-tRNA.2 Based on available experimental evidence, it was suggested that S7 might function as a gate keeper for tRNA,40 and the β-hairpin might function to separate the E-site codon-anticodon pair.41 Site-directed mutagenesis studies have indicated that the interaction region between S7 and S11, which connects the head of the small subunit to the platform, is important in translational fidelity.42 Ribosomal proteins S11 and S18 also closely interact with the S-D duplex and the 5’ end of the mRNA,30,43 and S18 is known to be crucial for cell survival.44
Normal mode analysis (NMA) is a powerful computational technique to elucidate the dynamics of large biological macromolecules.45–47 Especially, recent coarse-grained NMA methods48–54 have demonstrated the determination of functionally important transitional pathways of supramolecular assemblages revealed by a small number of low-frequency normal modes.55–59 The overall finding from many applications has been that the highest resolution structures are not generally required in order to compute the most important slowest modes, which are quite insensitive to the details of the structure. The conformational analysis of the ribosome by coarse grained normal mode computations has been investigated, although not sufficiently to extract all details of its mechanisms. The collective dynamics of the ribosome and its complexes have been analyzed with normal modes using a coarse-grained force field,58 and an elastic network model, namely the anisotropic network model,17,49,56 coarse-grained molecular dynamics (MD) up to 500ns,60 and full-atom targeted MD simulations in explicit water.61
In this study, the collective dynamics of the bacterial ribosome are investigated with the anisotropic network model (ANM)49 in order to understand the directions of conformational changes for various model systems based on 70S complex crystal structures. All models include 70S and mRNA, but tRNAs, EF-Tu, and ribosomal proteins are selectively excluded to analyze their effects on the ribosomal dynamics. The various 70S complex models used for our analysis are shown in Table 1. In contrast to recent computational studies that concentrate on the general dynamics of the mobile L1 and L7/L12 stalks, tRNAs and the ratchet-like rotations of the subunits, here we focus on (1) the changes in collective dynamics in the absence of some functional parts, and (2) the conformational rearrangements at the entry and exit channels of the mRNA on the ribosome, including ribosomal proteins S3, S4, S5, S7, S11, S18 and the 3’ end of the 16S rRNA in the 30S subunit. Finally, we propose a possible mechanism for the translocation of mRNA in the ribosome during the elongation phase.
Table 1.
Various ribosome models used in this study.
| Model name | Model Details | PDB | Number of Nodes |
Figure |
|---|---|---|---|---|
| 70S | 70S with A-, P-, E-tRNAs and mRNA | 1JGO (47), 1GIY (48) | 9808 | 2(a) |
| 70S_no_A | 70S with P-, E-tRNAs and mRNA | 1JGO, 1GIY | 9732 | 2(a) |
| 70S_no_E | 70S with A-, P-tRNAs and mRNA | 1JGO, 1GIY | 9734 | 2(a) |
| 70S_no_A_P_E | 70S without A-, P-, E-tRNAs, with mRNA | 1JGO, 1GIY | 9582 | 2(a) |
| 70S+EF-Tu | 70S with A-, P-, E-tRNAs, mRNA and EF-Tu | 1JGO, 1GIY, 1MJ1 (49) | 10213 | 2(b) |
| 70S_no_proteins | 70S with A-, P-, E-tRNAs, mRNA, without ribosomal proteins | 1JGO, 1GIY | 4763 | 2(c) |
| 70S_no_L1 | 70S with A-, P-, E-tRNAs, mRNA, without ribosomal protein L1 | 1JGO, 1GIY | 9584 | 2(a) |
| 70S_no_L7/L12 | 70S with A-, P-, E-tRNAs, mRNA, without ribosomal protein L7/L12 | 1JGO, 1GIY | 9552 | 2(a) |
2. Materials and methods
2.1 Structures studied
In order to observe the effect of some functional members of the complex using the elastic network model, various structures of the bacterial ribosome have been constructed for the purpose of simulations. The model systems are 70S-mRNA complexes with the mRNA and (i) with the A-, P-, E-tRNAs, called 70S; (ii) with the P-, E-tRNAs, called 70S_no_A; (iii) with the A-, P-tRNAs, called 70S_no_E; (iv) without the tRNAs, called 70S_no_A_P_E; (v) with the A-, P-, E-tRNAs and EF-Tu, called 70S+EF-Tu, (vi) without the ribosomal proteins, called 70S_no_proteins, (vii) without the ribosomal protein L1, called 70S_no_L1 and (viii) without the ribosomal proteins on the L7/L12 stalk, called 70S_no_L7/L12. These model details are listed in Table 1. These model systems are all generated from the same PDB files 1JGO-1GIY and 1MJ1 containing three tRNAs, a 27 nucleotide long mRNA and the EF-Tu.tRNA, respectively, so any distortions originating from deleted parts will not be accounted for in these calculations.
2.2 Anisotropic Network Model
The elastic network models utilize the protein, RNA, or protein-RNA structures in their native states. The simplest elastic network model is the Gaussian network model48 (GNM) that was originally developed for the theory of rubberlike elasticity of random polymer networks. It assumes that network nodes fluctuate randomly around their mean positions and that the distribution of fluctuations is isotropic, i.e. fluctuations are represented by spheres. The anisotropic network model49 (ANM) is an extension of GNM and assumes that fluctuations are anisotropic and are represented by ellipsoids. In these original models, ANM and GNM, the residues were represented by a coarse-grained node located at each Cα position or each residue’s center of mass. The close neighboring nodes within a cutoff distance rc are connected by harmonic springs with a force constant γ to define the three-dimensional network. Consequently, the total energy of the system of N residues is the sum over all harmonic interactions of i,j interconnected residue pairs given as
| (1) |
Here, Ri and ΔRi are the position and fluctuation vectors of node i (1≤i≤N); Rij is the distance between i,j pairs; and h(x) is the Heaviside step function [h(x)=1 if x≥0, and zero otherwise]. The adjustable harmonic spring constant γ is identical for all bonded and non-bonded interactions within the elastic network.
Because the starting structure is assumed to be the lowest in energy, the potential energy of the network in its distorted forms can be expressed as
| (2) |
where ΔR is the 3N-dimensional fluctuation vector, ΔRT its transpose and H is the (3N×3N) Hessian matrix, or force constant matrix, whose elements are the second derivatives of the total potential energy with respect to the Cartesian coordinates of the ith and jth nodes.
To calculate the normal modes, the Hessian matrix H is diagonalized in the canonical form
| (3) |
Here, Λ is a (3N × 3N) diagonal matrix with diagonal elements corresponding to eigenvalues (λ1,…, λ3N) or inverse squared normal mode frequencies. S is an orthogonal (3N × 3N) matrix (i.e. STS = I) whose columns are the eigenvectors.
After H is diagonalized, the overall motion can be obtained as the summation over (3N) normal modes. The frequencies of the translational and rotational motions of the structure are zero resulting in (3N - 6) normal modes. The mean-square fluctuation of residue i can be expressed in terms of coordinates as a summation over all normal modes as
| (4) |
where 〈ΔRi2〉 is the mean-square fluctuation of residue i, kB is the Boltzmann constant, T is the absolute temperature, and ωl is the normal mode frequency for the non-zeroth lth mode. The parameter γ is generally adjusted by scaling the theoretical fluctuations (cumulative of 3N-6 normal modes) according to those obtained from the experimental B-factors, but this adjustment is not essential to understand the relative importance of individual ribosomal motions.
In our simulations, the diagonalization of the Hessian matrix is the most computationally challenging task. For example, the final elastic network for the ribosome complex contains 9808 nodes for the 70S-mRNA-tRNAs complex and the diagonalization of a (29424 × 29424) Hessian matrix is required. The 3D displacement vectors of the nodes are calculated for a specified number of the slowest modes (k=10 in our case) using the computationally efficient software package BLZPACK62 using a block Lanczos algorithm63. This algorithm facilitates deriving the eigenmodes and allows us to obtain the desired eigenvectors of this large matrix within a reasonably short time.
In general, the low frequency collective dynamics relate to the biological function of the macromolecules and can be obtained with high computational efficiency compared with conventional full-atom molecular dynamics simulations.49,50,52,54,55,57,64 Our calculations on ribosome are based on the first ten slow modes, which correspond to the most collective motions of the ribosome complex, dominantly the ratchet-like rotation of the subunits explained in detail in the following section. We report the cumulative fluctuations of the subset of k slowest modes as
| (5) |
It should be noted that fluctuations reported in this work do not represent the absolute values in Å2 and specify the relative motions.
3. Results and discussion
We perform an ANM analysis on the bacterial ribosome structure 70S of T. Thermophilus in its complex with the A-, P-, E-tRNAs, and mRNA, using protein data bank structures (PDB)65 1JGO and 1GIY2,43 as an extension of an earlier work.56 Additionally, the crystal structure of EF-Tu in its complex with tRNA (1MJ1)66 originally fit to the 1GIY structure, is used to investigate the ribosome dynamics. The crystal structure of the entire 70S ribosome is at low-resolution (5.5 Å) and contains only the P and Cα atoms of the respective 30S and 50S subunits together with all heavy atoms of the tRNAs and mRNA. It is one of the most complete ribosome structures except for the notable disorder in the L7/L12 stalk. On the other hand, there are recently higher resolution crystal structures at 2.8 Å 67 and 3.5 Å;11 but the former lacks some proteins of the L7/L12 stalk together with the A-tRNA, and the latter does not contain the ribosomal protein L1 together with any of the tRNAs or the mRNA. Moreover, because we are focusing on the interactions of the mRNA with its surroundings at the entrance and exit channel and because these cannot be studied based on a short (11 nucleotide long) or completely absent mRNA in the higher-resolution structures; therefore, we consider the lower-resolution structure to be most suitable for our purpose since it has a 27 nucleotide long mRNA. Also since the ribosome structure will be coarse-grained, the best crystal structures for our purposes (i.e. structures with PDB codes 1JGO and 1GIY) have been chosen for our ribosome model.
In this crystal structure, two copies of L7/L12 proteins could be placed on the L7/L12 stalk.18,43 No electron density could be assigned to the protein L10 and the location of L7/L12 proteins was later viewed to have been erroneous in the original 5.5 Å crystal structure by more recent studies.22,68 Still the structure of the L7/L12 stalk will be retained in some of the present models to maintain some electron density for this stalk, which may possibly influence the collective dynamics. However, we observe essentially no changes in the dominant motions of the remainder of the structure, when this L7/L12 stalk is deleted, so the most likely gain from including this stalk is to learn about its likely directions of its motions.
A more globular structure of the large subunit could be obtained by removing the extended ribosomal protein L9 (Fig 1 (a)), similar to our previous work,56 which may undergo large amplitude motions and dominate the collective motions for the slow modes, the so-called “tip effect”.69 So far, different node densities have been mostly implemented in the coarse-grained elastic network models of protein-DNA/RNA structures such as placing a single node at the P position,56 two nodes at the P and O4’ positions,70 or three nodes at the P, C2 and C4’ positions71 of the nucleotides while retaining a single node at the Cα position for amino acids. Such complexes are also investigated as a full-atom elastic network using mechanical force fields, where the Hessian matrix is first reduced and then the eigenvectors are projected back.72 In the current elastic network model, we use the one-node per interaction site, i.e. P for nucleotides and Cα for amino acids since our crystal structures contain coordinates for these nodes only. Similar to our previous study,56 the cutoff distance to determine interacting Cα-Cα residue pairs is taken as 15Å, whereas it is 24Å for Cα-P and P-P pairs in order to have a sufficient number of interactions to constrain the structure.50 The harmonic force linking all these nodes is taken to be uniform throughout the elastic network, so there is no specificity to the interactions, and all the spring constants are identical.
3.1 General conformational changes
For the various model systems in this study, the presence of tRNAs, EF-Tu (or EF-G) and the ribosomal proteins really affect the overall shape of ribosome only slightly, as can be seen in Figure 2. Since we73 and Lu and Ma74 demonstrated the dominant effect of shape in determining the slowest most important motions of large structures, it can be anticipated that all of these structures are likely to have similar slowest motions. In Figure 3, the displacement vectors of the nodes are displayed for three of the model systems (70S, 70S+EF-Tu and 70S_no_proteins) for some of their comparable slowest frequency modes. In the collective dynamics of the different models of the ribosome, the ratchet-like rotation of the two subunits and the mobility of the L1 and L7/L12 stalks are the two main global motions consistently observed in the slowest modes.
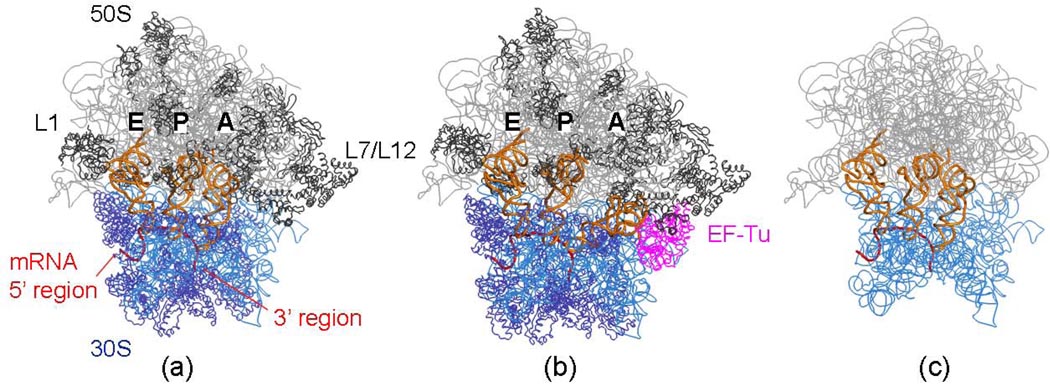
Figure 2.
Ribbon diagrams of some models for the 30S subunit, rRNA is represented in blue and ribosomal proteins in dark blue and for the 50S subunit, its rRNA is in grey and the ribosomal proteins in dark grey. (a) 70S and its complex with the A-, P- and E-tRNAs, together with the mRNA (b) the same 70S ribosome but also with EF-Tu and tRNA complex present in the hybrid A/T-site on 30S, and (c) rRNA of 70S and its complex with the A-, P-, E-tRNAs and mRNA, but without the proteins. PDB codes for the 30S and 50S subunits and EF-Tu are 1JGO,43 1GIY43 and 1MJ1,66 respectively.
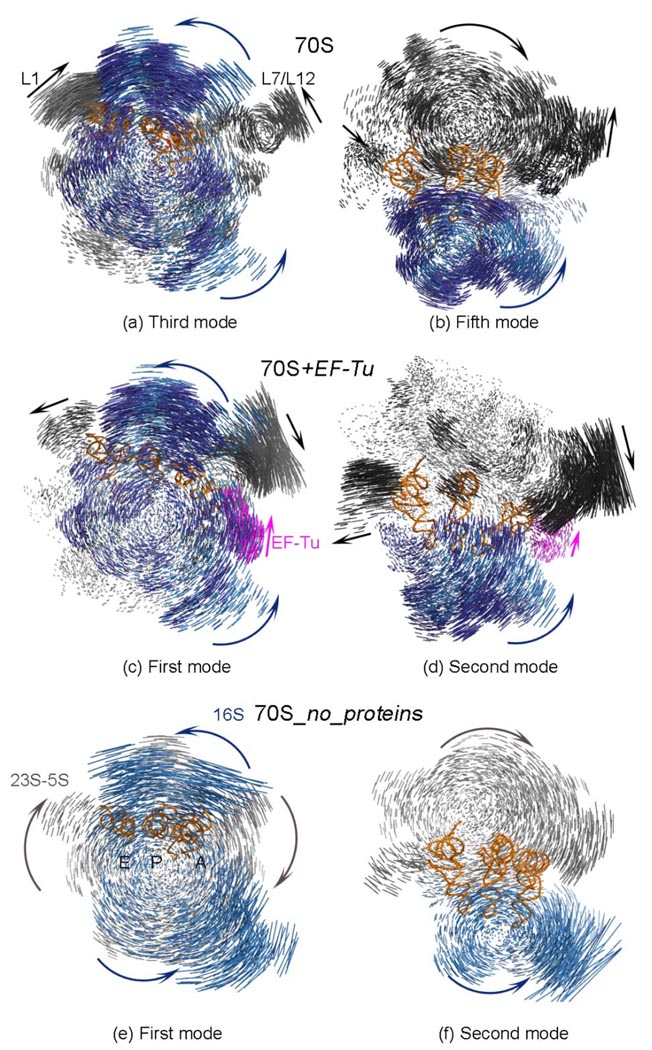
Figure 3.
Displacement vectors between alternative conformations of different model systems for slow modes. One alternative displacement direction is shown with the parts of the structure in different colored blue/dark blue (30S rRNA/ proteins), gray/ dark gray (50S rRNA/ proteins) and magenta (EF-Tu) arrows. For clarity, the displacements of tRNAs are not shown.
Ratchet-like rotation of the 30S and 50S subunits
The ratchet-like motion of the 30S and 50S subunits and the high mobilities of the L1 and L7/L12 stalks were clearly reported for the 70S-tRNAs-mRNA complex in previous computational studies.17,56,58,60 In addition, here, our elastic network analyses reveal that these motions are not only dominant for the 70S, but also for all of the different model systems studied here, i.e. the 70S with/ without tRNAs or ribosomal proteins, and even together with the EF-Tu.
To estimate the similarity between the directions manifested in the k normal modes of motion for the different models, the average overlap value or inner products of the normalized eigenvector sets vi and wj can be calculated75
| (6) |
An overlap value equal to 1 indicates a perfect match between the directions of displacement vectors for the nodes in two models. However, some normal modes from the reference model 70S exhibit high dot product values with multiple modes from the other set, i.e. there is not always a strict one-to-one correspondence. Still, each low frequency mode from 70S may be satisfactorily represented by a linear combination of the slowest ten modes from the other set. Table 2 provides the overlap values between different models with 70S used as a reference, where all values compared are averaged over the first 10 normal modes excluding the 6 modes corresponding to rigid body translation and rotation.
Table 2.
Overlap values averaged over 10 slowest modes
| Model | Overlapa |
|---|---|
| 70S_no_A | 0.99 |
| 70S_no_E | 0.97 |
| 70S_no_A_P_E | 0.92 |
| 70S+EF-Tu | 0.96 |
| 70S_no_proteins | 0.87 |
| 70S_no_L1 | 0.98 |
| 70S_no_L7/L12 | 0.98 |
Overlap values are calculated from equation 1 with the reference structure 70S.
Even in the absence of ribosomal proteins constituting only about half of the actual ribosome complex, the ratchet motion of the subunits (Fig3 (a), (c), (e)) and the small subunit head motion (Fig3 (b), (d), (f)) remains dominant. The ribosome, with its huge size of 2.5 MDa and high level of molecular complexity, probably evolved in a complex evolutionary process,65 but nonetheless at each stage in its evolution the functional motions were necessarily preserved. One piece of evidence supporting the viability and activity of the rRNA by itself is the set of in vitro experiments showing that the peptidyl transferase activity is preserved in 50S-like particles even in the absence of ribosomal proteins;76–78 this may imply that rRNA, mostly carrying out the ribosome functions, was present first and the ribosomal proteins were added later during its evolution.79 The ANM simulations of the system 70S_no_protein have shown that since the ribosome preserves its overall 3D shape; it remains capable of fulfilling its functional motions including the characteristic ratchet-like rotation of subunits, the small subunit head rotation, and the L1 and L7/L12 stalk fluctuations, with a resultant relatively high overlap value of 0.87 (Table 2).
Effect of the stalks and tRNAs on collective dynamics
The mobility of the two stalks -L1 and L7/L12- were observed experimentally with cryo-EM5,14 and reported to be dominant and important in the collective motions of ribosome during translation by the ribosome. In vitro experiments showed that the lack of proteins L7/L12 and L1 decreases the rate of protein synthesis.80,81 Previous computational studies on the ribosome complex with coarse-grained molecular dynamics,60 coarse-grained NMA,58 and coarse-grained ANM17,56 have also pointed out the large conformational rearrangements in the L1 and L7/L12 stalks, as well as the biologically functional importance of their anticorrelated movements.
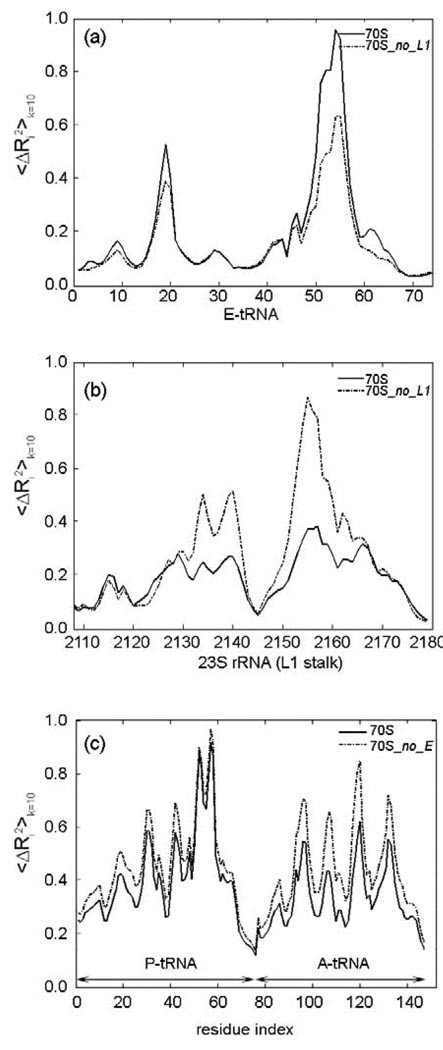
Next, the ribosome model structures lacking ribosomal proteins on L1 and L7/L12 stalks, namely 70S_no_L1 and 70S_no_L7/L12 respectively, are studied with ANM in order to observe the effect of these two regions on the collective dynamics of the ribosome complex. Extremely high overlap values (0.98, Table 2) are observed for both models indicating that some functional rearrangements such as the ratchet-like rotation of the two subunits, which is the dominant motion of the complex, together with the 30S head rotation and mobility of stalk-like regions are preserved. In this study, all mean-square deviation plots are obtained from the average sum over the slowest ten modes for the corresponding regions, and the motions are scaled between 0 and 1 for clarity. This permits us to focus on the relative motions of the different parts of the structure. The fluctuation graphs shown in Fig4(a)–(b) indicate that the absence of the ribosomal protein L1 especially affects the dynamics of the L1 stalk’s rRNA, which gains greater mobility, as well as the elbow of the E-tRNA which loses mobility. These changes may have important effects on the stability of this region especially for the removal of the E-tRNA, which is known to interact with ribosomal protein L1.2 On the other hand, the absence of proteins on the L7/L12 stalk does not make any significant changes in the fluctuations of tRNAs and mRNA.
Figure 4.
Cumulative mean-square fluctuations over 10 slowest modes 〈ΔRi2〉k=10 for the (a) E-tRNA, (b) 23S rRNA region of L1 stalk in models 70S and 70S_no_L1, and (c) A- and P-tRNAs in models 70S and 70S_no_E. The motions are scaled between 0 and 1 for clarity.
The root mean-square fluctuations for both the 70S and the 70S_no_A_P_E structures (also for 70S_no_A and 70S_no_E) show that the relative fluctuation magnitudes of each part of the structure (rescaled between 0 and 1), especially for the 30S head as well as for the L1 and L7/L12 stalks, increase in the absence of tRNAs. Hence, the binding of the tRNAs stabilizes several regions: the 30S head, especially ribosomal proteins S3, S4, S5, and their interactions with the mRNA, neck, spur, and the L1 and L7/L12 stalks of 50S (not shown). In addition, binding of elongation factor stabilizes 30S even further, as will be discussed in the following sections, for the entrance region of the mRNA strand. On the other hand, even though the presence of the E-tRNA does not affect the overall motion of the complex (showing an overlap of 0.97, Table 2), it does stabilize the fluctuations of the A-tRNA and the densely packed P-tRNA, and the codon-anticodon pair at the A-site in the slowest modes (Fig 4(c)). This finding may be related to the previous observations of the influence of the E-tRNA on the accuracy of A-site decoding.82
Helix 44 acts as a dynamic anchor for translocation
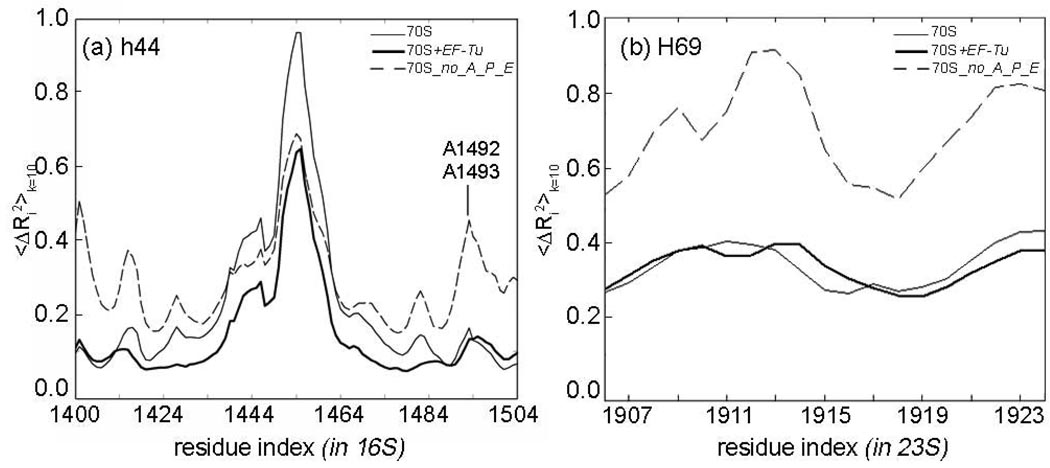
Helix 44 (h44) (C1399-G1504 in the T. Thermophilus 30S subunit, Fig 1(b)), which carries the universally conserved decoding center A1492 and A1493, and Helix 69 (H69) (G1906-C1924 in the T. Thermophilus 50S subunit) are known to be highly dynamic during translocation and ribosome recycling.13 Our simulations on the model 70S+EF-Tu suggest that including the EF-Tu (or EF-G due to similarity of the overall shapes of EF-G and Ef-Tu.tRNA complex)83 reduces somewhat the fluctuations of h44 carrying the decoding center A1492 and A1493; whereas the absence of the tRNAs may increase the motions of h44 and H69 to a significant extent (Fig5 (a), (b)). In addition, the lower end of h44 (nucleotides 1440–1460) is the most mobile part of h44 in the low frequency motions, contrary to the observations by VanLoock.13 This discrepancy can be explained by the size of the cavity in which h44 is confined. In contrast to the work of VanLoock,13 the elastic network models allow a larger dynamic space around h44, which is created as a result of the ribosome global motions. In this way, the lower part of h44 may still function as an “anchoring point” as suggested by VanLoock;13 yet not as a static anchor, but rather as a mobile anchor moving as part of a large supporting base (formed by its surroundings).
Figure 5.
Cumulative mean-square fluctuations over 10 slowest modes 〈ΔRi2〉k=10 for the regions (a) h44 and (b) H69 in models 70S, 70S+EF-Tu and 70S_no_A_P_E. The fluctuations are scaled between 0 and 1 for clarity.
3.2 mRNA and its interactions with ribosomal proteins
The ribosome as an mRNA helicase
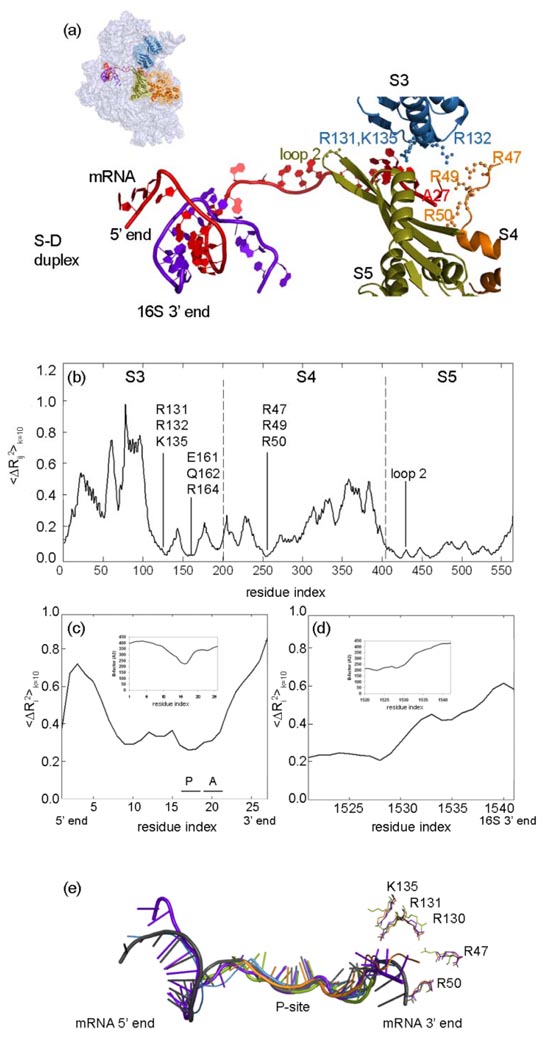
In a recent experimental study, it was proposed that the ribosome behaves as an mRNA helicase to unwind its helical structure for correct frame reading, and that ribosomal proteins S3 and S4 in the 30S, mediate residues to fulfill this task by acting like a ‘clamp’ around the mRNA.34 In addition, the helicase can function even in the absence of GTP and elongation factors34 implying that the collective motions of the complex are critical for this activity. In Figure 6, mean square fluctuations of the distance between the ith and jth residues 〈ΔRij2〉 = 〈ΔRi2〉 + 〈ΔRj2〉 −2 〈ΔRi ·ΔRj〉 averaged over the first ten modes are plotted for S3, S4 and S5 (the mean-square fluctuations shown have been rescaled between 0 and 1). The results are shown for distances between parts of the ribosomal proteins S3 (i=1, …, 207), S4 (i=1, …, 208), S5 (i=1, …, 150) and the closest nucleotide (j=A27 in 1JGO) on the mRNA shown in Fig.6 (a). The nodes at Arg131, Arg132, and Lys135 on S3, and also Arg47 and Arg50 on S4 (Fig.6 (a)), which are known to have helicase activity,34 are close to the mRNA and move in coordination with the mRNA for the 10 slowest modes summed cumulatively (Fig6 (b)). During this global motion, S3 also comes into close proximity with the mRNA strand in such a way that its polar and hydrophilic side chains Glu161, Gln162 and Arg164, as well as Arg49 of S4, are close to the mRNA and can interact. Our normal mode analysis strongly agrees with the experimental observations on the Arg and Lys residues on S3 and S4, and hence we suggest that these reported residues or some closely nearby regions might be playing an important role in the helicase activity of the ribosome. Unfortunately A27 is the end of the mRNA and if the mRNA were longer it might show further details of the mRNA interactions.
Figure 6.
(a) The mRNA (red) 3’ end surrounded by ribosomal proteins S3 (blue), S4 (orange) and S5 (green) and its 5’ end interacting with 16S 3’ end (purple). Residues reported to be important for the helicase activity and the translation fidelity of the ribosome,34,36 are shown on the S3, S4 and S5. (b) Mean-square fluctuations between S3, S4, S5 proteins and the closest nucleotide A27 on mRNA cumulatively over the slowest 10 modes. Cumulative mean-square fluctuations summed over the slowest 10 modes 〈ΔRi2〉k=10 are shown for (c) mRNA and (d) the 3’ end of the 16S rRNA. It should be noted that all mean-square fluctuation values are relative and not scaled to match experimental B-factors. (e) Comparison of mRNA strands observed in various crystal structures; color codes for pdb files: 1JGO (blue),43 2B64 (orange),85 2J00 (green),67 1YL4 (purple),86 2HGR (black).30 When the different ribosome crystal structures (color coded as mentioned above) are compared, it is noteworthy that the side chains of the residues functioning in the helicase activity are very mobile.
S5 orients the mRNA strand for translation fidelity
The ribosomal protein S5 in the small subunit (Fig 6(a)), despite its closeness to the helicase active site and its forming the ring around the entrance channel, was proposed not to play any role in unwinding the mRNA strand.34 However, ambiguous ribosome mutations on S5 (Fig6 (a)) have revealed that this protein has functional importance for translation fidelity, and specifically, loop 2 is involved in RNA binding.36 Mean-square distance fluctuations between S5 residues and the nucleotide A27 (Fig6 (b)) show that the entire S5 ribosomal protein remains closer to the mRNA than do S3 and S4. This suggests that the tight positional alignment of S5 with the mRNA is functionally important to orient the strand for correct frame reading at the A-site, confirming the previous work.
EF-Tu binding controls helicase activity
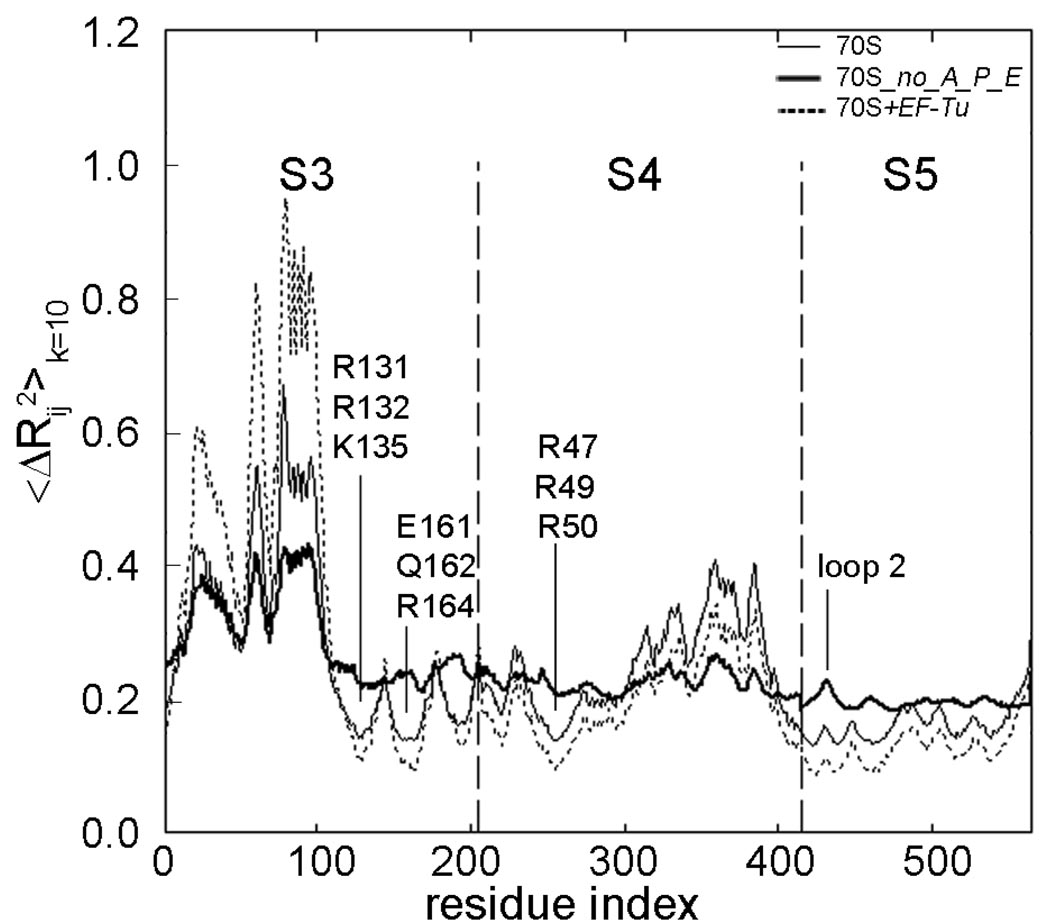
The presence of bound tRNAs and EF-Tu affects the fluctuations of the small subunit head significantly. Based on the cumulative effects of the sum of the slowest ten modes, the mean-square distance fluctuations between ribosomal proteins S3, S4, S5 and nucleotide A27 for the model systems 70S, 70S_no_A_P_E and 70S+EF-Tu are shown in Figure 7. The ratio of 〈ΔRij2〉 values averaged over residues of the specific region is 5.6: 1.6: 1 Å2 for 70S_no_A_P_E, 70S and 70S+EF-Tu, respectively. For purposes of comparison, the area under the curves has been normalized to one, for the S3, S4 and S5 proteins, and the motions are scaled between 0 and 1 to simplify comparisons of the relative motions of the different parts of the structure. In the absence of the tRNAs, the positional stabilities of helicase active site residues on S3–S4, as well as some residues on S5 are disturbed, whereas the stability of this region is enhanced by the presence of EF-Tu (with the tRNAs present). This finding indicates that the binding of the elongation factor may help the helicase active site to grasp the mRNA more strongly and S5 to further stabilize the structure by assuring correct codon reading at the A-site.
Figure 7.
Mean-square distance fluctuations 〈ΔRij2〉k=10 based on the cumulative sum of the slowest ten modes plotted for S3, S4, S5 between the closest mRNA nucleotide A27 for the model systems 70S, 70S_no_A_P_E and 70S+EF-Tu. The curves have been normalized and the motions are rescaled for purposes of comparison among the different parts of the structure.
S7, S11 and S18 control mRNA mobility
The ribosomal protein S7 is located near the decoding center, the mRNA and the tRNAs2,40 and has the particular role of initiating the 30S assembly process.38 Sequence analysis of S7 indicates that Arg75-Gly82 and Tyr86-Val91 around a β-hairpin are highly conserved among different species,40 and structural analysis implies that this arm may act as a gate keeper for the E-tRNA.2 Moreover, the β-hairpin is highly flexible as observed over various crystal structures.84 The presence of the ribosomal protein S18 in plastids is known to be essential for the survival of plant cells44 and the amino terminus of this protein is observed to be in close contact with the S-D duplex at the initiation step.30
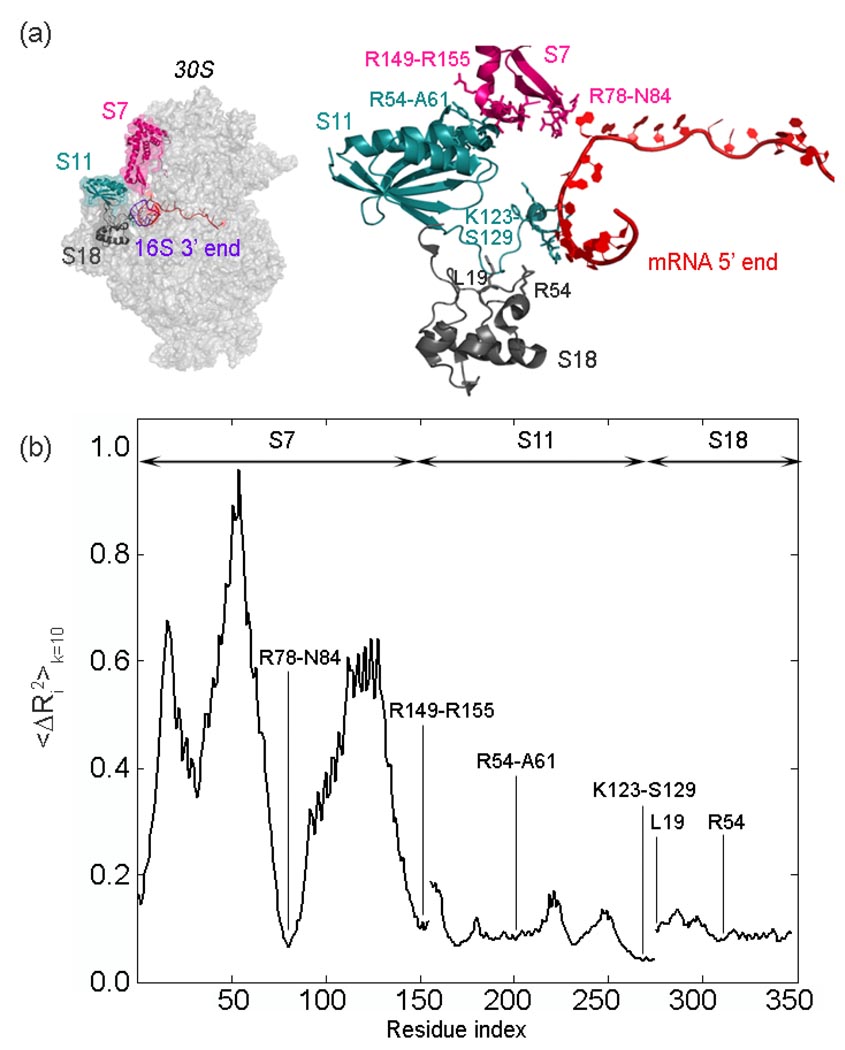
In order to investigate the dynamics of the proteins S7, S11 and S18 located at the exit of mRNA channel, the mean-square fluctuations averaged over the ten slowest modes are considered (Fig 8). The mean-square fluctuations of ribosomal proteins S11 and S18 have a lower amplitude compared with S7, indicating that the platform of the small subunit maintains its stability during collective motions. Similar to dynamics of the ribosomal proteins S3, S4, and S5 closely interacting with the mRNA strand, the low fluctuating residues of ribosomal proteins S7, S11 and S18 are lined up around the mRNA channel exit. This finding is significant since the flexibility of the small subunit head should be controlled for translational fidelity as has been previously suggested.42
Figure 8.
(a) The 5’ end of the mRNA (red) is surrounded by ribosomal proteins S7 (magenta), S11 (turquoise) and S18 (gray) at the exit channel, shown in two views. The important residues reported in various studies30, 42,43, 84 are shown on the right in stick form. (b) Cumulative mean-square fluctuations summed over the slowest 10 modes 〈ΔRi2〉 k=10 plotted for S7, S11 and S18, and rescaled between 0 and 1 . It is clear that the residues indicated in (a) and (b) surrounding the mRNA undergo extremely small fluctuations in ten slowest most important modes.
One of the regions fluctuating least on the small subunit is comprised of the highly conserved interaction residues in bacteria, between proteins S7 and S11, i.e. including polar residues Arg149-Arg155 on S7 and Arg54-Ala61 on S11.42 Additionally, residues Arg78-Asn84 on a β-hairpin of S7, residues Lys123-Ser129 on S11 and Lys19 on S18 barely fluctuate with to the mRNA strand, as well as Arg54 on S18 with the 16S rRNA, as indicated earlier.30,43 A consistent picture arises in the case of 〈ΔRij2〉 plots not shown here. This observation might be related to how the ribosome’s unusual structure controls the translational fidelity by stabilizing the shifting speed of mRNA during the elongation phase, besides requiring several critical interactions. In summary, the motion of the mRNA may be controlled both at the entrance by S3, S4 and S5, and at the exit by S7, S11 and S18, along with 3’ end of the 16S rRNA, based on the analysis of 10 slowest normal modes in this study.
The mRNA shows some limited range of motions at the P-site
To investigate the motions of the mRNA and the 3’ end of the 16S rRNA, which form the Shine-Dalgarno duplex with the 5’ mRNA in the initiation phase, the rescaled mean-square fluctuations 〈ΔRi2〉 over the 10 slowest modes are plotted in Figure 6 (c) and (d) where the inserts are the experimental B-factors from 2HGR (pdb) of the 30S T. Thermophilus crystal structure at 4.5Å 30 (there is no B-factor information for 1JGO). It is clear, both from the cumulative 10 modes and the B-factor plots that the A- and P-site codons are the least mobile sites in the low frequency motions while the 5’ and 3’ regions of the mRNA are more flexible, as previously reported from the difference Fourier maps.30 In addition, the 3’ end of the 16S rRNA, which has functional importance in the initiation complex to help the start codon be accommodated into the correct position,30 shows higher mobility, possibly to facilitate the movement of the ribosome along the mRNA after initiation.32,79
In order to investigate any coupling of motion between the entrance and the exit of the mRNA channel, the inner dot product between the displacement vectors of the 3’ end of the 16S rRNA and the helicase residues on S3 and S4 are calculated for the slowest 10 and 100 normal modes. Although in individual low frequency modes, these regions display correlated motions (e.g. in the second normal mode ~0.7) and anti-correlated motions (e.g. in the third normal mode ~−0.7), the average value of the correlation over the first 10 modes (first 100 modes) is ~ −0.2 (0.0) indicating a variety of motions of the entrance and exit of the mRNA channel.
To further validate our findings, the available crystal structures of the mRNAs (1JGO (blue),43 2B64 (orange) 85, 2J00 (green) 67, 1YL4 (purple)86 and 2HGR30) are superimposed in Figure 6 (e). The superposition shows that both the 5’ and 3’ regions of the mRNA are more flexible and that the single stranded mRNA shows flexibility by assuming a range of conformations except where it is bound to the P-site due to codon-anticodon pairing and strong interactions, and to a lesser extent at the A site, recently studied in detail.31 In addition, side chains of the helicase active site residues are flexible and adjust their conformations in coordination with those of the mRNA.
3.3 A model mechanism for translocation
By combining the hybrid-state model, the experimental data and the normal mode analysis for the elastic network models, one possible sequence of events for mRNA translocation may be summarized as a reach-catch-pull-release-reach cycle: (i) During the elongation phase, the ribosome collective motions cause the mRNA helical structure to be disrupted by being ‘clamped’ by the small subunit proteins S3 and S4, while S5 orients mRNA for correct frame reading at the A-site. (ii) Symmetrical to this site, the mRNA strand mobility is hindered by ribosomal proteins S7, S11 and S18 of the small subunit. (iii)The mRNA strand is held particularly tightly at the P-site due to dense packing and strong interactions critical for peptide bond synthesis, but after the head and ratchet-like rotation of 30S, the mRNA is forced to translocate one codon, and to A- and P-tRNAs adopt their hybrid-states. Then the CCA ends of the tRNAs translocate. (iv) The translocated mRNA strand finds enough space after leaving the P-site to the E-site and adjusts its conformation so that E-tRNA can detach itself with the help of the L1 stalk. (v) Meanwhile, due to the 30S head motion, interactions between the 3’ end of the 16S rRNA and the 5’ region of the mRNA as well as proteins S7, S11, S18 are already weakened following initiation, but the intrinsically mobile 3’ end of 16S acts like a hook to reel the folded mRNA into the inter-subunit region. (vi) The S3 and S4 clamp releases the shifted mRNA strand and seizes again the incoming mRNA strand to unwind it.
4. Conclusions and outlook
Recent elastic network models of large proteins coarse-grained at different hierarchical levels, i.e. at different resolutions, all exhibit extremely similar collective motions.50,51,53,87 Thus it is well established that the overall shape of the molecule rather than the details governs its dynamics. Simulating different model systems of the ribosome with the coarse-grained elastic network model has shown that the ratchet-like rotation of the two major subunits, rotation of the 30S head, and high fluctuation movements of the L1 and L7/L12 stalks are the motions most intrinsic to the structure of the ribosome. Our results support the view a specific shape has been selected for a system to fulfill some specific function, and once more points out the fact that similar motions can be found in structures with similar shapes, regardless of whether they are made up of RNA or RNA and proteins together. Translocation6 occurs even in the absence of EF-G.GTP and the helicase activity of the ribosome does not require GTP hydrolysis,34 indicating that the collective motions are important for translocation, the helicase activity of the ribosome, and translation fidelity.
The mean-square fluctuations in the slow modes reveal that the absence of ribosomal proteins L1, L7/L12 and the E-tRNA does not inhibit the ratchet-like rotation of the two subunits. However, in the model structure lacking the ribosomal protein L1, i.e. 70S_no_L1, the rRNA region of this stalk becomes more mobile while the elbow of E-tRNA loses mobility. On the other hand, the absence of E-tRNA (i.e. in model structure 70S_no_E) affects specifically the A- and P- tRNAs where the fluctuations increase. Our results have further revealed the stabilization of helix 44 carrying the decoding center, in the presence of the elongation factor.
We have proposed a possible sequence of events (a reach-catch-pull-release-reach cycle) for translocation during the elongation phase. In the small 30S subunit, the residues Arg131, Arg132, Lys135 in S3 and Arg47 and Arg50 in S3 were reported to be the important residues for the helicase activity of the ribosome.34 In addition to these residues, our results suggest that Glu161, Gln162 and Arg164 on S3, and Arg49 on S4 may also be important to disrupt the helical structure of the mRNA. While the mRNA is constrained by the clamp at the entrance channel, at the exit site the 3’ end of 16S rRNA remains intrinsically mobile, forming the S-D interaction with the 5’ end of the mRNA during initiation acting like a hook to help the mRNA to exit the ribosome during elongation.
Another important observation concerns the exit site of the mRNA channel, which is surrounded by ribosomal proteins S7, S11 and S18. This region is observed to be one of the most stable parts of the ribosome complex while undergoing large collective motions such as the ratchet-like rotation of two subunits. Our results indicate that interaction region between proteins S7–S11 (i.e. Arg149-Arg155 on S7 and Arg54-Ala61 on S11),42 residues Arg78-Asn84 on β-hairpin of S7, residues Lys123-Ser129 on S11, and residues Lys19 and Arg54 on S18 undergo limited motions with the mRNA compared to the remainder of the complex. These low amplitude fluctuations controlled by the overall structure of the complex are likely related to the control mechanism of translational fidelity during protein synthesis.
Many results observed in these simulations and in our recent calculations88 conform closely to what was previously known about the ribosome and its motions, which is a significant accomplishment and which serves to validate the elastic ribosome models. Nonetheless there are a large number of testable roles of specific residues interacting with mRNA that have been suggested by the present calculations, as mentioned in the previous paragraphs. Finally, an intriguing question remains to be investigated whether the motions of the entrance and the exit channels of the mRNA are synchronized during protein synthesis.
Acknowledgments
The Scientific and Technological Research Council of Turkey BIDEB-No: 2214 fellowship supported a visit of OK to Iowa State University where most simulations were performed. RLJ and AK acknowledge the financial support provided by NIH grants 1R01GM073095, 1R01GM072014, and 1R01GM081680. RLJ acknowledges the financial support provided by NIH grant R33GM066387 and NSF grant CNS-0521568. PD and OK acknowledge partial support by TUBITAK Project 104M247 and EU-FP6-ACC-2004-SSA-2 contract no. 517991.
Abbreviations list
- rRNA
ribosomal RNA
- tRNA
transfer RNA
- mRNA
messenger RNA
- GDP
guanosine diphosphate
- GTP
guanosine triphosphate
- A
aminoacyl
- P
peptidyl
- E
exit
- EF
elongation factor
- S-D
Shine-Dalgarno
- MD
molecular dynamics
- ANM
anisotropic network model
- NMA
normal mode analysis
- cryo-EM
cryo-electron microscope
- msf
mean-square fluctuation.
Footnotes
'This is an author-created, un-copyedited version of an article accepted for publication Physical Biology. IOP Publishing Ltd is not responsible for any errors or omissions in this version of the manuscript or any version derived from it. The definitive publisher authenticated version is available online at http://www.iop.org/EJ/journal/physbio'
References
- 1.Agrawal RK, Penczek P, Grassucci RA, Li Y, Leith A, Nierhaus KH, Frank J. Direct visualization of A-, P-, and E-site transfer RNAs in the Escherichia coli ribosome. Science. 1996;271:1000–1002. doi: 10.1126/science.271.5251.1000. [DOI] [PubMed] [Google Scholar]
- 2.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 3.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 4.Agrawal RK, Penczek P, Grassucci RA, Frank J. Visualization of elongation factor G on the Escherichia coli 70S ribosome: The mechanism of translocation. Proc Natl Acad Sci U.S.A. 1998;95:6134–6138. doi: 10.1073/pnas.95.11.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat. Struct. Biol. 1999;6:643–647. doi: 10.1038/10695. [DOI] [PubMed] [Google Scholar]
- 6.Joseph S. After the ribosome structure: how does translocation work? RNA. 2003;9:160–164. doi: 10.1261/rna.2163103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;334:362–364. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 8.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 9.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 10.Lata KR, Agrawal RK, Penczek P, Grassucci R, Zhu J, Frank J. Three-dimensional reconstruction of the Escherichia coli 30 S ribosomal subunit in ice. J. Mol. Biol. 1996;262:43–52. doi: 10.1006/jmbi.1996.0497. [DOI] [PubMed] [Google Scholar]
- 11.Schurwith BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 12.Frank J, Spahn CMT. The ribosome and the mechanism of protein synthesis. Rep. Prog. Phys. 2006;69:1383–1417. [Google Scholar]
- 13.VanLoock MS, Agrawal RK, Gabashvili IS, Qi L, Frank J, Harvey SC. Movement of the decoding region of the 16 S ribosomal RNA accompanies tRNA translocation. J. Mol. Biol. 2000;304:507–515. doi: 10.1006/jmbi.2000.4213. [DOI] [PubMed] [Google Scholar]
- 14.Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 15.Agrawal RK, Linde J, Sengupta J, Nierhaus KH, Frank J. Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G dependent translocation. J. Mol. Biol. 2001;311:777–787. doi: 10.1006/jmbi.2001.4907. [DOI] [PubMed] [Google Scholar]
- 16.Bocharov EV, Sobol AG, Pavlov KV, Korzhnev DM, Jaravine VA, Gudkov AT, Arseniev AS. From structure and dynamics of protein L7/L12 to molecular switching in ribosome. J. Biol. Chem. 2004;279:17697–17706. doi: 10.1074/jbc.M313384200. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y, Jernigan RL. Comparison of tRNA Motions in the free and ribosomal bound structures. Biophys. J. 2005;89:3399–3409. doi: 10.1529/biophysj.105.064840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ilag LL, Videler H, McKay AR, Sobott F, Fucini P, Nierhaus KH, Robinson CV. Heptameric (L12)6/L10 rather than canonical pentameric complexes are found by tandem MS of intact ribosomes from thermophilic bacteria. Proc. Natl. Acad. Sci. USA. 2005;102:8192–8197. doi: 10.1073/pnas.0502193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malhotra A, Penczek P, Agrawal RK, Gabashvili IS, Grassucci RA, Junemann R, Burkhardt N, Nierhaus KH, Frank J. E. Coli 70S ribosome at 15 Å by cryo-electron microscopy: localization of fMet-tRNA(fMet) and fitting of L1 protein. J. Mol. Biol. 1998;280:103–115. doi: 10.1006/jmbi.1998.1859. [DOI] [PubMed] [Google Scholar]
- 20.Montesano-Rhoditis L, Litz DG, Traut RB, Stewart PL. Cryo-electron microscopic localization of protein L7/L12 within the Escherichia coli 70 S ribosome by difference mapping and nanogold labeling. J. Biol. Chem. 2001;276:14117–14123. doi: 10.1074/jbc.M008430200. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Q, Ofverstedt LG, Skoglund U, Isaksson LA. Morphological variation of individual Escherichia coli 50S ribosomal subunits in situ, as revealed by cryo-electron tomography. Exp. Cell Res. 2004;300:190–201. doi: 10.1016/j.yexcr.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Diaconu M, Kothe U, Schlunzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 23.Gomez-Lorenzo MG, Spahn CMT, Agrawal RK, Grassucci RA, Penczek P, Chakraburtty K, Ballesta JPG, Lavandera JL, Garcia-Bustos JF, Frank J. Three-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80S ribosome at 17.5 Å resolution. EMBO J. 2000;19:2710–2718. doi: 10.1093/emboj/19.11.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 25.Frank J, Sengupta J, Gao H, Li W, Valle M, Zavialov A, Ehrenberg M. The role of tRNA as a molecular spring in decoding, accommodation, and peptidyl transfer. FEBS Lett. 2005;579:959–962. doi: 10.1016/j.febslet.2004.10.105. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalo P, Reboud JP. The puzzling lateral flexible stalk of the ribosome. Biol. Cell. 2003;95:179–193. doi: 10.1016/s0248-4900(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 27.Matadeen R, Patwardhan A, Gowen B, Orlova EV, Pape T, Cuff M, Mueller F, Brimacombe R, van Heel M. The Escherichia coli large ribosomal subunit at 7.5 Å resolution. Structure. 1999;7:1575–1583. doi: 10.1016/s0969-2126(00)88348-3. [DOI] [PubMed] [Google Scholar]
- 28.Spahn CMT, Blaha G, Agrawal RK, Penczek P, Grassucci RA, Trieber CA, Connell SR, Taylor DE, Nierhaus KH, Frank J. Localization of the tetracycline resistance protein Tet(O) on the ribosome and the inhibition mechanism of tetracycline. Mol. Cell. 2001;7:1037–1045. doi: 10.1016/s1097-2765(01)00238-6. [DOI] [PubMed] [Google Scholar]
- 29.Shine J, Dalgarno L. The 3′-terminal sequence of escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. U. S. A. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 31.Jenner L, Rees B, Yusupov M, Yusupova G. Messenger RNA conformations in the ribosomal E site revealed by X-ray crystallography. EMBO Reports. 2007;8:846–850. doi: 10.1038/sj.embor.7401044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uemura S, Dorywalska M, Lee TH, Kim HD, Puglisi JD, Chu S. Peptide bond formation destabilizes Shine–Dalgarno interaction on the ribosome. Nature. 2007;446:454–457. doi: 10.1038/nature05625. [DOI] [PubMed] [Google Scholar]
- 33.Korostelev A, Trakhanov S, Asahara H, Laurberg M, Lancaster L, Noller HF. Interactions and dynamics of the Shine–Dalgarno helix in the 70S ribosome. Proc. Natl. Acad. Sci. U. S. A. 2007;104:16840–16843. doi: 10.1073/pnas.0707850104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 35.Kuriyan J, O’Donnell M. Sliding clamps of DNA polymerases. J. Mol. Biol. 1993;234:915–925. doi: 10.1006/jmbi.1993.1644. [DOI] [PubMed] [Google Scholar]
- 36.Kirthi N, Roy-Chaudhuri B, Kelley T, Culver GM. A novel single amino acid change in small subunit ribosomal protein S5 has profound effects on translational fidelity. RNA. 2006;12:1–12. doi: 10.1261/rna.302006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wen JD, Lancaster L, Hodges C, Zeri AC, Yoshimura SH, Noller HF, Bustamante C, Tinoco I. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nowotny V, Nierhaus KH. Assembly of the 30S subunit from Escherichia coli ribosomes occurs via two assembly domains which are initiated by S4 and S7. Biochemistry. 1988;27:7051–7055. doi: 10.1021/bi00418a057. [DOI] [PubMed] [Google Scholar]
- 39.Brodersen DE, Nissen P. The social life of ribosomal proteins. FEBS J. 2005;272:2098–2108. doi: 10.1111/j.1742-4658.2005.04651.x. [DOI] [PubMed] [Google Scholar]
- 40.Hosaka H, Nakagawa A, Tanaka I, Harada N, Sano K, Kimura M, Yao M, Wakatsuki S. Ribosomal protein S7: a new RNA-binding motif with structural similarities to a DNA architectural factor. Structure. 1997;5:1199–1208. doi: 10.1016/s0969-2126(97)00270-0. [DOI] [PubMed] [Google Scholar]
- 41.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 42.Robert F, Brakier-Gingras L. A functional interaction between ribosomal roteins S7 and S11 within the bacterial ribosome. Biol. Chem. 2003;278:44913–44920. doi: 10.1074/jbc.M306534200. [DOI] [PubMed] [Google Scholar]
- 43.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 44.Rogalski M, Ruf S, Bock R. Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Research. 2006;34:4537–4545. doi: 10.1093/nar/gkl634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tama F, Brooks CL. Symmetry, form and shape: Guiding principles for robustness in macromolecular machines. Annu. Rev. Biophys. Biomol. Struct. 2006;35:115–133. doi: 10.1146/annurev.biophys.35.040405.102010. [DOI] [PubMed] [Google Scholar]
- 46.Ma JP. Usefullness and limitations of normal mode analysis in modeling dynamics of biomolecular complexes. Structure. 2005;13:373–380. doi: 10.1016/j.str.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 47.Bahar I, Rader AJ. Coarse-grained normal mode analysis in structural biology. Current Opinion in Structural Biology. 2005;15:1–7. doi: 10.1016/j.sbi.2005.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bahar I, Atilgan AR, Erman B. Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold Des. 1997;2:173–181. doi: 10.1016/S1359-0278(97)00024-2. [DOI] [PubMed] [Google Scholar]
- 49.Atilgan AR, Durell SR, Jernigan RL, Demirel MC, Keskin O, Bahar I. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 2001;80:505–515. doi: 10.1016/S0006-3495(01)76033-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doruker P, Jernigan RL, Bahar I. Dynamics of large proteins through hierarchical levels of coarse-grained structures. J. Comput. Chem. 2002;23:119–127. doi: 10.1002/jcc.1160. [DOI] [PubMed] [Google Scholar]
- 51.Ming D, Kong Y, Lambert MA, Huang Z, Ma JP. How to describe protein motion without amino-acid sequence and atomic coordinates. Proc. Natl. Acad. Sci. USA. 2002;99:8620–8625. doi: 10.1073/pnas.082148899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haliloglu T, Bahar I, Erman B. Gaussian dynamics of folded proteins. Phys. Rev. Lett. 1997;79:3090–3093. [Google Scholar]
- 53.Kurkcuoglu O, Jernigan RL, Doruker P. Mixed levels of coarse-graining of large proteins using elastic network model succeeds in extracting the slowest motions. Polymer. 2004;45:649–657. [Google Scholar]
- 54.Eom K, Baek SC, Ahn JH, Na S. Coarse-graining of protein structures for the normal mode studies. J. Comput. Chem. 2007;28:1400–1410. doi: 10.1002/jcc.20672. [DOI] [PubMed] [Google Scholar]
- 55.Keskin O, Bahar I, Flatow D, Covell DG, Jernigan RL. Molecular mechanisms of chaperonin GroEL-GroES function. Biochemistry. 2002;41:491–501. doi: 10.1021/bi011393x. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y, Rader AJ, Bahar I, Jernigan RL. Global ribosome motions revealed with elastic network model. J. Struct. Biol. 2004;147:302–314. doi: 10.1016/j.jsb.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 57.Chennubhotla C, Bahar I. Markov methods for hierarchical coarse-graining of large protein dynamics. J. Comput. Biol. 2007;14:765–776. doi: 10.1089/cmb.2007.R015. [DOI] [PubMed] [Google Scholar]
- 58.Tama F, Valle M, Frank J, Brooks CL. Dynamic reorganization of the functionally active ribosome explored by normal mode analysis and cryo-electron microscopy. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9319–9323. doi: 10.1073/pnas.1632476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tama F, Wriggers W, Brooks CL. Exploring global distortions of biological macromolecules and assemblies from low-resolution structural information and elastic network theory. J. Mol. Biol. 2002;321:297–305. doi: 10.1016/s0022-2836(02)00627-7. [DOI] [PubMed] [Google Scholar]
- 60.Trylska J, Tozzini V, McCammon JA. Exploring global motions and correlations in the ribosome. Biophys. J. 2005;89:1455–1463. doi: 10.1529/biophysj.104.058495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanbonmatsu KY, Tung CS. Large-scale simulations of the ribosome: a new landmark in computational biology. J. Phys.: Conference Series. 2006;46:334–342. [Google Scholar]
- 62.Marques O, Sanejouand YH. Hinge-bending motion in citrate synthase arising from normal mode calculations. Proteins. 1995;23:557–560. doi: 10.1002/prot.340230410. [DOI] [PubMed] [Google Scholar]
- 63.Grimes RG, Lewis JG, Simon HD. A shifted block Lanczos algorithm for solving sparse symmetric eigenvalue problems. Seattle, WA: Boeing Computer Services; 1991. [Google Scholar]
- 64.Doruker P, Nilsson L, Kurkcuoglu O. Collective dynamics of EcoRI-DNA complex by elastic network model and molecular dynamics simulations. J. Biomol. Struct. Dyn. 2006;24:1–15. doi: 10.1080/07391102.2006.10507093. [DOI] [PubMed] [Google Scholar]
- 65.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stark H, Rodnina MV, Wieden HJ, Zemlin F, Wintermeyer W, van Heel M. Ribosome interactions of aminoacyl-tRNA and elongation factor TU in the codon recognition complex. Nat. Struct. Biol. 2002;9:849–854. doi: 10.1038/nsb859. [DOI] [PubMed] [Google Scholar]
- 67.Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 68.Datta PP, Sharma MR, Qi L, Frank J, Agrawal RK. Interaction of the G’ domain of elongation factor G and the C-terminal domain of ribosomal protein L7/L12 during translocation as revealed by Cryo-EM. Molecular Cell. 2005;20:723–731. doi: 10.1016/j.molcel.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 69.Lu M, Poon B, Ma JP. A New Method for Coarse-Grained Elastic Normal-Mode Analysis. J. Chem. Thy. Comput. 2006;2:464–471. doi: 10.1021/ct050307u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bahar I, Jernigan RL. Vibrational dynamics of transfer RNAs: comparison of the free and synthetase-bound forms. J. Mol. Biol. 1998;281:871–884. doi: 10.1006/jmbi.1998.1978. [DOI] [PubMed] [Google Scholar]
- 71.Delarue M, Sanejouand YH. Simplified normal mode analysis of conformational transitions in DNA-dependent polymerases: the elastic network model. J. Mol. Biol. 2002;320:1011–1024. doi: 10.1016/s0022-2836(02)00562-4. [DOI] [PubMed] [Google Scholar]
- 72.Van Wynsberghe, Cui Q. Comparison of mode analyses at different resolutions applied to nucleic acid systems. Biophys J. 2005;89:2939–2949. doi: 10.1529/biophysj.105.065664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Doruker P, Jernigan RL. Functional motions can be extracted from on-lattice construction of protein structures. Proteins. 2003;53:174–181. doi: 10.1002/prot.10486. [DOI] [PubMed] [Google Scholar]
- 74.Lu M, Ma JP. The role of shape in determining molecular motions. Biophys. J. 2005;89:2395–2401. doi: 10.1529/biophysj.105.065904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Amadei A, Ceruso MA, Nola AD. On the convergence of the conformational coordinates basis set obtained by the essential dynamics analysis of proteins’ molecular dynamics simulations. Proteins. 1999;36:419–424. [PubMed] [Google Scholar]
- 76.Polacek N, Mankin AS. The ribosomal peptidyl transferase center: structure, function, evolution, inhibition. Crit. Rev. Biochem. Mol. Biol. 2005;40:285–311. doi: 10.1080/10409230500326334. [DOI] [PubMed] [Google Scholar]
- 77.Noller HF, Hoffarth V, Zimniak L. Unusual resistence of peptidyl transferase to protein extraction procedures. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- 78.Khaitovich P, Mankin AS, Green R, Lancester L, Noller HF. Characterization of functionally active subribosomal particles from Thermus aquaticus. Proc. Natl. Acad. Sci. U. S. A. 1999;96:85–90. doi: 10.1073/pnas.96.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Korostelev A, Noller HF. The ribosome in focus: new structures bring new insights. TRENDS in Biochem. Sci. 2007;32:434–441. doi: 10.1016/j.tibs.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 80.Pettersson I, Kurland CG. Ribosomal protein L7/L12 is required for optimal translation. Proc. Natl. Acad. Sci. U. S. A. 1980;77:4007–4010. doi: 10.1073/pnas.77.7.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Subramanian AR, Dabbs ER. Functional studies on ribosomes lacking protein L1 from mutant Escerichia coli. Eur. J. Biochem. 1980;112:425–430. doi: 10.1111/j.1432-1033.1980.tb07222.x. [DOI] [PubMed] [Google Scholar]
- 82.Geigenmüller U, Nierhaus KH. Significance of the third tRNA binding site, the E Site, on E. coli ribosomes for the accuracy of translation : an occupied E site prevents the binding of non-cognate aminoacyl-transfer RNA to the A site. EMBO J. 1990;9:4527–4533. doi: 10.1002/j.1460-2075.1990.tb07904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BFC, Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 84.Hosaka H, Yao M, Kimura M, Tanaka I. The structure of the archaebacterial ribosomal protein S7 and its possible interaction with 16S rRNA. J Biochem. 2001;130:695–701. doi: 10.1093/oxfordjournals.jbchem.a003036. [DOI] [PubMed] [Google Scholar]
- 85.Petry S, Brodersen DE, Murphy FV, Dunham CM, Selmer M, Tarry MJ, Kelley AC, Ramakrishnan V. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell. 2005;123:1255–1266. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 86.Jenner L, Romby P, Rees B, Schulze-Briese C, Springer M, Ehresmann B, Moras D, Yusupova G, Yusupov M. Translational operator of mRNA on the ribosome: how repressor proteins exclude ribosome binding. Science. 2005;308:120–123. doi: 10.1126/science.1105639. [DOI] [PubMed] [Google Scholar]
- 87.Sen TZ, Feng Y, Garcia J, Kloczkowski A, Jernigan RL. The extent of cooperativity of protein motions observed with elastic network models is similar for atomic and coarser-grained models. J. Chem. Theory & Comput. 2006;2:696–704. doi: 10.1021/ct600060d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yan A, Wang Y, Kloczkowski A, Jernigan RL. Effects of protein subunits removal on the computed motions of partial 30S structures of the ribosome. J. Chem. Theory & Comput. 2008;4:1757–1767. doi: 10.1021/ct800223g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]