Abstract
Digital brain atlas is a kind of image database that specifically provide information about neurons and glial cells in the brain. It has various advantages that are unmatched by conventional paper-based atlases. Such advantages, however, may become disadvantages if appropriate cares are not taken. Because digital atlases can provide unlimited amount of data, they should be designed to minimize redundancy and keep consistency of the records that may be added incrementally by different staffs. The fact that digital atlases can easily be revised necessitates a system to assure that users can access previous versions that might have been cited in papers at a particular period. To inherit our knowledge to our descendants, such databases should be maintained for a very long period, well over 100 years, like printed books and papers. Technical and organizational measures to enable long-term archive should be considered seriously. Compared to the initial development of the database, subsequent efforts to increase the quality and quantity of its contents are not regarded highly, because such tasks do not materialize in the form of publications. This fact strongly discourages continuous expansion of, and external contributions to, the digital atlases after its initial launch. To solve these problems, the role of the biocurators is vital. Appreciation of the scientific achievements of the people who do not write papers, and establishment of the secure academic career path for them, are indispensable for recruiting talents for this very important job.
Keywords: brain, atlas, image database, molecular marker, neuron, curator
Introduction
Atlases of the brain provide images and information about neurons and glial cells, brain regions, fascicles, and arborizations, transmitters and other brain-related chemicals, antibodies, and strains for labeling specific cell types, known functions, and developmental origins of the neurons and brain regions, etc. The term “digital brain atlas” has been used for referring to several kinds of neuroanatomical projects. In the simplest meaning, it refers to the electronic version of the conventional atlases that have been published in the form of books. More sophisticated digital atlases feature computer-based three-dimensional visualization systems to generate maps of the brain, which are used as tools and platforms for analyzing neurons and brain structures (Toga and Thompson, 2001; Rein et al., 2002; Van Essen, 2002; Brandt et al., 2005; Kurylas et al., 2008; El Jundi et al., 2009a,b; Kvello et al., 2009; Dreyer et al., 2010; Huetteroth et al., 2010; Jahrling et al., 2010; Lofaldli et al., 2010; Peng et al., 2010).
Digital atlases are not only useful for conducting researches to be published in the form of research articles. Many digital atlases serve as publicly accessible information resources, in which data are stored in host server computers and provided over the Internet upon users’ request. Such atlases can be regarded as a specific kind of image databases, and the number of such online atlases is increasing (Table 1). They will become useful platforms for providing comprehensive information about the entire projection patterns of all the neurons – called the projectome (Kasthuri and Lichtman, 2007), and connectivity data of all the neurons – called the connectome (Sporns et al., 2005; Seung, 2009).
Table 1.
Examples of web-based digital atlases.
| Vertebrates | ||||
|---|---|---|---|---|
| Humans | The Human Brain Atlas | https://www.msu.edu/∼brains/brains/human/ | ||
| The Whole Brain Atlas | http://www.med.harvard.edu/AANLIB/home.html | |||
| The HumanBrain.Info | http://www.thehumanbrain.info/ | |||
| Monkeys | Marmoset Brain Atlas | http://marmoset-brain.org:2008/ (Tokuno et al., 2009) | ||
| Mice | The Allen Brain Atlas | http://www.brain-map.org/ (Ma et al., 2005; Lein et al., 2007) | ||
| Blue Brain Project | http://bluebrain.epfl.ch/ (Markram, 2006) | |||
| Birds | AvianBrain.org | http://avianbrain.org/atlases.html | ||
| Avian Brain Circuitry Database | http://www.behav.org/abcd/ | |||
| Zebra Finch MRI Atlas | http://webh01.ua.ac.be/biomag/zebrafinch_mri_atlas.htm (Poirier et al., 2008) | |||
| Fish | Zebrafish Atlas | http://zfatlas.psu.edu/index.php | ||
| VARIOUS ORGANISMS INCLUDING THE ABOVE | ||||
| The Brainmaps | http://brainmaps.org/ (Mikula et al., 2008) | |||
| Comparative Mammalian Brain Collections | http://www.brainmuseum.org/ | |||
| INVERTEBRATES | ||||
| Flies | Flybrain | http://www.flybrain.org/ (Armstrong et al., 1995) | ||
| Flytrap | http://www.fly-trap.org/ (Armstrong and van Hemert, 2009) | |||
| Virtual Insect Brain Lab for Drosophila Standard Brain | http://132.187.25.13/ (Rein et al., 2002) | |||
| Flybrain at Stanford | http://flybrain.stanford.edu/ (Jefferis et al., 2007) | |||
| Bees | The Virtual Atlas of the Honeybee Brain | http://www.neurobiologie.fu-berlin.de/beebrain/ (Brandt et al., 2005) | ||
| Moths | Moth Standard Brain | http://www.ntnu.no/biolog/english/neuroscience/brain/ (Kvello et al., 2009) | ||
| Manduca Standard brain | http://online-media.uni-marburg.de/biologie/3d_brain/manduca/standardbrain/ (El Jundi et al., 2009b) | |||
| Locusts | Schistocerca Standard Brain | http://online-media.uni-marburg.de/biologie/3d_brain/schistocerca/ (Kurylas et al., 2008) | ||
| Nematodes | Wormatlas | http://www.wormatlas.org/ | ||
| Database of Synaptic Connectivity of C. elegans for Computation | http://ims.dse.ibaraki.ac.jp/ccep/ | |||
| INDIVIDUAL IDENTIFIED NEURONS IN THE BRAIN | ||||
| The NeuroMorpho.org | http://neuromorpho.org/ (Ascoli et al., 2007; Halavi et al., 2008) | |||
| Neuron Bank | http://neuronbank.org/ (Katz et al., 2010) | |||
| Flybrain Neuron Database | http://ndb.flybrain.org/ (Shinomiya et al., in preparation) | |||
Atlases of the brain have traditionally been provided as printed books (Strausfeld, 1976; Wullimann et al., 1996; Valverde, 1998; Woolsey et al., 2003; Mai et al., 2004; Paxinos and Watson, 2005; Puelles et al., 2007; Franklin and Paxinos, 2008; Schambra, 2008). Compared to web-based digital atlases, conventional paper-based atlases still have several advantages. First, it is easier to browse high-resolution images. Whereas even the best computer monitor can display images at only less than 110 dots per inch (DPI), typical commercial printing offer the resolution of 350 DPI for photographs and 2,540 DPI for text and line drawings. A page of a printed atlas can present several high-resolution images and detailed text explanation at the same time. On the other hand, even a large computer monitor can display only a single high-resolution image. To view more than one full-resolution image or to display an image and its complete text explanation, users have to scroll the screen or switch between different windows, which significantly decreases readability.
And second, a paper-based atlas has a much more reliable value as an archive. Because the hard copies of the atlas are stored in public libraries, they will be accessible for a long time in the future even if the authors would die, the books would become out of press, or the publishers may go out of business. Web-based atlases, on the other hand, are highly unstable. Useful web sites may disappear when the person who established the server moves or retires, or the financial support that covers its operational cost is cut out. Even during the period when the database is maintained actively, some of the contents may be revised or even deleted in the future so that original version becomes inaccessible.
In spite of these shortcomings, computer-based digital atlases gain increasingly popular support, because they have also various advantages that are unmatched by conventional printed atlases. First, digital atlases can in principle provide much more data. Whereas the amount of information that can be put in a printed atlas is limited by the practical size and page number of a book, the volume of a digital atlas is limited only by the size of the hard disk storage, which is practically infinite (Howe et al., 2008; Van Horn and Toga, 2009).
Second, computer-based approach offers more flexible ways of navigation. Though paper-based atlases are easier to read through, locating pages that mention a particular item is difficult, especially if it is not listed in the index. Digital atlases equipped with adequate search engines enable users to locate whatever terms they are interested in. In addition, hyperlinks provide a convenient way to jump to other parts of the atlas that offer relevant information.
Third, digital atlases can present images in more versatile ways. Unlike printed atlases, they can provide not only still images but also movies and interactive images, with which users may view particular parts of the brain sections of different samples at various depths, or watch three-dimensional structure of neurons from different viewing angles with various visualization parameters. Combined with hyperlinks embedded in the graphics, interactive images offer easy-to-use navigation that is useful especially for the users who are not familiar with brain anatomy.
And fourth, contents of the digital atlases can be updated dynamically. Paper-based atlases remain stable once they are published. Though this ensures their value as archives, some of the information may become obsolete as the time goes by. Because of the relatively limited readership and the high cost of printing, it is financially not feasible to publish revised editions at regular intervals except for the atlases of human and a few clinical model animals. On the contrary, revision is very easy with computer-based atlases. The first version of an atlas could be started with a minimum amount of contents just to present the outline of the brain structure of that organism, and new data will be added incrementally thereafter. This is especially suited for modern molecular-oriented neuroanatomy, where new techniques for visualizing neurons are being developed rapidly.
These advantages, however, mean that people working for a digital atlas have to be engaged in the project for a much longer period than those who write a printed atlas. Though it may take several years to prepare for the figures and manuscripts of a paper-based atlas, authors are freed from the tasks for its distribution and archival once it is published. Publishers and libraries will take care of these issues. On the other hand, people who develop a digital atlas are responsible for its maintenance, archival, and regular updates for tens of years after it is first established. There are various technical and organizational issues that are keys to the steady long-term maintenance and development of the web-based databases. Compared to the technical issues for developing novel visualization and navigation tools, however, such organizational issues are not discussed very often.
The Flybrain1 is one of the first digital brain atlases operating on the Internet. It was established in 1995 by a consortium of a few laboratories working on the Drosophila brain (Armstrong et al., 1995). My laboratory joined soon after its establishment as one of the organizers and is contributing to its maintenance since then. Based on this 15-year experience, in this semi-review paper I will discuss various problems that may affect the development of digital atlases and web-based databases after it is first established.
Documenting the positions in the brain
In the first seven sections I will discuss issues concerning the design of the digital atlas that is easy to maintain and expand. A digital atlas is in a manner similar to the databases of genes and proteins, because they all provide comprehensive information about important aspects of an organism. Describing the structure of a gene or a protein is easy, because it is essentially a one-dimensional chain of clearly defined units (four nucleotides or 20 amino-acid residues, respectively). Three-dimensional morphology of a protein can also be documented unambiguously using the coordinates of each atom. Documenting the structure of a neuron is much more demanding, because it has a complex three-dimensional architecture spanning various parts of the brain. The location of the cell body, trajectories, and branching points of the neural fibers, and areas of dendritic and terminal arborizations could in principle be documented using the coordinates of the labeled areas (pixels/voxels) of the image of the neuron. However, because of the inter-individual variability in the overall size, shape, and relative positions of brain regions, coordinates of a corresponding part of the brain may not be identical from sample to sample, making simple coordinates useless for describing the positions in the brain.
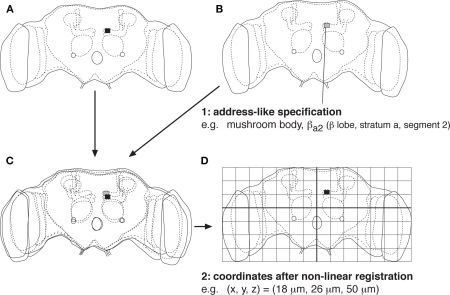
There have been two approaches to address this issue. One approach is to divide the brain into morphologically and functionally meaningful regions, such as cortical areas and nuclei in the mammalian brain and neuropils of the insect brain. Each region can further be subdivided into smaller parts, such as glomeruli in the insect antennal lobe (Laissue et al., 1999), layers in the optic lobe neuropils (Fischbach and Dittrich, 1989; Otsuna and Ito, 2006), and strata and segments in the central complex and mushroom body lobes (Hanesch et al., 1989; Tanaka et al., 2008). The names of these subregions can be used as unambiguous descriptors of the neural position, because such subregions should exist in all the brain samples even if their respective locations may slightly vary (Figures 1A,B).
Figure 1.
Specification of the position in the brain. (A,B) Illustrations of two brain samples, whose shape are slightly different because of individual variability. Position in the brain can nevertheless be specified by the area of the brain region and distinct subregions within it. An example is shown for a particular subregion of the mushroom body lobe of the Drosophila brain (Tanaka et al., 2008). (C) Comparison of the two brain samples after linear affine transformation. Best match cannot be attained, because individual brain region may have slightly different size, shape, and orientation. (D) Comparison of the two brain samples after non-linear registration (so-called morphing or warping). All the brain samples can be fit into a standardized framework, in which the position in the brain can be specified using coordinates.
Another approach is to standardize the shape of each dataset of the brain into a standard framework using a computer program. In the simplest form, the width, height, thickness, and axes of the brain samples are transformed using linear expansion and rotation. Coordinates of all the parts of the brain cannot usually be matched with such simple affine transformation, however, because some parts of a brain sample may be twisted or disproportionally larger or smaller than the corresponding structure in other samples (Figure 1C). Non-linear registration, so-called morphing or warping, is necessary to attain a better match (Tanaka et al., 2004; Guetat et al., 2006; Jenett et al., 2006; Jefferis et al., 2007; Kurylas et al., 2008; Yap et al., 2009). Because all the brain samples can be fit into a standard shape, positions in the brain can be described using the coordinates in the standardized brain (Figure 1D).
These two approaches are comparable to the two ways we use for specifying the location on the earth, using either address (name of the country, state, city, and street) or latitude/longitude coordinates. The former address-like specification system is more intuitive and convenient for text-based search: Users can easily recognize the position of the brain from the region name, and all the neurons that project to a particular area can be looked up by simply using the region name as the search key. The latter coordinate-based specification is less intuitive for humans but advantageous for quantitative analyses: The length of the neural fibers, distance, and potential contact between two neurons, etc., can be calculated using the standardized coordinates. The resolution of the positional specification is higher, because fine differences within a defined brain subregion can be distinguished by the coordinate values. The two specification systems are therefore complementary. Ideally, documentation in both ways should better be provided in a digital atlas.
A potential problem of the address-like approach is that there has been some inconsistency in the names of the brain regions: Apparently comparable brain regions are given different names depending on the researchers and species. In addition, the areas of the brain that have not been attracted extensive research were not given clearly defined names and boundaries. Discrepancy in the nomenclature of the brains of various avian species has been resolved by a proposal of a coordinated naming scheme (Reiner et al., 2004a,b)2. A similar attempt is going way for the insect brain; a working group of neuroanatomists of various insect species is discussing the issue, and the controlled nomenclature system and the definition of neuropil boundaries using the Drosophila brain as a model will be proposed in the near future, which is going to be adopted by various databases of the fly brain. Such a system will enable the description of the locations of the cell bodies, branching points of neural fibers, and synaptic arborizations in a consistent manner, making it easier to compile information reported in various papers by various researchers.
Keeping consistency of the information about neurons, images, and molecular markers
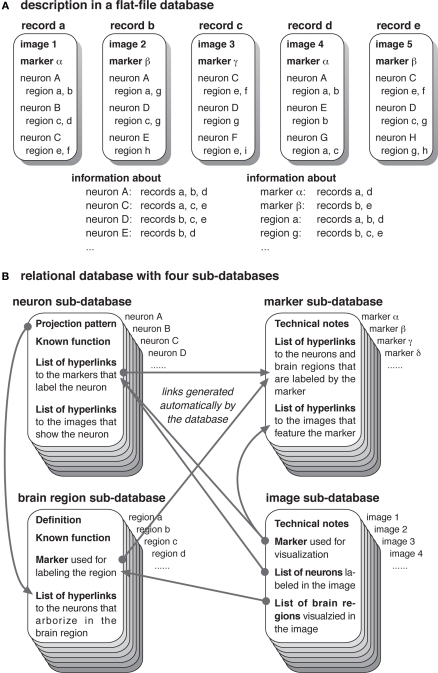
An atlas usually provides two types of images. The overall architecture of the brain will be presented with the serial sections of the entire brain, visualized with histological labeling methods such as Bodian, Ethylgallate, Nissl, and hematoxylin–eosin stain or with antibodies against ubiquitous molecules associated with synapses, membranes, cytoskeletons, etc. The structure of specific types of neurons and glial cells will be shown with the images visualized with various techniques including Golgi impregnation, dye filling, cell-specific antibodies, and transgenic expression driver strains. It is important to note that multiple types of neurons tend to be labeled in a single image, and that a single neuron type may be visualized in several images (Figure 2A). Thus, the relationship between neurons and the images in which they are presented is not one-to-one but many-to-many. The texts that describe the structure of a particular neuron would therefore appear in multiple parts of the atlas.
Figure 2.
Composition of the records for ensuring consistency. (A) A schematic example of a typical image database. Each record presents an image, or a set of images, visualizing particular groups of neurons with a particular molecular marker, showing particular regions of the brain. Different images may visualize the same neuron type with different combinations, and the same molecular marker is used for visualizing multiple types of neurons. As each record describes particular area of the brain, only a subset of the arborization targets of a neuron may be mentioned. Thus, description of a specific neuron type, marker, or brain region is scattered in the database, causing redundancy and possible inconsistency. (B) An example of a relational database with the sub-databases dedicated for the records of neurons, markers, brain regions, and images. Information about the complete projection patterns of the neurons, definition, and known functions of the brain regions, and technical details of the molecular markers and image preparation, are documented in respective records. When a new record is added, respective information in other records will automatically be updated.
In addition, a molecular marker like an antibody or an expression driver often labels more than one type of neuron in the brain. Thus, a particular marker is likely to be mentioned in multiple parts of the atlas. Each part of the atlas describes a distinct subset of the neurons labeled by the marker, without mentioning the labeling patterns in other neurons (Figure 2A). A comprehensive list of all the neurons that are labeled by that marker is not directly available in many cases. Likewise, a particular brain region receives projections from many neurons and is visualized by various molecular markers. Thus, many parts of the atlas mention a particular brain region, while none of them provides comprehensive information about all the neurons that project to the area.
In case of paper-based atlases, only a few images are usually presented to describe each neuron type, molecular marker, and brain region because of the space limitation of a book. This has effectively reduced the problem of redundancy. Because there is no limiting factor to restrict the overall amount of database records, on the contrary, digital atlases may contain much more data that describe a particular neuron, marker, and brain region than paper-based atlases do. The number of such records, and the number of the texts that describe various aspects of the same neurons, markers, and brain regions is likely to increase over time as new data are added.
It may seem easy to keep track of such distributed and redundant information in digital atlases, because electronic text search systems will provide the list of all the records that mention a particular item. However, this approach runs into difficulty as the number of records increases. Because (1) different images reveal different aspects of the same neuron, (2) a molecular marker is utilized in a particular record to describe only a small subset of the neurons it labels, and (3) a brain region is mentioned in the context of diverse neurons and molecular markers, each record is likely to lack certain information that is described in various other records of the database. Though users may be able to search all the records that mention particular neuron, marker, and brain region, comprehensive information can only be obtained by reading through all these documents scattered in the database, many of which may partially be redundant. This is not very efficient.
Moreover, having such redundancy increases the likelihood of inconsistent documentation between records. Images of a single neuron, marker, or brain region may appear slightly differently in different samples, either by technical differences or by inter-individual variations. Developmental and experience-dependent plasticity would also affect the appearance of the same neuron in different data (Livneh and Mizrahi, 2010). Because explanatory texts of different records may be written at different times and possibly by different staffs of the database organization, documentation in some records might become inconsistent with those in other records. Users are faced to resolve such contradicting information.
To avoid such situation, a database should feature a system to help keeping the consistency of neuron, marker, and brain region data. This is best achieved by making a relational database that keeps description of each neuron type, molecular marker, brain region, and image separately (Figure 2B). A relational database consists of a set of sub-databases each of which keeps records of particular items like neurons, markers, brain regions, and images. Relevant data field in one sub-database is linked (or related) with the corresponding field in another sub-database, providing hyperlinks between the information provided in each sub-database.
In this approach, each record of the image sub-database will provide information about its technical feature, the marker used for visualization, and a list of all the neuron types that are shown in the image. Relevant images, such as those of the same brain sample before and after registration described in the previous section (Figure 1), and still images and movies generated from the same three-dimensional dataset, will be kept in separate records of the sub-database and mutually linked.
Description of each neuron type, such as the location of the cell bodies, projection targets, brain regions in which pre- and postsynaptic sites are distributed, physiological properties such as transmitters and electrophysiological responses to stimuli, and known functions in behavioral experiments, will be provided in a single record of the neuron sub-database (Figure 2B). A record in the marker sub-database will provide its technical specification such as antigens of the antibody or genetic characteristics of the expression driver strain. A record in the brain region sub-database will explain the definition of the region boundary, methods to visualize the area, general architecture of its neural circuits, and known functions. Hyperlinks generated by the relational database engine will provide the list of all the neurons and images that feature a particular marker, the list of all the images and markers that visualize a particular neuron type and brain region, as well as the list of all the neurons that arborize in a particular brain region. These lists will be updated automatically whenever new records are added.
Such separation of neuron, marker, brain region, and image data enables keeping all the information of a particular neuron type, marker, and brain region in a single place of the database, avoiding redundancy, and possible inconsistency. When writing descriptive documentation of a new image data, novel information about the neurons visualized in the newly added image should be appended to the documentation of the neuron record rather than noted sporadically in the image records. By doing so, it becomes easy for the database organizers to check whether there is any inconsistency between the description in the neuron record and the structure visualized in the new image data. This approach also enables users and database organizers to keep consistency of the information provided. Because of the automatic hyperlinks between relational sub-databases, documentation of the neurons labeled with a particular molecular marker will automatically be reflected in the list of the labeled cells in the marker sub-database, and addition of new data about the arborization areas of a particular neuron will automatically update the list of contributing neurons in the respective record of the brain region sub-database.
Keeping track of revisions
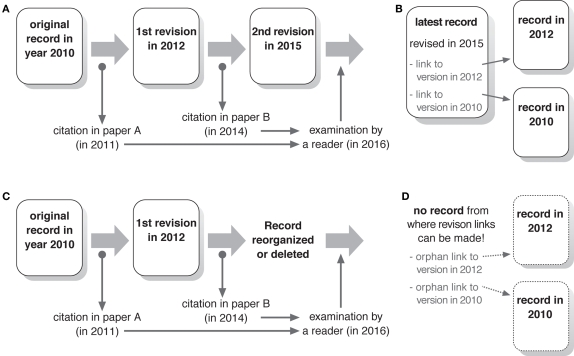
As mentioned earlier, the fact that it is easy to revise contents of digital atlases can be regarded as both advantage and disadvantage. There are two occasions when a digital atlas is cited in literature. First, the existence of the digital atlas itself may be cited, just like citing a paper-based atlas in its entirety. Second, information provided in a particular part of the database may be cited, just like citing a page or a chapter of a book atlas. In the latter case, it is important that the readers of the literature can access the same information as the author had obtained when she/he wrote it. Though revisions of the digital atlases enable incorporation of latest observations and correction of inadequate descriptions, they would modify, or erase, certain contents that users of the database may have cited in their literature. In case of books, older editions remain available in the library and are therefore citable, even if some of the contents are revised in the following editions. In the digital database, however, previous versions of the documents usually become inaccessible from the database server.
This causes a serious problem as the reliability of digital atlases as reference sources. If a user of the database cites a document in the database in her/his paper, and if the document is revised after the paper is published, readers of the paper can no longer access the document in the form it was cited (Figure 3A). It would especially cause a trouble, if the revision involves correction or deletion of a particular aspect of the documentation that the user wanted to cite. For example, a neuron may be documented as excitatory in a digital atlas, and later it turned out to be wrong and the digital atlas organizer revised the relevant documentation to be inhibitory. If a user of the atlas cites the document before this revision, and a reader of the literature accesses the atlas after the revision, confusion should occur. Because scientific papers rely on the references that readers can examine by themselves, digital atlases should consider that the previous versions of the documents should remain accessible after they are revised.
Figure 3.
Importance of the revision tracking system. (A) A schematic example of the citation of the database at different periods. Two papers refer to the same database record at different periods before and after revision. This may cause inconsistency between what are cited. If a reader of these papers examines the database at a later period, the person may not be able to get the same information cited in these papers. (B) Record with a revision tracking system. Links to the previous versions ensure examination of the data cited in previous papers. Indication of the date of revision enables the identification of the versions cited at a particular period. This is not possible if only version numbers are provided. (C) Situation when the entire database is reorganized, causing the deletion of the record that has been cited in literature. (D) Because there is no record from which previous versions can be traced, previous cited data become inaccessible.
Presenting latest and older versions of the same record in the database, on the other hand, may cause problems because users might accidentally access older versions even though they are interested only in the latest information. Thus, it is important to design the database so that the latest version of the record is presented usually, while providing a way to access older versions if users want. The simplest way to achieve this is to attach a list of hyperlinks to older versions of the document at the bottom of each revised record (Figure 3B). The revisions should be designated by the date of modification rather than version numbers, so that users can easily identify the version that was cited in a paper published at a particular time. To avoid human-related error and inconsistency, such a revision tracking links should better be generated automatically by the database.
Access to the older version of records is even more difficult, when the entire architecture of the digital atlas is modified to accommodate latest database technologies or when the atlas database is integrated into another database. Because contents of all the database records might be reorganized, each record of the old database may not have corresponding record in the new database (Figure 3C). This makes revision tracking between old and new databases practically impossible (Figure 3D). In such cases, the old database should better be kept accessible for the archival purpose, so that users who specifically want to read the contents of the old version can reach the information in its original form.
Providing a reliable way to cite records
It is not straightforward to cite particular contents of a digital atlas. Information source in the Internet is in general referred to with the uniform resource locator (URL; the string that starts with “http://”), which consists of the location of the host server (indicated by the characters before the first slash in the string) and the location of the file within it (characters after the first slash). The documents indicated in this way, however, tend to become inaccessible over time. There are two problems concerning this issue: the “file not found” problem and “host not found” problem.
The former problem occurs if the server cannot find the file requested by the user. A record of the digital atlas is typically constructed as a computer file. Writing the directory path of the file in the URL is the easiest way to make it accessible via the Internet. The directory path, however, is a highly vulnerable way to describe the location of information. A file becomes inaccessible even if just a single character of a file or directory name is changed. Connections may also be lost easily, if the directory that contains certain data files is moved within the host computer. Thus, using directory paths is not recommended for citing documents.
Many databases therefore assign numbers to each record, which is often called the accession number. Because this logical identifier is independent from the actual name and location of the file, it is robust against reorganization of the database. The records of all the sub-databases (e.g., neurons, images, brain regions, and markers) should be given different accession numbers. In addition, each identified neuron may also be given unique identification number, like the digital object identifier (DOI) of journal publication and ID numbers in genomic databases. To make use of these accession numbers, a digital atlas has to provide an easy way to search its contents with the accession number.
It would also be necessary to distinguish different versions of the data, especially when the original data were modified when putting into the digital atlas. For example, the dataset of the original microscopy data may be modified by non-linear registration for standardization, or signals and noises of the images may be enhanced or cleaned. In such cases, the original and modified data should be given different accession numbers.
Keeping the database accessible for a very long period
The latter “host not found” problem occurs in two cases. The first case is that the database exists but the URL has become invalid. This may occur because (1) the name of the host computer has been changed, (2) the institution that hosts the database was reorganized or renamed so that it acquired a new URL, or (3) the laboratory or organization that maintains the digital atlas has moved to a different institution. In these cases, users have to look for the atlas database that is operational with a new URL. This problem is relatively easy to address, because it is rather likely that the new URL can be spotted using popular Internet search services.
The second case is much more difficult to address: the digital atlas has stopped its operation because of the personnel, financial, or technical problems. Many web-based database sites have disappeared because the people who made the database have moved or retired. Only very large, public databases are run by organizations that can sustain their activity over a long period. Many scientific databases are maintained by a single laboratory or a group of only a few laboratories. Few if any institutions have a system to take over the maintenance of such databases after the people who established them have left the institute.
A database web site may also cease to exist, if the grant that has supported its operation is cut out. Many databases are funded by competitive money source. Such grants are unlikely to be awarded to the efforts that simply keep operation of old databases tens of years after they were established.
Even if the people responsible for the digital atlas remain active with enough financial support, maintenance of atlases may become difficult, when the computer system that runs the database becomes obsolete. Unlike printed atlases or their online versions provided as simple electronic files like the portable document format (PDF), a digital atlas is often a complex database with specialized software running on a computer. During the last decades, many computer platforms have disappeared from the market, making it impossible to use the software and even data that are made for older systems. Companies developing commercial database software continuously make updates and stop support for older versions. Old software often does not run on new computers, and new versions of the software that do run on the latest computers often fail to read data of their older versions properly. Considering that computer hardware must break sooner or later, it is a serious problem that long-term backward compatibility is hardly respected by computer and software companies.
In all, most digital atlases are destined to disappear within less than a couple of decades after they are established. I am confident that people in the twenty-second century will be able to read the article I am writing now, because it will be archived in many libraries, and because its electronic version in a popular industry-standard PDF format will remain accessible for a long period. On the other hand, it is not likely that someone would maintain the digital atlas I have been making to keep it accessible through the next century, long after I die. It is ironical that, whereas we can read and cite anatomical books and papers written even more than 150 years ago to access the knowledge and thoughts of our predecessors, most of the effort we are putting into computer-based database will not be seen by the people who live 150 years later; we cannot inherit our knowledge to our descendants. All the scientists who work with computer-based knowledge accumulation system should seriously think about this problem.
There would be several ways to avoid, or at least improve, this scenario. The best, but the most demanding, solution would be to establish a public organization that archive databases, just like libraries archive books and journals. Some university libraries and computer centers provide services for hosting databases in their servers, but the scope is usually limited to help the people who are currently working in the institute, not to keep these databases functional for the next 100 years.
Long-term maintenance will not be a problem for popular databases that are used widely by the research community, because there will be strong incentive to keep them functional. On the other hand, more specialized databases that are accessed only occasionally but yet provide important information are destined to disappear, because there would not be enough incentive to update them once they become obsolete. Libraries keep old books and journals even if they are accessed rarely. Only a very few scientific literature written before the nineteenth century are cited rather often. But information provided in other, less popular articles remain available: They just sit idle in the bookshelves of the libraries until someone want to read them. Likewise, old databases should be maintained even if they would attract few accesses per year.
Considerable financial support and political decision would be necessary to make such venture possible. For example, besides providing a portal site for cross-searching various life science databases, the Integrated Database Project by the Ministry of Education, Culture, Sports, Science and Technology of Japan3 is attempting to take over the operation of useful existing databases that suffer from bad maintenance. Such a project could in principle serve as a library-like archive of old databases, but the Japanese government recently decided to restructure this project to be merged with another bioinformatics-associated agency. Whether this archival activity can be sustained through the next century is not yet clear.
Another approach would be to establish a business model for maintaining databases by commercial companies such as journal publishers. Because publishers already have the systems to store and distribute electronic text, image, and supplementary data of their journals, publishing databases is technically feasible in a similar framework. The operating cost of the database could be passed on to users, but ideally such scientific databases should be available free of charge like open access journals. Though the help by the publishers will significantly reduce the workload of the scientists who organize the databases, however, private companies may not be as stable as public library-like archive in the long term.
A more decent solution would be to establish a system that fellow scientists of the relevant field should take over the maintenance of the database. Though this may be done on a private basis for some databases, a more systematic approach is required to ensure that all the databases in the field should be maintained, even if no people is interested in the further development of the database. This, however, is not easy under current circumstances, because it is very difficult to get financial support for the people who inherit the database just to keep it operational.
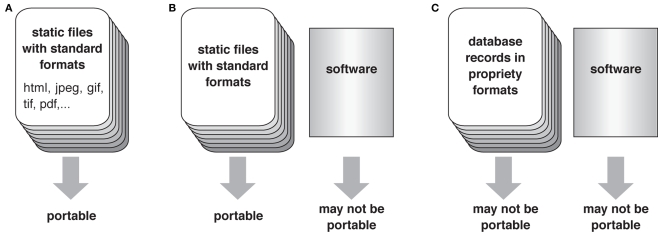
Archiving image databases is much more difficult than archiving electronic documents and web pages. If a digital atlas consists of only static data files, such as html and PDF documents and industry-standard graphic and movie files, it is easy to copy the atlas to other computers while keeping it accessible (Figure 4A). However, more sophisticated atlases that feature complex relational databases, search engines and interactive navigation systems are more vulnerable, because the associated software that enables such functions may not run on different computer platforms and may conflict with the programs of other databases that are archived in the same computer (Figures 4B,C). Even if the software successfully runs in the new server, it may become incompatible when the current computer hardware would get broken and replaced in the future.
Figure 4.
Portability of the database. (A) A database with only files of standard formats is easy to port to other computers. (B) If a database consists of standard format files and software for navigation, visualization, and search system, the software part may not function properly when copied to different computers. (C) If the documents are composed as relational records of a database program, even the data may not be accessible when copied to a platform that is not compatible with the database software used in the original system.
When the data transfer rate of the Internet was still rather slow, we set up mirror sites of the original Flybrain digital atlas4 in the United States, Germany, and Japan to ensure speedy access from respective local community. Because the Flybrain atlas is essentially a set of static html, jpeg, and movie files, it was easy to port most of its contents to mirror servers. The dedicated search engine and graphic navigation system, however, did not run properly in the mirror sites. The platform on which the original Flybrain was established, the Silicon Graphics workstation with the Irix operating system, has long been out of production.
Recently we are building a new atlas, the Flybrain Neuron Database5 (Shinomiya et al., in preparation), which provides information about the projection patterns and known functions of all the Drosophila brain neurons reported so far in a unified format. It is a true relational database but requires two servers to run, one for the database engine and the other for the interactive visualization tool. All the contents are stored as database records rather than static html files. Though this makes the new atlas much more versatile, maintenance of the servers and porting of its contents to other institutes are much more demanding than in the case of the classic Flybrain. It is a dilemma for digital atlas designers: The more sophisticated the atlas database is, the more difficult to maintain it for a long period, especially after the people who made the database leave or retire.
One approach to address this problem is to establish a common sophisticated digital atlas platform architecture, i.e., the same set of relational sub-databases, data fields, methods to specify the locations in the brain, revision tracking system, accession number system, etc. If digital brain atlases of diverse organisms provided by various organizations can share such a platform, long-term maintenance and integration of databases will be easier. Unfortunately, industry standard does not yet exist in this field of electronic databases; researchers are exploring the best solutions. Imposing common database architecture at this stage may be too preliminary, because it may disturb further development of better alternatives. Establishment of a compatible database architecture and eventual integration to it, nevertheless, should be considered as one of the long-term goals of the digital atlas projects.
Coordination of diverse digital brain atlas projects is going on under the initiative of the International Neuroinformatics Coordinating Facility (INCF6; Bjaalie and Grillner, 2007; De Schutter, 2009). Besides organizing workshops and promoting communication between the people working in the field, INCF provides useful research tools and aims at developing standard techniques for neural data archival and presentation. Such activity would serve as a basis for establishing a common digital atlas platform in the future.
Selection of data formats
A digital brain atlas transfers various types of files to users, which should be displayed properly on their computers. Selection of the data formats is therefore an important issue to ensure accessibility. Whereas formats of the text and two-dimensional image files have been standardized and can be decoded by most of the computers and web browsers provided by diverse venders, formats of the movies and three-dimensional image datasets are not yet standardized. Many of these data formats require so-called decoders, helper applications, or plug-in files to be installed in the users’ computer. Such plug-ins are not always provided for all kinds of computer platforms; they often require specific combinations of the operation system and web browsing software. To make things worse, not all of such propriety data formats survive the competition. Development of the plug-in software for the formats that do not gain enough support may be stopped, and the existing plug-ins may not run in the newer versions of operation systems and web browsers.
This causes a severe problem for the digital atlas. For example, the classic Flybrain features a section that provides description of the three-dimensional brain structure using the virtual reality modeling language (VRML). The VRML was established in 1997 as ISO/IEC 14772-1:1997 standard and enables interactive visualization and manipulation of three-dimensional data, a seemingly ideal format at that time. Because most web browsers did not handle VRML files, users had to download and install the VRML plug-in files. However, such plug-in files ran smoothly only on just a few computer platforms. In other computers they were either very slow or tended to crash. Because of the low usability, the VRML format never got strong support, and few plug-in files are provided for the latest computers and browsers. Thus, the section of the Flybrain that features VRML has effectively become inaccessible to most users. The efforts that were paid for generating this part of the atlas essentially came to naught.
To avoid such unfortunate consequences, it is important to stick to the file formats that have already become industry standards. This sometimes contradicts with the intention of the digital atlas designers, who want to resort to cutting-edge computer technology for offering so far unprecedented functions. However, a digital atlas is not a test bench of computer science but a venue for conveying anatomical information to as many users as possible for a very long period in the future. Careful consideration is therefore required when using latest data formats. In recent years, Adobe Acrobat 3D7 and Cortona3D Viewer8 have evolved as possible alternatives to VRML. Whether they would survive as industry standards or not, however, remains to be seen.
Problems in attracting the depository of external data: differences between journals and digital atlases
In the following sections, I will discuss less technical issues that might hamper steady accumulation of data once a digital atlas is first established. The first problem concerns the recruitment of data from the people who are not directly working on the database.
The laboratories that run digital brain atlases may not have all sorts of data that are suitable for the atlas, and other laboratories that have useful image data may not publish their own digital atlases. Users will feel it more convenient, if one or just a few digital atlases provide a comprehensive set of image data rather than many independent digital atlases each describes fragmental sets of information. Thus, it seems effective to ask a broad group of scientists for contributing their data to the atlas. This, however, is actually not so easy, or, practically unrealistic to be honest.
To recruit data from other laboratories, the classic Flybrain database set up a submission and review system similar to those of scientific journals in year 1995. People were in principle willing to provide data, but in reality few contributions were materialized. There were two problems underlying this consequence.
Scientists are generally very busy, and we have to allocate our resource (money, time, and personnel) to achieve the best outcome per effort. Preparing data for contributing to an atlas requires certain effort by the contributors. It is not just sending image files away by email; it requires detailed explanation of the technical procedure for the preparation of images, precise description of the neurons visualized in them, and conversion of the original propriety microscope data to standard web formats. Contributing to an image database is essentially similar to writing a mini paper. As it demands time and labor of the contributor, a strong incentive is needed to justify the effort. Because publishing papers in journals is considered mandatory for scientists, people are willing to spend time and energy to this aim. Giving data away to a database run by other scientists, on the contrary, is not regarded as an equally important job of scientists. To make the situation worse, the data that are put into a database are regarded as already published and are no longer suitable for publication as an original research article. This effectively gives a negative incentive not to contribute original data to digital atlases.
A more delicate issue is that a digital atlas like Flybrain is run by fellow scientists, whereas journals are run by public organizations. Advancement of the Flybrain is more often regarded as the achievements of its organizers rather than the achievements of the people who contribute their data. Considering this, it is understandable that people other than the database organizers would not want to spend time for the task that would not receive due appreciation from the research community.
To solve these problems, we once considered putting Flybrain under the aegis of certain public organization that issues scientific journals. This, however, could result in the restriction of the control by the original organizers, because the journal publisher gets rights on the management of the database. Also, there is a significant difference between journals and digital atlases. A journal is a venue for publishing various documents that are essentially independent from each other. It can, for example, publish papers that report contradicting observation in the same issue, or papers that describe similar contents using different terminology. Documents in a digital atlas should be more integrated with each other. It is not considered adequate that an atlas provides contradicting reports of a single neuron in different pages. Terminology should be consistent throughout the database. Organizers of a digital atlas therefore have more power of influence on its contents. This, however, may not always match the interests of the potential contributors who want to report their findings in the form they prefer.
To expect extensive contribution of data by the people other than the database organizers, a database should (1) be regarded mandatory by the scientific community to submit data to it in order to publish papers, and (2) collect simple information that does not require modification by the organizers. Databases that collect DNA and protein sequences, such as GenBank, fall in this category. It is difficult for anatomical databases to fulfill such conditions at this moment.
Lack of the system for evaluating the achievements of the people working with database
Because it is difficult to attract external contributions, contents of a digital atlas should be prepared mostly by the people who run the database. This seemingly simple task, however, is actually not easy to achieve.
When the Flybrain database was first established in 1995, its development was reported in a rather popular journal (Armstrong et al., 1995), even though the provided data were still in an initial, rather preliminary state. New data and better navigation tools were implemented extensively during the following years. Such subsequent efforts, however, could not be published as papers. As described earlier, new contents added in the database are regarded as already published and are no longer suitable for publication in a journal. However, whereas publication of a paper is commonly accepted as a scientific achievement of the person, addition of data to an existing database is almost always not evaluated as scientific achievements. This causes serious problems when students and postdoctoral fellows who are engaged in the digital atlas project write their curriculum vitae. If we consider that the efforts of our laboratory members should result in maximum rewards to them, reporting precious data in the database instead of in papers is suicidal.
Acknowledging this problem, the classic Flybrain shifted the strategy to provide contents that are already published elsewhere. But preparation of data for the database is by itself a laborious task, which requires a comparable amount of time and effort as those required for writing a paper. Considering that such effort would not be regarded as distinct scientific achievements once the original paper has been published, it is difficult to ask students and postdoctoral fellows for spending time for such unappreciated task. It is better for their carrier, if they instead use their precious time for doing new experiments for writing another paper. Indeed, even though we have identified many neurons in the fly brain and reported them in papers during the past decade, I did not ask my laboratory members for putting the obtained information in the Flybrain database.
The same evaluation holds true for the potential external contributors of the digital atlas. Scientists generate a lot of data when writing a paper, but many of them are eventually not presented in the final publication. It is often proposed that such data, e.g., the screening data of the materials that are not used in the study and the experimental data that are not put in the final manuscript because they do not fit in the context of the paper, should be donated to databases so that other people can access them to avoid the duplication of work. This, however, is essentially unrealistic as a matter of practice. To publish papers in the competitive field of neuroscience, research should be performed as efficiently as possible, concentrating only to the things that are publishable. Screening is practiced to select promising materials and discard others as quickly as possible, not to document observations of all the materials examined. Spending any effort to prepare such unused data to make them accessible to other people is a waste of time and decreases the competitiveness of the research group. Preparation of data for papers and preparation of data for databases require different approaches and, in reality, often pose a conflict of interest.
It is an ironical situation that development of a new a database system and development of novel visualizing or data mining tools are regarded as scientific achievements, whereas adding concrete contents to the existing database is not. Though the latter is in principle considered as an important contribution, we have to accept the hard fact that such effort does not materialize in the form of a journal paper and that the activity of a person or a laboratory is measured primarily by the number of published papers, not the amount of the data put into databases.
Given this trend, the most effective way to help the carrier development of the laboratory members and to raise the chance of getting grants for the laboratory leaders is as follows: First, write a paper that reports the establishment of a new database or the development of a technical tool, with a minimum amount of raw data to show its functionality. Once the paper is published, it is not wise to spend time to add new contents to the database. Instead, it is much more rewarding to develop yet another database or tools to write a new paper, leaving the initial database untouched to let it become incomplete and obsolete. Though this may sound bizarre, it is the only strategy to generate the maximum number of papers per the effort of the laboratory members.
Importance of the biocurators
In the previous section I explained the inherent problem that discourages paying due effort to add new contents to the databases once they are established and a paper announcing its development is published. Our new Flybrain Neuron Database9 is trying to address a popular demand: providing information about all the known neurons of the Drosophila brain. A tremendous amount of work was necessary for extracting information from previous publications to make the core contents of the database. Because it is made as an entirely new database, a paper describing its system and possible application is publishable (Shinomiya et al., in preparation). If the contents had been put into the records of the existing classic Flybrain database instead of the new database, however, the effort could not be considered worthy of journal publication. Likewise, the effort for adding further information to the new Flybrain Neuron Database would hardly lead to another publication. It would be convenient, if such information could be sampled and imported to the atlas automatically by knowledge database software. However, as computers cannot read papers and write review articles by themselves, database records cannot be prepared automatically by computers. Human brain is required for the job.
If researchers cannot spend enough time for this very important task of adding new contents to the existing database, someone else have to do this. Contribution of the people called biocurators is vital in this respect (Howe et al., 2008; St Pierre and McQuilton, 2009). The curators originally mean the people who work in the museums to maintain the collections, prepare annotations to them, and collect new items to be exhibited. These are exactly what are needed for the online database.
Unlike technicians, biocurators are academic staffs, because they require specialized scientific knowledge about the collection they maintain. In case of the digital brain atlas, biocurators should have detailed knowledge about brain anatomy and computer technology. They will perform experiments to generate data that will be presented in the atlas database, and ask laboratory members to provide data that can be published in the database. They will then convert these data to the formats that can be distributed on the Internet. They will also read latest papers in the field and update relevant parts of the digital atlas to keep the documentation up to date. They will ask people of other laboratories to provide relevant data. If digital data are provided, biocurators will perform all the tasks for converting them to the form that can be put into the atlas, thereby minimizing the burden of the contributors. If raw samples or original drawings are provided, they will photograph or scan them. They may also scan the figures of old publications that have not been digitized. They will then put all these materials into database records with sufficient annotations. They will also design the database and develop navigation and visualization tools to optimize its function and usability. Such diverse tasks may not be performed by a single person. A team of biocurators with different types of expertise would often be required.
Large databases are employing such biocurators. For example, the FlyBase10, a central database of Drosophila biology that collects all the data about genome sequences, known genes and molecules, and natural, mutant and transgenic strains, hires around 13 biocurators as of now (D. Sutherland, personal communication). Most of them are postdoctoral fellows with Drosophila background. These people not only gather and compile data to be incorporated into the database but also actively ask individual scientists of the community for providing information that is missing in their published materials. Without their contribution, the FlyBase cannot sustain its activity. The primary reason why FlyBase can successfully accumulate extensive data, in my view, is that it does not rely on the good intentions of external contributors but instead relies on the exhaustive efforts of its team of biocurators to actively collect information by itself.
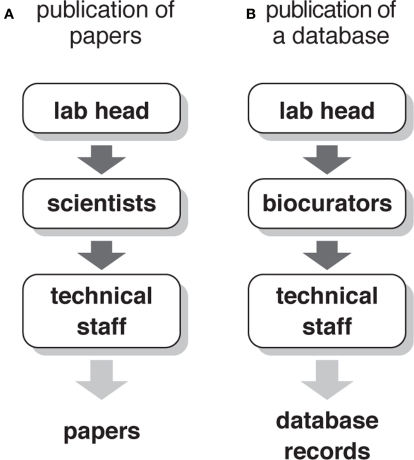
Unfortunately, biocurators are often confused with competent technical staffs. Even laboratory leaders who conduct large-scale neuroscience projects often think that well-trained technicians will be sufficient for the job. Problem of such misappreciation will be clear, if we compare the people who are required for writing papers and for making digital atlases. In many cases, a scientific research to be published in papers cannot be conducted with only a laboratory head and technicians (Figure 5A). Active scientists such as postdoctoral fellows and senior PhD students make detailed research plans, gather information that is vital for the project from publications and fellow scientists, organize the work that should be performed by the technical staff and train them, perform key experiments to generate data for the paper, and write the draft of the manuscripts. Likewise, a digital atlas cannot be materialized if there are only a laboratory head and technical staff. Someone has to make detailed plans of the atlas, gather information that is vital for the database, organize the work that should be performed by the technical staff and train them, perform key experiments to generate data for the database, and write the documents of the database records (Figure 5B). The role of the biocurators is indeed comparable to that of scientists.
Figure 5.
Role of scientists and biocurators. (A) People who are involved in a publication of journal papers. Scientists (e.g., postdoctoral fellows and senior PhD students) play decisive roles in materializing the overall project envisaged by the laboratory head. Technical staff helps scientists but cannot by themselves complete the study for writing up the papers. (B) People who are involved in a publication of databases such as digital brain atlases. Biocurators perform the roles that are equivalent to that of scientists for writing papers. Without them, technical staff cannot complete the database.
Many brain databases including our Flybrain have not yet reached the state to finance such employee. However, contribution of the biocurators is indispensable for ensuring substantial and continuous addition of contents after the database is first launched. The role of the biocurators is especially decisive for the so-called “omics” projects. In case of the genome analysis, for example, the initial release of the Drosophila genome sequence was published in year 2000 (Adams et al., 2000) and attracted a broad interest, despite the fact that the data contained numerous gaps where sequencing was incomplete. These gaps are subsequently filled, and extensive annotation and association with other data have been continuously performed during the last entire decade. Though successive releases of improved genome data (now release 5.27) have been vital for the promotion of the fly genome science, few of these efforts were reported in the form of journal publications; refining previous data is not considered qualified as independent research papers. It is likely that the anatomical omics projects (e.g., projectome and connectome for documenting the entire neural projections and connectivity, respectively; Sporns et al., 2005; Kasthuri and Lichtman, 2007; Seung, 2009) will also require extensive refining efforts after the announcement of their initial versions is published in journals. Hiring and training of expert biocurators are therefore keys to the success of such large-scale digital atlas projects.
A demanding problem in this respect is the difficulty of recruiting able person for the job. As explained above, the tasks of biocurators generally requires the scientific capability that is equivalent to that of the experienced postdoctoral fellows. Keeping track of newly published papers and incorporating relevant information to the database with adequate explanatory text requires the same level of academic capability as those for writing review articles or textbooks. In spite of this, they can hardly write papers by themselves, because the vast amount of their activity should be published in the form of database records. If they spend their time writing papers instead of database records, it conflicts with their expected duty. Therefore, the performance of the biocurators should never be measured by the number of their paper publications, but instead by the quality and quantity of the database they prepared. However, as discussed earlier such activity has not been receiving appreciation as scientists.
There will be two ways to address this apparent contradiction. A practical but passive approach is to look for the people who are trained as scientists but for some reason chose not to pursue the carrier. Because the number of people who obtain PhD degree largely exceeds the number of academic positions available each year, many people end up working as non-scientists. In addition, some researchers perform better compiling the data obtained by other people rather than doing experiments by themselves. The job of biocurators will become an attractive choice for such people, if it is possible to provide appropriate salary and future carrier path. The latter is especially important, because people would not want to jump into a job that does not offer the prospect until the age of retirement. A job market of the biocurators should be established, so that they can move from one database project to another depending on varying requirement of work force. Experienced biocurators should be awarded with higher salary and authority, so that there is enough incentive for personal development.
A more proactive approach is to change the way we evaluate scientific performance. It is in principle possible to integrate the job of biocurators in the academic carrier path: A student or postdoctoral fellow may work as a biocurator for several years and then becomes a senior postdoctoral fellow or professor after that. The task of an able biocurator may actually be more academic than some of the researchers who spend most of the time doing routine experiments. It is not fair that the latter type of people should be evaluated better simply because their results are published in the form of journal papers. Biocurators’ task is in a manner similar to that of the principal investigators in that they may not spend their time in benches but compose documents based on the results of other laboratory members.
Until the twentieth century, most of the scientific information is provided in the form of books and papers. This is already a thing of the past. In the twenty-first century, databases serve as an equally important venue of publication as journals. In spite of this, we are yet to establish a suitable system to appreciate this type of scientific publication. It is not a matter of politicians and grant agencies but a matter of scientists ourselves. If peer reviewers of grant applications, search committees of faculty members, and individual principal investigators who look for their laboratory staff, change their attitudes to acknowledge the task of biocurators as an important aspect of scientific activity even though they do not publish papers, and regard them as fellow scientists rather than trained technicians, it would become easy to recruit competent biocurators who would add unprecedented functions and values to the digital atlases before they are promoted as an academic faculty personnel.
Conclusion
When we see cutting-edge visual databases like Google Earth, we tend to praise the technical achievements of the people who developed amazing database engines and visualization tools, but often forget about the people who made and provided the core contents of the database. Fancy map databases would make no sense without the data obtained through the quiet and persistent efforts of the people in the geographical survey institutes of each country. Development and maintenance of the digital brain atlases confront the same problem of this unconscious neglect to the unostentatious effort for generating core data. Even we ourselves, who work on the development of visual databases, tend to allocate more time and effort to develop and discuss techniques for manipulating data, assuming that the data themselves can be acquired somehow routinely. This is not the case. Whereas Google Earth can buy geographical data from mapping institutes, we have to generate the geographical data of the brain by ourselves. Systematic approach to enable the generation of consistent and unambiguous database records and organizational consideration to pay due appreciation to the effort of the people doing this task would be prerequisites to establish and maintain comprehensive digital brain atlases.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I am grateful to David Osumi Sutherland of the FlyBase, Yoshiki Hotta of the Integrated Database Project, and Shiro Usui of the International Neuroinformatics Coordinating Facility Japan-node for discussion and providing information, and Karl-Friedrich Fischbach, Nicholas J. Strausfeld, J. D. Armstrong and late Kim Kaiser for the organization of and discussion about the Flybrain project.
Footnotes
References
- Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D., Amanatides P. G., Scherer S. E., Li P. W., Hoskins R. A., Galle R. F., George R. A., Lewis S. E., Richards S., Ashburner M., Henderson S. N., Sutton G. G., Wortman J. R., Yandell M. D., Zhang Q., Chen L. X., Brandon R. C., Rogers Y. H., Blazej R. G., Champe M., Pfeiffer B. D., Wan K. H., Doyle C., Baxter E. G., Helt G., Nelson C. R., Gabor Miklos G. L., Abril J. F., Agbayani A., An H. J., Andrews-Pfannkoch C., Baldwin D., Ballew R. M., Basu A., Baxendale J., Bayraktaroglu L., Beasley E. M., Beeson K. Y., Benos P. V., Berman B. P., Bhandari D., Bolshakov S., Borkova D., Botchan M. R., Bouck J., Brokstein P., Brottier P., Burtis K. C., Busam D. A., Butler H., Cadieu E., Center A., Chandra I., Cherry J. M., Cawley S., Dahlke C., Davenport L. B., Davies P., de Pablos B., Delcher A., Deng Z., Mays A. D., Dew I., Dietz S. M., Dodson K., Doup L. E., Downes M., Dugan-Rocha S., Dunkov B. C., Dunn P., Durbin K. J., Evangelista C. C., Ferraz C., Ferriera S., Fleischmann W., Fosler C., Gabrielian A. E., Garg N. S., Gelbart W. M., Glasser K., Glodek A., Gong F., Gorrell J. H., Gu Z., Guan P., Harris M., Harris N. L., Harvey D., Heiman T. J., Hernandez J. R., Houck J., Hostin D., Houston K. A., Howland T. J., Wei M. H., Ibegwam C. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195 10.1186/gb-2002-3-12-research0079 [DOI] [PubMed] [Google Scholar]
- Armstrong J. D., Kaiser K., Müller A., Fischbach K. -F., Merchant N., Strausfeld N. J. (1995). Flybrain, an on-line atlas and database of the Drosophila nervous system. Neuron 15, 17–20 10.1016/0896-6273(95)90059-4 [DOI] [PubMed] [Google Scholar]
- Armstrong J. D., van Hemert J. I. (2009). Towards a virtual fly brain. Philos. Transact. A Math Phys. Eng. Sci. 367, 2387–2397 10.1098/rsta.2008.0308 [DOI] [PubMed] [Google Scholar]
- Ascoli G. A., Donohue D. E., Halavi M. (2007). NeuroMorpho.org: a central resource for neuronal morphologies. J. Neurosci. 27, 9247–9251 10.1523/JNEUROSCI.2055-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjaalie J. G., Grillner S. (2007). Global neuroinformatics: the International Neuroinformatics Coordinating Facility. J. Neurosci. 27, 3613–3615 10.1523/JNEUROSCI.0558-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt R., Rohlfing T., Rybak J., Krofczik S., Maye A., Westerhoff M., Hege H. C., Menzel R. (2005). Three-dimensional average-shape atlas of the honeybee brain and its applications. J. Comp. Neurol. 492, 1–19 10.1002/cne.20644 [DOI] [PubMed] [Google Scholar]
- De Schutter E. (2009). The International Neuroinformatics Coordinating Facility: evaluating the first years. Neuroinformatics 7, 161–163 10.1007/s12021-009-9054-1 [DOI] [PubMed] [Google Scholar]
- Dreyer D., Vitt H., Dippel S., Goetz B., El Jundi B., Kollmann M., Huetteroth W., Schachtner J. (2010). 3D standard brain of the red flour beetle Tribolium castaneum: a tool to study metamorphic development and adult plasticity. Front. Syst. Neurosci. 4, 3. 10.3389/neuro.06.003.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Jundi B., Heinze S., Lenschow C., Kurylas A., Rohlfing T., Homberg U. (2009a). The locust standard brain: a 3D standard of the central complex as a platform for neural network analysis. Front. Syst. Neurosci. 3, 21. 10.3389/neuro.06.021.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Jundi B., Huetteroth W., Kurylas A. E., Schachtner J. (2009b). Anisometric brain dimorphism revisited: Implementation of a volumetric 3D standard brain in Manduca sexta. J. Comp. Neurol. 517, 210–225 10.1002/cne.22150 [DOI] [PubMed] [Google Scholar]
- Fischbach K. F., Dittrich A. P. M. (1989). The optic lobe of Drosophila melanogaster. – I. a golgi analysis of wild-type structure. Cell Tissue Res. 258, 441–475 10.1007/BF00218858 [DOI] [Google Scholar]
- Franklin K. B. J., Paxinos G. (2008). The Mouse Brain in Stereotaxic Coordinates, 3rd edn.San Diego, CA: Academic Press; 10.1016/0165-0270(80)90021-7 [DOI] [Google Scholar]
- Guetat G., Maitre M., Joly L., Lai S. L., Lee T., Shinagawa Y. (2006). Automatic 3-D grayscale volume matching and shape analysis. IEEE Trans. Inf. Technol. Biomed. 10, 362–376 10.1109/TITB.2005.863875 [DOI] [PubMed] [Google Scholar]
- Halavi M., Polavaram S., Donohue D. E., Hamilton G., Hoyt J., Smith K. P., Ascoli G. A. (2008). NeuroMorpho.org implementation of digital neuroscience: dense coverage and integration with the NIF. Neuroinformatics 6, 241–252 10.1007/s12021-008-9030-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanesch U., Fischbach K. F., Heisenberg M. (1989). Neuronal architecture of the central complex in Drosophila melanogaster. Cell Tissue Res. 257, 343–366 10.1007/BF00261838 [DOI] [Google Scholar]
- Howe D., Costanzo M., Fey P., Gojobori T., Hannick L., Hide W., Hill D. P., Kania R., Schaeffer M., Pierre S. S., Twigger S., White O., Rhee S. Y. (2008). Big data: The future of biocuration. Nature 455, 47–50 10.1038/455047a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huetteroth W., El Jundi B., El Jundi S., Schachtner J. (2010). 3D-reconstructions and virtual 4D-visualization to study metamorphic brain development in the sphinx moth Manduca sexta. Front. Syst. Neurosci. 4, 7. 10.3389/fnsys.2010.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahrling N., Becker K., Schonbauer C., Schnorrer F., Dodt H. U. (2010). Three-dimensional reconstruction and segmentation of intact Drosophila by ultramicroscopy. Front. Syst. Neurosci. 4, 1. 10.3389/neuro.06.001.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferis G. S., Potter C. J., Chan A. M., Marin E. C., Rohlfing T., Maurer C. R., Jr., Luo L. (2007). Comprehensive maps of Drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell 128, 1187–1203 10.1016/j.cell.2007.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenett A., Schindelin J. E., Heisenberg M. (2006). The virtual insect brain protocol: creating and comparing standardized neuroanatomy. BMC Bioinformatics 7, 544. 10.1186/1471-2105-7-544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasthuri N., Lichtman J. W. (2007). The rise of the ‘projectome’. Nat. Methods 4, 307–308 10.1038/nmeth0407-307 [DOI] [PubMed] [Google Scholar]
- Katz P. S., Calin-Jageman R., Dhawan A., Frederick C., Guo S., Dissanayaka R., Hiremath N., Ma W., Shen X., Wang H. C., Yang H., Prasad S., Sunderraman R., Zhu Y. (2010). NeuronBank: a tool for cataloging neuronal circuitry. Front. Syst. Neurosci. 4, 9. 10.3389/fnsys.2010.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurylas A. E., Rohlfing T., Krofczik S., Jenett A., Homberg U. (2008). Standardized atlas of the brain of the desert locust, Schistocerca gregaria. Cell Tissue Res. 333, 125–145 10.1007/s00441-008-0620-x [DOI] [PubMed] [Google Scholar]
- Kvello P., Lofaldli B. B., Rybak J., Menzel R., Mustaparta H. (2009). Digital, three-dimensional average shaped atlas of the Heliothis virescens brain with integrated gustatory and olfactory neurons. Front. Syst. Neurosci. 3, 14. 10.3389/neuro.06.014.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laissue P. P., Reiter C., Hiesinger P. R., Halter S., Fischbach K. F., Stocker R. F. (1999). Three-dimensional reconstruction of the antennal lobe in Drosophila melanogaster. J. Comp. Neurol. 405, 543–552 [DOI] [PubMed] [Google Scholar]
- Lein E. S., Hawrylycz M. J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A. F., Boguski M. S., Brockway K. S., Byrnes E. J., Chen L., Chen T. M., Chin M. C., Chong J., Crook B. E., Czaplinska A., Dang C. N., Datta S., Dee N. R., Desaki A. L., Desta T., Diep E., Dolbeare T. A., Donelan M. J., Dong H. W., Dougherty J. G., Duncan B. J., Ebbert A. J., Eichele G., Estin L. K., Faber C., Facer B. A., Fields R., Fischer S. R., Fliss T. P., Frensley C., Gates S. N., Glattfelder K. J., Halverson K. R., Hart M. R., Hohmann J. G., Howell M. P., Jeung D. P., Johnson R. A., Karr P. T., Kawal R., Kidney J. M., Knapik R. H., Kuan C. L., Lake J. H., Laramee A. R., Larsen K. D., Lau C., Lemon T. A., Liang A. J., Liu Y., Luong L. T., Michaels J., Morgan J. J., Morgan R. J., Mortrud M. T., Mosqueda N. F., Ng L. L., Ng R., Orta G. J., Overly C. C., Pak T. H., Parry S. E., Pathak S. D., Pearson O. C., Puchalski R. B., Riley Z. L., Rockett H. R., Rowland S. A., Royall J. J., Ruiz M. J., Sarno N. R., Schaffnit K., Shapovalova N. V., Sivisay T., Slaughterbeck C. R., Smith S. C., Smith K. A., Smith B. I., Sodt A. J., Stewart N. N., Stumpf K. R., Sunkin S. M., Sutram M., Tam A., Teemer C. D., Thaller C., Thompson C. L., Varnam L. R., Visel A., Whitlock R. M., Wohnoutka P. E., Wolkey C. K., Wong V. Y., Wood M., Yaylaoglu M. B., Young R. C., Youngstrom B. L., Yuan X. F., Zhang B., Zwingman T. A., Jones A. R. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- Livneh Y., Mizrahi A. (2010). A time for atlases and atlases for time. Front. Syst. Neurosci. 3, 17. 10.3389/neuro.06.017.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofaldli B. B., Kvello P., Mustaparta H. (2010). Integration of the antennal lobe glomeruli and three projection neurons in the standard brain atlas of the moth Heliothis virescens. Front. Syst. Neurosci. 4, 5. 10.3389/neuro.06.005.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Hof P. R., Grant S. C., Blackband S. J., Bennett R., Slatest L., McGuigan M. D., Benveniste H. (2005). A three-dimensional digital atlas database of the adult C57BL/6J mouse brain by magnetic resonance microscopy. Neuroscience 135, 1203–1215 10.1016/j.neuroscience.2005.07.014 [DOI] [PubMed] [Google Scholar]
- Mai J. K., Assheuer J. K., Paxinos G. (2004). Atlas of the Human Brain, 2nd edn.San Diego, CA: Academic Press; 10.1145/1452392.1452434 [DOI] [Google Scholar]
- Markram H. (2006). The blue brain project. Nat. Rev. Neurosci. 7, 153–160 10.1038/nrn1848 [DOI] [PubMed] [Google Scholar]
- Mikula S., Stone J. M., Jones E. G. (2008). BrainMaps.org – Interactive high-resolution digital brain atlases and virtual microscopy. Brains Minds Media 3, bmm1426. 10.1016/j.neuroimage.2006.11.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuna H., Ito K. (2006). Systematic analysis of the visual projection neurons of Drosophila melanogaster. I. Lobula-specific pathways. J. Comp. Neurol. 497, 928–958 10.1002/cne.21015 [DOI] [PubMed] [Google Scholar]
- Paxinos G., Watson C. (2005). The Rat Brain in Stereotaxic Coordinates – The New Coronal Set, 5th edn.San Diego, CA: Academic Press; 10.1016/0165-0270(80)90021-7 [DOI] [Google Scholar]
- Peng H., Ruan Z., Long F., Simpson J., Myers E. (2010). V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nat. Biotechnol. 28, 348–353 10.1038/nbt.1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier C., Vellema M., Verhoye M., Van Meir V., Wild J. M., Balthazart J., Van Der Linden A. (2008). A three-dimensional MRI atlas of the zebra finch brain in stereotaxic coordinates. Neuroimage 41, 1–6 10.1016/j.neuroimage.2008.01.069 [DOI] [PubMed] [Google Scholar]
- Puelles L., Martinez-de-la-Torre M., Paxinos G., Watson C., Martinez S. (2007). The Chick Brain in Stereotaxic Coordinates: An Atlas Featuring Neuromeric Subdivisions and Mammalian Homologies San Diego, CA: Academic Press; 10.1016/S0891-0618(98)00046-5 [DOI] [Google Scholar]
- Rein K., Zockler M., Mader M. T., Grubel C., Heisenberg M. (2002). The Drosophila standard brain. Curr. Biol. 12, 227–231 10.1016/S0960-9822(02)00656-5 [DOI] [PubMed] [Google Scholar]
- Reiner A., Perkel D. J., Bruce L. L., Butler A. B., Csillag A., Kuenzel W., Medina L., Paxinos G., Shimizu T., Striedter G., Wild M., Ball G. F., Durand S., Gunturkun O., Lee D. W., Mello C. V., Powers A., White S. A., Hough G., Kubikova L., Smulders T. V., Wada K., Dugas-Ford J., Husband S., Yamamoto K., Yu J., Siang C., Jarvis E. D. (2004a). Revised nomenclature for avian telencephalon and some related brainstem nuclei. J. Comp. Neurol. 473, 377–414 10.1002/cne.20118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A., Perkel D. J., Bruce L. L., Butler A. B., Csillag A., Kuenzel W., Medina L., Paxinos G., Shimizu T., Striedter G., Wild M., Ball G. F., Durand S., Guturkun O., Lee D. W., Mello C. V., Powers A., White S. A., Hough G., Kubikova L., Smulders T. V., Wada K., Dugas-Ford J., Husband S., Yamamoto K., Yu J., Siang C., Jarvis E. D. (2004b). The avian brain nomenclature forum: terminology for a new century in comparative neuroanatomy. J. Comp. Neurol. 473, E1–E6 10.1002/cne.20118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schambra U. (2008). Prenatal Mouse Brain Atlas New York, NY: Springer; 10.1016/0014-4886(91)90034-A [DOI] [Google Scholar]
- Seung H. S. (2009). Reading the book of memory: sparse sampling versus dense mapping of connectomes. Neuron 62, 17–29 10.1016/j.neuron.2009.03.020 [DOI] [PubMed] [Google Scholar]
- Sporns O., Tononi G., Kotter R. (2005). The human connectome: a structural description of the human brain. PLoS Comput. Biol. 1, e42. 10.1371/journal.pcbi.0010042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Pierre S., McQuilton P. (2009). Inside FlyBase: biocuration as a career. Fly (Austin) 3, 112–114 10.1149/1.3043419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld N. J. (1976). Atlas of an Insect Brain Berlin: Springer-Verlag; 10.1063/1.3047786 [DOI] [Google Scholar]
- Tanaka N. K., Awasaki T., Shimada T., Ito K. (2004). Integration of chemosensory pathways in the Drosophila second-order olfactory centers. Curr. Biol. 14, 449–457 10.1016/j.cub.2004.03.006 [DOI] [PubMed] [Google Scholar]
- Tanaka N. K., Tanimoto H., Ito K. (2008). Neuronal assemblies of the Drosophila mushroom body. J. Comp. Neurol. 508, 711–755 10.1002/cne.21692 [DOI] [PubMed] [Google Scholar]
- Toga A. W., Thompson P. M. (2001). Maps of the brain. Anat. Rec. 265, 37–53 10.1002/ar.1057 [DOI] [PubMed] [Google Scholar]
- Tokuno H., Tanaka I., Umitsu Y., Akazawa T., Nakamura Y. (2009). Web-accessible digital brain atlas of the common marmoset (Callithrix jacchus). Neurosci. Res. 64, 128–131 10.1016/j.neures.2009.02.003 [DOI] [PubMed] [Google Scholar]
- Valverde F. (1998). Golgi Atlas of the Postnatal Mouse Brain Wien: Springer-Verlag; 10.1063/1.3047786 [DOI] [Google Scholar]
- Van Essen D. C. (2002). Windows on the brain: the emerging role of atlases and databases in neuroscience. Curr. Opin. Neurobiol. 12, 574–579 10.1016/S0959-4388(02)00361-6 [DOI] [PubMed] [Google Scholar]
- Van Horn J. D., Toga A. W. (2009). Is it time to re-prioritize neuroimaging databases and digital repositories? Neuroimage 47, 1720–1734 10.1016/j.neuroimage.2009.03.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolsey T. A., Hanaway J., Gado M. H. (2003). The Brain Atlas: A Visual Guide to the Human Central Nervous System, 2nd edn.Hoboken, NK: John Wiley & Sons [Google Scholar]
- Wullimann M. F., Rupp B., Reichert H. (1996). Neuroanatomy of the Zebrafish Brain: A Topological Atlas Basel: Birkhäuser; 10.1007/BF00195012 [DOI] [PubMed] [Google Scholar]
- Yap P. T., Wu G., Zhu H., Lin W., Shen D. (2009). TIMER: tensor image morphing for elastic registration. Neuroimage 47, 549–563 10.1016/j.neuroimage.2009.04.055 [DOI] [PMC free article] [PubMed] [Google Scholar]