Abstract
Genome-wide association studies (GWAS) focus on relatively few highly significant loci while less attention is given to other genotyped markers. Employing pathway analysis to existing GWAS data may shed light on relevant biological processes, and illuminate new candidate genes. We employed a pathway-based approach to the breast cancer GWAS data of the National Cancer Institute (NCI) Cancer Genetic Markers of Susceptibility (CGEMS) project that includes 1145 cases and 1142 controls. Pathways were retrieved from three databases: KEGG, BioCarta, and the NCI’s Protein Interaction Database (PID). Genes were represented by their most strongly associated SNP, and an enrichment score (ES) reflecting the overrepresentation of gene-based association signals in each pathway was calculated using a weighted Kolmogorov-Smirnov procedure. Finally, hierarchical clustering was used to identify pathways with overlapping genes, and clusters with excess of association signals were determined by the adaptive rank-truncated product (ARTP) method. A total of 421 pathways containing 3962 genes were included in our study. Of these, three pathways (‘Syndecan-1-mediated signaling ‘, ‘Signaling of Hepatocyte Growth Factor Receptor’ and ‘Growth Hormone Signaling’) were highly enriched with association signals (PES < 0.001, False Discovery Rate (FDR) = 0.118). Our clustering analysis revealed that pathways containing key components of the RAS/RAF/MAPK canonical signaling cascade, were significantly more likely to have excess of association signals than expected by chance (PARTP = 0.0051, FDR = 0.07). These results suggest that genetic alterations associated with these three pathways and one canonical signaling cascade may contribute to breast cancer susceptibility.
Keywords: Pathways, GWAS, Breast cancer, Susceptibility, Genetics
Introduction
Breast cancer is a complex disease with a well established genetic component (1-4). Different genetic methodologies have been employed in the last twenty years to identify multiple susceptibility loci that vary in population frequency, relative risk and potential functional role. For example, familial linkage and positional cloning studies found that rare mutations in the genes BRCA1, BRCA2, TP53, and PTEN, are associated with a 10-fold to 20-fold increase in breast cancer risk (5-8). Variations in these genes are thought to contribute to breast cancer susceptibility through different cellular mechanisms. While BRCA1 and BRCA2 belong to the DNA repair mechanism in the cells (9, 10), TP53 and PTEN are tumor suppressor genes that participate in processes related to cell cycle control and cell proliferation (11, 12). Further interrogation of candidate genes associated with these cellular processes has led to the discovery of additional rare genetic variants conferring moderate relative risks (2-3 fold) of breast cancer (13-16).
Recently, Genome-wide association studies (GWAS) have become a key paradigm in genetic studies of complex diseases. These studies are particularly powerful in identifying common-low penetrance risk alleles. Indeed, several novel markers with low (< 2) relative risk for breast cancer have been detected by this approach (17-21). However, these reported loci are only those that reached a stringent statistical “genome-wide” significance criterion, while less attention has been given to the other hundreds of thousands of genotyped markers. Therefore, employing new methods to the existing GWAS data may provide additional biological insights and highlight new candidate loci. To this end, a pathway-based approach is particularly appealing. This method examines whether the cumulative contribution of genes with a common biological denominator is greater than expected by chance. This approach has recently been applied to GWAS of several non-cancer complex diseases (22-25).
In this study we applied pathway analysis to the breast cancer GWAS of the Cancer Genetic Markers of Susceptibility (CGEMS) project of the National Cancer Institute (NCI). This study has recently identified significant association of single-nucleotide polymorphisms (SNPs) in the FGFR2 gene with breast cancer susceptibility (18). We used the modified gene-set enrichment analysis (GSEA) of Wang et al. (22) to identify an excess of genotype-phenotype association signals in pathways from different resources. Finally, we examined whether the excess of association signals in different pathways is driven by the same subset of genes comprising a common biological module. These analyses illuminated three pathways and one canonical cascade that are possibly involved in genetic susceptibility to breast cancer.
Materials and Methods
Pathway data construction
We collected pathway data from three widely-used resources: the BioCarta pathway database (26), the Kyoto Encyclopedia of Genes and Genomes (KEGG) (27), and the NCI’s Protein Interaction Database (PID) (28). Genes belonging to these pathways were associated with SNPs included in the Illumina’s Sentrix HumanHap550 genotyping BeadChip that was used in the CGEMS GWAS. SNPs were mapped to genes if they were located within a genomic region encompassing 20kb 5′ upstream and 10kb 3′ downstream of the gene’s coding region (NCBI’s human genome build 36.3). Since FGFR2 is highly associated with breast cancer in CGEMS (18), we excluded all SNPs mapped to this gene from our analysis. Finally, we restricted our analysis to pathways with 10-100 genes so as to alleviate the multiple testing problem by avoiding testing too narrowly- or too broadly- defined functional categories. A complete list of the studied pathways is available in supplementary Table 1.
Gene-set enrichment analysis
The CGEMS breast cancer GWAS includes 1145 postmenopausal women of European ancestry with invasive breast cancer and 1142 controls. Associations between single nucleotide polymorphisms (SNPs) and breast cancer were determined using the Cochran-Armitage test for trend (29). Each gene Gj(j = 1, …, N, where N is the total number of gene in our dataset) was assigned the chi-square trend test statistic of the SNP with the highest statistical significance among all genotyped SNPs that mapped to its region (rj). Next, for each pathway (S) we ordered its gene’s test statistics (rj ∊ S) from highest to lowest, and used a weighted Kolmogorov-Smirnov procedure to calculate enrichment score (ES):
| (1) |
where WS = ΣGj*∈S|rj*| and NH is the number of genes in a pathway. This statistics reflects the relative overrepresentation of genotype-phenotype association signals in a particular set of genes (30). We used a permutation procedure (10,000 permutations, permuting the case-control status) to assess the significance of the pathway-based ES. This procedure generates a null distribution for the ES of each pathway based on its real genotype data and therefore accounts for the variable SNP count and the intrinsic correlation between SNPs within the different genes. Finally, a normalized enrichment score (NES) was calculated for each pathway in the observed and permutated data to allow a direct comparison of pathways of different sizes as suggested by Wang et al. (22). The calculation of the normalized enrichment score for each pathway NESS relied on the ESS, and the mean and standard deviation (SD) of in all permutations for a given pathway (S):
| (2) |
These data were then used to calculate a false discovery rate (FDR) (31) that controls for the proportion of expected false positive findings to be under a certain threshold. For a pathway with an NES value of NESS*, the FDR is calculated as:
| (3) |
Pathways with FDR < 0.2 were considered noteworthy.
Pathway overlap
To explore the relationships between pathways in our database, we defined the overlap between the set of genes in pathway A and the set of genes in pathway B as the percentage of genes common to both pathways (A and B) and calculated it as:
| (4) |
Next, we used a hierarchical clustering algorithm to assemble pathways with similar gene content. This algorithm iterates over all pathways, finds the most similar pair of pathways and groups them together. This process repeated until all pathways are grouped in one cluster in a hirarchial manner. To determine clusters of significantly related pathways, we used the same guidelines of the confidence interval algorithm originally developed for LD block determination in the genome described by Gabriel et al. (32). In short, clusters were defined as containing significantly related pathways if they included ≥ 5 pathways, and if ≥ 95% of all the pairwise overlap scores were greater than 9.6% (the 95th percentile of all pairwise overlap scores in our database).
Finally, we applied the ‘adaptive rank truncated product’ method (33) using the 10,000 permutated data sets, to assess whether pathways within the defined clusters had higher gene-set enrichment scores than expected by chance. This method assesses the null hypothesis that a cluster of pathways do not contain excess of pathways with high enrichment scores. Denote the ordered P-values of enrichment scores of pathways belonging to a particular cluster as (p1 ≤ … ≤ pL), with p1 being the smallest P-value. To test for the global null hypothesis, the ARTP procedure calculate the RTP statistics over all possible truncation points (k), and assess the significance of the best RTP using the permutation data.
Results
Overall, 421 pathways and 3962 genes (164/996, 155/3088 and 102/1317 pathways/genes from BioCarta, KEGG and PID respectively) were included in our study. These were represented by 69,525 of the 528,173 SNPs (13.2%) that were originally genotyped in this GWAS. A significant variation was seen between pathway resources. Over 90% of the pathways from BioCarta in our database had <30 genes, while pathways from KEGG and PID were generally larger and more variable in size (mean gene count = 44 and 36, SD = 24 and 16 respectively). This variation between databases was evident even in pathways nominally representing the same biological processes. For example, the mTOR signaling pathway included 19, 24 and 49 genes in BioCarta, PID and KEGG respectively. Interestingly, pathways from BioCarta tended to be ranked higher (higher enrichment scores) than KEGG pathways but not than PID pathways (Kruskal-Wallis test P = 0.013 and P = 0.151 respectively). No rank differences were seen between pathways from PID and KEGG.
Of the 421 examined pathways, twenty one were significantly enriched with association signals at the p-value < 0.05 level (Table 1). Of these, ‘Syndecan-1-mediated signaling events‘ from the PID database (Figure 1A; PES = 0.00053), ‘Signaling of Hepatocyte Growth Factor Receptor’ from BioCarta (Figure 1B; PES = 0.00063), and ‘Growth Hormone Signaling’ from BioCarta (Figure 1C; PES = 0.00084), had distinctly smaller p-values than the rest of the pathways, with a noteworthy False Discovery Rate (FDR) value of 0.118. For brevity, we will further refer to these three pathways as ‘syndecan-1 signaling’, ‘c-Met signaling’ and ‘GH signaling’ respectively.
Table 1.
A list of pathways associated with breast cancer susceptibility at the p ≤ 0.05 level
| Rank | Source | Pathway name | # genes | *PES | †FDR |
|---|---|---|---|---|---|
| 1 | PID | Syndecan-1-mediated signaling events | 13 | 0.00053 | 0.118 |
| 2 | BioCarta | Signaling of hepatocyte growth factor receptor | 33 | 0.00063 | 0.118 |
| 3 | BioCarta | Growth hormone signaling | 22 | 0.00084 | 0.118 |
| 4 | BioCarta | Insulin signaling | 19 | 0.00642 | 0.442 |
| 5 | KEGG | Basal cell carcinoma | 53 | 0.00674 | 0.487 |
| 6 | BioCarta | Eicosanoid metabolism | 19 | 0.00695 | 0.515 |
| 7 | PID | Signaling events mediated by Stem cell factor receptor (c-Kit) |
50 | 0.00779 | 0.515 |
| 8 | KEGG | Tryptophan metabolism | 57 | 0.01516 | 0.664 |
| 9 | BioCarta | ADP-ribosylation factor | 16 | 0.01800 | 0.842 |
| 10 | PID | FGF signaling pathway | 52 | 0.02189 | 0.871 |
| 11 | KEGG | ErbB signaling | 86 | 0.02379 | 0.871 |
| 12 | BioCarta | ALK in cardiac myocytes | 33 | 0.02747 | 0.871 |
| 13 | BioCarta | IL-2 receptor beta chain in T cell activation | 34 | 0.03389 | 0.881 |
| 14 | BioCarta | Intrinsic prothrombin activation | 16 | 0.03600 | 0.881 |
| 15 | KEGG | Streptomycin biosynthesis | 10 | 0.03600 | 0.881 |
| 16 | BioCarta | Angiotensin II mediated activation of JNK pathway via Pyk2 dependent signaling |
27 | 0.03663 | 0.881 |
| 17 | BioCarta | Sprouty regulation of tyrosine kinase signals | 15 | 0.04095 | 0.881 |
| 18 | KEGG | Maturity onset diabetes of the young | 24 | 0.04547 | 0.881 |
| 19 | BioCarta | Regulation of hematopoiesis by cytokines | 15 | 0.04568 | 0.881 |
| 20 | KEGG | Phosphatidylinositol signaling system | 75 | 0.04674 | 0.881 |
| 21 | KEGG | Melanogenesis | 98 | 0.04747 | 0.881 |
p-values are computed for the pathway’s enrichment scores (ES) using 10,000 permutations.
False discovery rate for the 421 pathways examined in this study.
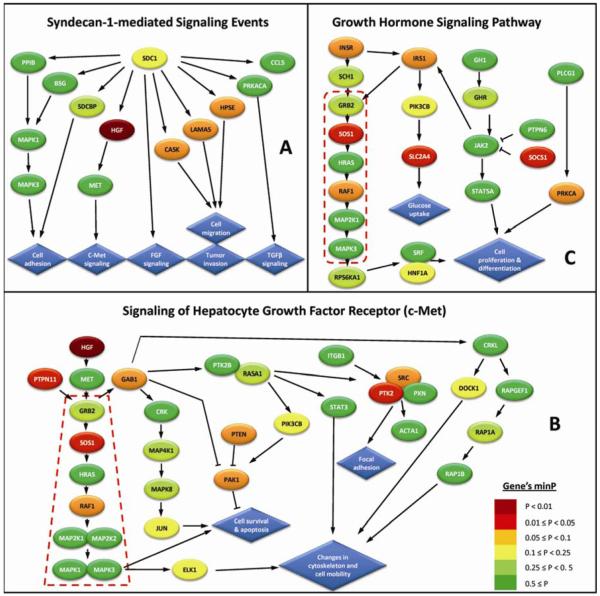
Figure 1.
Schematic illustration of the three top ranked pathways in this study.
‘Syndecan-1-mediated signaling events’ from PID; (B) ‘Signaling of hepatocyte growth factor (c-Met)’ from BioCarta; (C) ‘Growth hormone signaling pathways’ from BioCarta. Genes (ellipses) are color-coded according to their minP value that assesses the significance of the most strongly associated SNP in each gene, using a permutation-based resampling procedure (10,000 permutations) that takes into account the number of SNPs in a gene and their underlying linkage disequilibrium (LD) structure. Arrows indicate interaction or positive regulation and perpendicular lines indicate negative regulation. Blue diamonds represent the cellular processes induced by these pathways. Red broken boxes outline the genes of the RAS/RAF/MAPK canonical signaling cascade
Further examination of the gene content of these three pathways revealed some overlap. For example, the gene mitogen-activated protein kinase 3 [MAPK3] is involved in all three pathways, but does not appear to contribute to their significant enrichment scores due to its non-significant association with breast cancer risk in this study (p-value = 0.35). In contrast, the gene hepatocye growth factor [HGF] was the strongest associated gene in the ‘Syndecan-1 signaling‘ and the ‘c-Met signaling’ pathways (Ptrend = 0.0007, for rs975360) and therefore a major contributor for their high enrichment scores. Another two genes (mitogen-activated protein kinase 1 [MAPK1], and met proto-oncogene (hepatocyte growth factor receptor) [MET]) were shared by these two pathways. Similarly, 6 other genes (growth factor receptor-bound protein 2 [GRB2], v-Ha-ras Harvey rat sarcoma viral oncogene homolog [HRAS1], mitogen-activated protein kinase kinase 1 [MAP2K1], phosphoinositide-3-kinase, catalytic, beta polypeptide [PIK3CB], v-raf-1 murine leukemia viral oncogene homolog [RAF1] and son of sevenless homolog 1[SOS1]) were shared by the pathways: ‘c-Met signaling’ and ‘GH Signaling’.
In light of the considerable overlap between the three most significant pathways in our study, we hypothesized that common biological modules may underlie the enrichment signals in multiple biological pathways. To explore this hypothesis, we used hierarchical clustering to identify pathways with overlapping genes likely to constitute common biological process. Consequently, sixteen clusters of highly related pathways were characterized (Figure 2A). Of these, cluster C1 (Figure 2B) was significantly more likely to include pathways with high enrichment scores than expected by chance (adaptive rank-truncated product (ARTP) P = 0.0045, FDR = 0.07). Further examination of this cluster revealed that it contained 2/3 of the top three pathways (‘Signaling of Hepatocyte Growth Factor Receptor’, and ‘Growth Hormone Signaling’) and it was dominated by the genes GRB2, SOS1, HRAS, RAF1, MAP2K1, and MAPK3, which are major components of the RAS/RAF/MAPK canonical signaling cascade (Figures 1B,1C).
Figure 2.
Clustering analysis of pathway overlap. Pair-wise pathway overlap scores are depicted in a matrix color-coded in grayscale. (A) Clusters of highly significant related pathways are delineated by black triangles. (B) Details of the pathways included in cluster C1.
To assess the power of our analysis to identify true susceptibility pathways, we constructed an artificial positive control pathway from 11 genes that were previously associated with breast cancer risk in different GWAS of postmenopausal women of European ancestry (see details in Supplemental Table 2). Applying the pathway analysis to this pseudo-pathway result with a PES = 0.0126 that ranked it 8th out of 422 pathways. It is noteworthy that most of the individual genes in the pseudo pathway had unremarkable significance (Supplemental Table 2, last column) in the CGEMS study itself. Thus the high rank of this artificial positive control pathway demonstrates the ability of the gene-set enrichment analysis to detect pathways containing multiple genes that individually have weak association signals and may be missed by standard single-SNP analysis of GWAS data.
Discussion
Three pathways and one signaling cascade are highlighted in this study. The top ranked pathway in this study, ‘syndecan-1 signaling’, contains 13 genes involved in different cellular processes that are mediated by syndecan-1 [SDC1]. This gene encodes a transmembrane heparan sulfate proteoglycan which mediates signal transduction cascades leading to cell proliferation, cell migration and cell adhesion processes following interactions with extracellular matrix proteins. There are multiple lines of evidence for a potential role of syndecan-1 in breast cancer development. For example, altered syndecan-1 expression has been detected in several different tumor types and has been linked with unfavourable breast cancer prognosis (34). Additionally, expression of syndecan-1 by stromal fibroblasts, has been shown to promote breast carcinoma growth in vivo and stimulates tumor angiogenesis (35). In this GWAS, SDC1 was only moderately associated with breast cancer susceptibility (Ptrend = 0.019 for rs7563245) however, it is a key mediator of different pathways illuminated in this study (e.g. ‘c-Met Signaling’ and ‘Fibroblast Growth Factor Signaling’; see discussion below). Therefore, it can modulate breast cancer susceptibility through different biological mechanisms.
The second highest ranked pathway in our analysis, ‘c-Met signaling’ consists of 33 genes participating in signal transduction mechanisms induced by the tyrosine-kinase proto-oncogene c-Met [MET]. Stimulation of the c-Met pathway can lead to several different cellular processes related to tumor growth and progression such as proliferation, enhanced cell motility, invasion, and apoptosis (36). Moreover, both c-Met and its ligand, hepatocyte growth factor (HGF), have been shown to be dysregulated and correlated with poor prognosis in a number of human malignancies including breast cancer (37). Consequently, this pathway has served as an important therapeutic target for human cancers, particularly through the development of small-molecule that inhibit the c-Met/HGF-dependent signaling (37). Notably, co-expression of SDC1 and MET, the key players in the top two ranked pathways in our study, have been established as a marker signature associated with poor prognosic factors in ductal carcinoma in situ (DCIS) of the breast (38).
The third ranked pathway in our study, ‘GH Signaling’, contains 22 genes participating in cellular mechanisms induced by either growth hormone or insulin receptors. These two receptors as well as the insulin-like growth factor (IGF) receptor are all transmembrane tyrosine kinase receptors inducing cell growth and proliferation. Alteration in the activity of these receptors or their related pathways may lead to hyperplasia and eventually to the development of a tumor (39). Naturally, the ‘GH signaling’ pathway, had considerable overlap with both the ‘IGF-1 signaling’ and ‘insulin signaling’ pathways from BioCarta (32% and 41% respectively), however only the latter had a significant enrichment score in our study (PES = 0.0064). Examining the genes of these three closely related pathways revealed that what differentiates their respective enrichment scores is a SNP in the insulin receptor gene [INSR] with a small p-value (Ptrend = 0.0019 for rs12460755) that is absent from the ‘IGF-1 signaling pathway’.
An interesting pathway related to syndecan-1 signaling is the ‘fibroblast growth factor (FGF) signaling’. This pathway was ranked 10th in our study (Table 1; PES = 0.0219) in spite of the exclusion of the FGFR2 SNPs from our analyses (see Methods). Adding the FGFR2 SNPs to this pathway improved its enrichment signal (PES = 0.0053) that ranked it 4th out of the 421 pathways. This finding in combination with the FGFR2 signal in CEGEMS (18) and elsewhere (17) suggests that variations in other genes involved in FGFR2 signaling may modulate breast cancer susceptibility. An important extension to the gene-set enrichment analysis the clustering analysis that highlighted the RAS/RAF/MAPK canonical signaling cascade as the common denominator of pathways associated with breast cancer risk in this study. This cascade plays an essential role in transmitting extracellular signals from growth factors to promote the growth, proliferation, differentiation and survival of cells, and modification in its activity has been linked to multiple human malignancies (40). Notably, this cascade plays an important role in all three top pathways in this study. It is an integral component in the ‘Signaling of Hepatocyte Growth Factor Receptor’, and ‘Growth Hormone Signaling’ pathways, and a transducer for many of the signals initiated by the Syndecan-1 pathway.
An important limitation of the pathway-based approach for GWAS analysis is the incomplete annotation of the human genome. At present, the function of many human genes is unknown and therefore these genes cannot be assigned to known pathways. Moreover, susceptible loci in intergenic regions are also not included in a study of this kind. As a result, when employing this approach, only a small portion of the human genome variation can be studied and therefore it should only be used as a supplementary method to the standard single-SNP analysis of GWAS. Additionally, there is no gold standard for pathway definition, and different databases have different guidelines for their pathways construction and curation. Consequently, the gene content of pathways representing the same biological process may vary substantially between different databases, and this may have a major impact on the sensitivity and specificity of this approach. We aimed at minimizing this effect by selecting pathways from three commonly used and manually curate resources. Still, considerable differences were observed between similar pathways from different resources. For example, the ‘c-Met signaling’ pathway from BioCarta contained 33 genes and was ranked second in our analysis, while a pathway with the same name and presumably the same cellular function in the PID database included 52 genes but was ranked only 69th. Although 30 of the 33 genes in the BioCarta pathway were also included in the PID pathway, the remaining 22 genes likely attenuated the signal in the PID pathway. These differences emphasize the importance of further synthesizing the results of such pathway-based approach. The clustering analysis we applied in this study is one way to do so, as it helps in finding the common genes underlying enrichment signals in multiple pathways and allows one to focus on a limited number of candidate genes. Other methods aiming to improve the characterization of pathways and their overlap might be useful.
A second limitation of our study is that it does not include validation of the results using a completely independent dataset. The experiment we conducted using a positive-control-pathway with known breast cancer susceptibility genes provides us validation that our approach has high power to detect pathways containing multiple weakly associated susceptibility genes. Therefore, the top ranked pathways in our study should be prime targets for future analyses in independent datasets.
In conclusion, our results suggest that genetic alterations associated with the top three pathways and one canonical signaling cascade in our study may contribute to breast cancer susceptibility. Ultimately, additional studies would be needed to confirm and further explore the genetic variations underlying the association of these pathways with breast cancer. Moreover, this study highlights the potential insight that could be gained by pathway-based approach as a complimentary method to the primary single-SNP analysis of GWAS. Particularly, the organization of multiple association signals according to underlying biological processes may improve our understanding about the cellular mechanisms underlying this too common malignancy.
Supplementary Material
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH/National Cancer Institute.
Footnotes
The authors have no conflicts to disclose.
References
- 1.Cancer CGoHFiB Familial breast cancer: collaborative reanalysis of individual data from 52 epidemiological studies including 58,209 women with breast cancer and 101,986 women without the disease. Lancet. 2001;358:1389–99. doi: 10.1016/S0140-6736(01)06524-2. [DOI] [PubMed] [Google Scholar]
- 2.Ahlbom A, Lichtenstein P, Malmstrom H, Feychting M, Hemminki K, Pedersen NL. Cancer in twins: genetic and nongenetic familial risk factors. J Natl Cancer Inst. 1997;89:287–93. doi: 10.1093/jnci/89.4.287. [DOI] [PubMed] [Google Scholar]
- 3.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 4.Peto J, Mack TM. High constant incidence in twins and other relatives of women with breast cancer. Nature genetics. 2000;26:411–4. doi: 10.1038/82533. [DOI] [PubMed] [Google Scholar]
- 5.Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 6.Wooster R, Bignell G, Lancaster J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–92. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 7.Malkin D, Li FP, Strong LC, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–8. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 8.Nelen MR, Padberg GW, Peeters EA, et al. Localization of the gene for Cowden disease to chromosome 10q22-23. Nature genetics. 1996;13:114–6. doi: 10.1038/ng0596-114. [DOI] [PubMed] [Google Scholar]
- 9.Jasin M. Homologous repair of DNA damage and tumorigenesis: the BRCA connection. Oncogene. 2002;21:8981–93. doi: 10.1038/sj.onc.1206176. [DOI] [PubMed] [Google Scholar]
- 10.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–82. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 11.Tokino T, Nakamura Y. The role of p53-target genes in human cancer. Crit Rev Onco Hematol. 2000;33:1–6. doi: 10.1016/s1040-8428(99)00051-7. [DOI] [PubMed] [Google Scholar]
- 12.Keniry M, Parsons R. The role of PTEN signaling perturbations in cancer and in targeted therapy. Oncogene. 2008;27:5477–85. doi: 10.1038/onc.2008.248. [DOI] [PubMed] [Google Scholar]
- 13.Meijers-Heijboer H, van den Ouweland A, Klijn J, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nature genetics. 2002;31:55–9. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 14.Rahman N, Seal S, Thompson D, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nature genetics. 2007;39:165–7. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renwick A, Thompson D, Seal S, et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nature genetics. 2006;38:873–5. doi: 10.1038/ng1837. [DOI] [PubMed] [Google Scholar]
- 16.Seal S, Thompson D, Renwick A, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nature genetics. 2006;38:1239–41. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 17.Easton DF, Pooley KA, Dunning AM, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunter DJ, Kraft P, Jacobs KB, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nature genetics. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gold B, Kirchhoff T, Stefanov S, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci U S A. 2008;105:4340–5. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng W, Long J, Gao YT, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nature genetics. 2009;41:324–8. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas G, Jacobs KB, Kraft P, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nature genetics. 2009;41:579–84. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K, Li M, Bucan M. Pathway-Based Approaches for Analysis of Genomewide Association Studies. Am J Hum Genet. 2007;81:1278–83. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K, Zhang H, Kugathasan S, et al. Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn Disease. Am J Hum Genet. 2009;84:399–405. doi: 10.1016/j.ajhg.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baranzini SE, Galwey NW, Wang J, et al. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Human molecular genetics. 2009;18:2078–90. doi: 10.1093/hmg/ddp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elbers CC, van Eijk KR, Franke L, et al. Using genome-wide pathway analysis to unravel the etiology of complex diseases. Genet Epidemiol. 2009;33:419–31. doi: 10.1002/gepi.20395. [DOI] [PubMed] [Google Scholar]
- 26.BioCarta cited; Available from: http://www.biocarta.com/genes/allpathways.asp.
- 27.KEGG. Kyoto Encyclopedia of Genes and Genomes.
- 28.PID. Pathway Interaction Database.
- 29.Armitage P. Tests for Linear Trends in Proportions and Frequencies. Biometrics. 1955;11:375–86. [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–75. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 32.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 33.Yu K, Li Q, Bergen AW, et al. Pathway analysis by adaptive combination of P-values. Genet Epidemiol. 2009 doi: 10.1002/gepi.20422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leivonen M, Lundin J, Nordling S, von Boguslawski K, Haglund C. Prognostic value of syndecan-1 expression in breast cancer. Oncology. 2004;67:11–8. doi: 10.1159/000080280. [DOI] [PubMed] [Google Scholar]
- 35.Maeda T, Desouky J, Friedl A. Syndecan-1 expression by stromal fibroblasts promotes breast carcinoma growth in vivo and stimulates tumor angiogenesis. Oncogene. 2006;25:1408–12. doi: 10.1038/sj.onc.1209168. [DOI] [PubMed] [Google Scholar]
- 36.Maulik G, Shrikhande A, Kijima T, Ma PC, Morrison PT, Salgia R. Role of the hepatocyte growth factor receptor, c-Met, in oncogenesis and potential for therapeutic inhibition. Cytokine Growth Factor Rev. 2002;13:41–59. doi: 10.1016/s1359-6101(01)00029-6. [DOI] [PubMed] [Google Scholar]
- 37.Christensen JG, Burrows J, Salgia R. c-Met as a target for human cancer and characterization of inhibitors for therapeutic intervention. Cancer Lett. 2005;225:1–26. doi: 10.1016/j.canlet.2004.09.044. [DOI] [PubMed] [Google Scholar]
- 38.Gotte M, Kersting C, Radke I, Kiesel L, Wulfing P. An expression signature of syndecan-1 (CD138), E-cadherin and c-met is associated with factors of angiogenesis and lymphangiogenesis in ductal breast carcinoma in situ. Breast Cancer Res. 2007;9:R8. doi: 10.1186/bcr1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner K, Hemminki K, Forsti A. The GH1/IGF-1 axis polymorphisms and their impact on breast cancer development. Breast Cancer Res Treat. 2007;104:233–48. doi: 10.1007/s10549-006-9411-9. [DOI] [PubMed] [Google Scholar]
- 40.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.