Abstract
MicroRNAs (miRNAs) play important roles in many developmental processes, including cell differentiation and apoptosis. Transition of proliferative ovarian granulosa cells to terminally differentiated luteal cells in response to the ovulatory surge of luteinizing hormone (LH) involves rapid and pronounced changes in cellular morphology and function. MicroRNA 21 (miR-21, official symbol Mir21) is one of three highly LH-induced miRNAs in murine granulosa cells, and here we examine the function and temporal expression of Mir21 within granulosa cells as they transition to luteal cells. Granulosa cells were transfected with blocking (2′-O-methyl) and locked nucleic acid (LNA-21) oligonucleotides, and mature Mir21 expression decreased to one ninth and one twenty-seventh of its basal expression, respectively. LNA-21 depletion of Mir21 activity in cultured granulosa cells induced apoptosis. In vivo, follicular granulosa cells exhibit a decrease in cleaved caspase 3, a hallmark of apoptosis, 6 h after the LH/human chorionic gonadotropin surge, coincident with the highest expression of mature Mir21. To examine whether Mir21 is involved in regulation of apoptosis in vivo, mice were treated with a phospho thioate-modified LNA-21 oligonucleotide, and granulosa cell apoptosis was examined. Apoptosis increased in LNA-21-treated ovaries, and ovulation rate decreased in LNA-21-treated ovaries, compared with their contralateral controls. We have examined a number of Mir21 apoptotic target transcripts identified in other systems; currently, none of these appear to play a role in the induction of ovarian granulosa cell apoptosis. This study is the first to implicate the antiapoptotic Mir21 (an oncogenic miRNA) as playing a clear physiologic role in normal tissue function.
Keywords: apoptosis,; granulosa cells,; luteinizing hormone,; microRNA,; ovary
In vivo and in vitro loss of microRNA 21, an LH-induced microRNA, results in mouse granulosa cell apoptosis.
INTRODUCTION
Luteinizing hormone (LH) acts on ovarian granulosa cells to induce ovulation and luteinization, resumption of oocyte meiosis, and cumulus expansion, all of which are critical for female fertility. Luteinization is a terminal differentiation process characterized by the rapid transition in morphology and function of the granulosa cell, with phenotypical changes becoming evident within 5–7 h of the ovulatory surge of LH/human chorionic gonadotropin (hCG) [1, 2]. Proper luteinization and sustained progesterone secretion are necessary for maintenance of pregnancy, and thus survival of periovulatory granulosa cells is obligatory for pregnancy. Thus, in addition to its differentiative effects, LH also acts as a survival factor by preventing apoptosis of the granulosa cells [3–5]. Although several antiapoptotic LH-induced factors have been identified in periovulatory granulosa cells, including progesterone receptor, pituitary adenylate cyclase-activating polypeptide, and atrial natriuretic peptide, it is expected that a conglomeration of factors are necessary for the molecular changes that need to occur to ensure both the survival and differentiation of these cells [6–8].
MicroRNAs (miRNAs) are emerging as important mediators of differentiation, proliferation, and apoptotic events [9–14]. Studies have implicated miRNA in the differentiation of mouse embryonic stem cells [15], effector and memory T cells [16], and many other tissues/cells [17–19]. Recently, conditional deletion of Dicer1 within granulosa cells using the anti-Müllerian hormone receptor-2 promoter-Cre (Dicer1 cKO) demonstrated that miRNAs are essential for normal ovarian function [20, 21]. Female Dicer1 cKO mice exhibited reduced ovulation rates, had increased granulosa cell apoptosis, and were infertile [20, 21]. Our laboratory recently identified 13 differentially regulated miRNAs in murine granulosa cells collected before and 4 h after the hCG/LH surge [22]. Three of these miRNAs were highly upregulated, and two (Mir132 and Mir212) were transcriptionally coregulated and induced by cAMP in cultured granulosa cells [22]. The remaining highly upregulated miRNA, Mir21, increased 7.5-fold by microarray analysis in response to in vivo hCG treatment and is the subject of this manuscript.
Enhanced Mir21 expression is evident in multiple types of cancer, including breast, pancreatic, colorectal, and esophageal [23–27]. Therefore, it has received a substantial amount of scientific attention to determine the signaling pathways that dictate its regulation as well as to identify its target gene transcripts [28–30]. Studies using locked nucleic acid (LNA) or 2′ O-methyl-oligonucleotides complementary to Mir21 demonstrate that loss of Mir21 action leads to increased cell apoptosis in a variety of cell culture systems [31, 32]. Several Mir21 target transcripts have been identified to explain its antiapoptotic effect, including programmed cell death 4 (Pdcd4), tropomyosin 1 (Tpm1), phosphatase and tensin homologue (Pten), serine (or cysteine) peptidase inhibitor, clade B, member 5 (Serpinb5, previously maspin), reversion-inducing cysteine-rich protein with Kazal motifs (Reck), sprouty homolog 2 (Drosophila; Spry2), and heterogeneous nuclear ribonucleoprotein K (Hnrnpk) and transformation-related protein p63, Trp63 (Tap63) [30, 33–37]. Although many different cell types undergo apoptosis in response to inhibition of Mir21 action, the Mir21 targets implicated vary widely for different cells [34, 36].
In this study, we examine the temporal expression of Mir21 in murine granulosa cells in response to LH/hCG, and we examine the function of Mir21 in cultured granulosa cells and in vivo by depleting mature Mir21 function with LNA oligonucleotides complementary to Mir21. We provide evidence that Mir21 acts as an antiapoptotic factor in cultured murine granulosa cells and in ovarian tissue. Furthermore, we show that Mir21 expression is transcriptionally regulated in ovarian granulosa cells in vivo, and loss of Mir21 in vivo results in reduced ovulation rates.
MATERIALS AND METHODS
Animals and Isolation of Granulosa Cells
All procedures involving animals were reviewed and approved by the Institutional Animal Care and Use Committee at the University of Kansas Medical Center and were performed in accordance with the Guiding Principles for the Care and Use of Laboratory Animals. All experiments were performed using CF1 female mice from Charles River Laboratories (Wilmington, MA). Granulosa cell isolation was performed as described previously [22]. Briefly, for in vivo expression analysis, 19-day-old female CF1 mice were injected i.p. with 5 IU of equine chorionic gonadotropin (eCG; FSH analog; Calbiochem, San Diego, CA) to stimulate follicular development, and ovaries were collected at 12, 24, 36, and 48 h later. To stimulate ovulation and luteinization, mice were injected 48 h after eCG with 5 IU of hCG (LH analog; Sigma, St. Louis, MO) and then collected 1, 2, 4, 6, 8, or 12 h later. Granulosa cells were obtained from the ovarian tissue by follicular puncture into PBS. For each time point, granulosa cells from three mice were pooled and processed for RNA isolation or protein analysis, and this was independently repeated four times. For in vitro analyses, ovaries from 26-day-old CF1 female mice were collected, incubated for 15 min in M-199 media containing 0.5 M sucrose, and rinsed with M-199 media, and the granulosa cells were expressed by follicular puncture into M-199 media [38]. This protocol generates a high yield of preovulatory granulosa cells without the need for eCG treatment.
Ovarian bursal injections were completed as described previously [39]. Briefly, female mice (19 days of age) were injected i.p. with 2 IU of eCG; 24 h later, mice were anesthetized by i.p. injection of a 1:1 mixture of ketamine (0.75 mg per animal; Ketaset, Fort Dodge, IA) and xylazine (0.75 mg per animal; Anased; Lloyd Laboratories, Shenendoah, IA). The ovaries and associated fat pad were exteriorized through incisions in the dorsal abdominal wall. To deliver treatments, a Hamilton syringe (10 μl) outfitted with a 33-gauge needle (45° beveled tip) was passed through the fat pad into the ovarian bursa. Each animal (n = 4) served as its own control, with one ovary receiving the blocking oligonucleotide and the other vehicle control or a nonspecific oligonucleotide. Animals were then closed, and 20 h later (i.e., 44 h after eCG) they were injected i.p. with 2 IU of hCG. To not mask a knockdown effect with excessive ovarian stimulation, “physiologic” doses of 2 IU of eCG and hCG were administered to ovarian bursa-treated mice. Animals were killed 16 h after hCG injection, and the number of cumulus-oocyte complexes in the oviduct was counted to determine ovulation rate. Ovaries were removed and fixed overnight in freshly made 4% paraformaldehyde.
In Vitro Granulosa Cell Culture
Isolated granulosa cells were seeded at 2.5 × 104 cells per well into six-well fibronectin-coated tissue culture plates for gene expression and apoptosis analyses, and 1.5 × 106 cells per 10-cm tissue culture dish for protein analyses. An aliquot of the isolated granulosa cells was placed in Trizol (Invitrogen, Carlsbad, CA) prior to culture to determine “before-culture” expression values. Cells were cultured in Dulbecco modified Eagle medium/F12 (10% fetal bovine serum, 1% gentamicin) at 37°C with 5% CO2. To determine whether plating affects Mir21 expression, RNA from cells was collected in Trizol at 6, 12, 24, 48, and 72 h after plating. To mimic the culture conditions used during transfection, another group of cells were cultured for 48 h in serum-containing medium and then cultured for 24 h in serum-free medium prior to RNA collection. To determine whether serum affects Mir21 expression, a final group of cells were cultured in serum-free media from the initial plating. Granulosa cells treated with hCG (Sigma) or 8-bromo-cAMP (Sigma) were cultured for 48 h in medium containing serum, then with serum-free medium for 24 h prior to treatment with 8-bromo-cAMP (1 mM) for 1, 2, 4, 6, 8, or 12 h. Oligonucleotide transfections were performed 48 h after plating, with the medium containing serum replaced with serum-free medium directly before transfection.
Oligonucleotide Transfections
The nonspecific control scrambled (anti-NS) and anti-21 2′-O-methyl RNA oligonucleotides were purchased from Ambion (Houston, TX). Locked nucleic acid oligonucleotides were synthesized with a mix of LNA/DNA nucleotides: LNA-21 5′-TCA GTC TGA TAA GCT A-3′, and the nonspecific control (LNA-NS), 5′-CGG CAG TAT GCG AAT C-3′ (underlined bases denote the LNA nucleotides; Integrated DNA Technologies, Coralville, IA). The in vivo LNA oligonucleotides, LNA-21ps and LNA-NSps, (Integrated DNA Technologies), were identical in sequence and LNA base location to the in vitro oligonucleotides, the only difference being the addition of a nuclease-resistant phospho thioate backbone. The nonspecific scrambled control, LNA-NSps, was synthesized with a rhodamine attached to the 5′ end to allow for visualization of injection efficiency and tissue incorporation.
Oligonucleotides (42 nM) were mixed with Lipofectamine 2000 reagent (Invitrogen) and then added to the isolated granulosa cells 48 h after culture at a final concentration of 42 nM in serum-free media. Mock transfected (Lipofectamine alone) and nontransfected controls were also performed, and cells were collected after 24 h. Oligonucleotide-transfected cells were collected after 4, 8, 12, and 24 h of transfection for protein, RNA, or apoptosis assays. To determine whether serum withdrawal affected LNA results, serum was reintroduced 4 h after transfection in a subset of cells, and then cells were collected 24 h later. Granulosa cell protein lysates were obtained by resuspending cells in 200 μl of cell lysis buffer (Cell Signaling Technology, Danvers, MA) with 1 mM phenylmethylsulphonyl fluoride. Cells were centrifuged at 1000 × g for 5 min to pellet the cellular membrane debris, and supernatants were stored at −80°C until use. For RNA, cells were resuspended in 500 μl of Trizol, and RNA was isolated following the manufacturer's instructions.
For in vivo transfections, the LNA-21ps and LNA-NSps oligonucleotides (0.4 nM) were mixed with Lipofectamine 2000 prior to bursal injection. Saline plus Lipofectamine was used as the vehicle control.
Apoptosis Assay
Granulosa cells were trypsinized, washed twice with cold PBS, and resuspended in 1× binding buffer (BD Pharmingen, San Jose, CA) at a concentration of 1 × 106 cells/ml. A total of 100 μl (1 × 105 cells) of the solution was transferred to a 5-ml polystyrene tube and incubated with 5 μl of phycoerythrin-conjugated annexin A5 (BD Pharmingen) and 1 μl of live/dead fixable violet dead cell stain (Invitrogen) for 10 min at room temperature in the dark. A total of 1 ml of 1× binding buffer was added to the cells, centrifuged at 1000 × g for 5 min, and the supernatant was removed. Cells were resuspended in 1% paraformaldehyde/PBS. The samples were analyzed by flow cytometry (BD Biosciences LSR II, San Jose, CA) using FACSDiva research software (BD Biosciences). Annexin A5-positive cells were considered apoptotic.
RNA Isolation and Quantitative RT-PCR
Total RNA was isolated using Trizol following the manufacturer's instructions. Total RNA (250 ng) was reverse transcribed using a stem-loop primer specific to Mir21 (Applied Biosystems, Carlsbad, CA) [22]. The resulting cDNA was used in the quantitative PCR reactions using Mir21 Taqman primers and probe (Applied Biosystems). For analysis of primary-Mir21 transcript (pri-Mir21), Tmem49, small nuclear RNA U6 (Rnu6), and Gapdh expression, 1 μg of total RNA was reverse transcribed using random hexamer primers, and with the exception of Gapdh, which used Taqman primers and probe (Applied Biosystems), quantitative PCR was performed with the appropriate forward and reverse primer pairs and Power Sybr Green Master Mix (Applied Biosystems). The primer pairs used are as follows: pri-Mir21, forward: 5′-GAC ATC GCA TGG CTG TAC CA-3′; reverse: 5′-CCA TGA GAT TCA ACA GTC AAC ATC A-3′; Tmem49, forward: 5′-TGG CAT CGT CAA AGC ATT GT-3′, reverse: 5′-CCG CTG CAC ATA CTG TTG GT-3′; Rnu6, forward: 5′-CTC GCT TCG GCA GCA CA-3′, reverse: 5′-AAC GCT TCA CGA ATT TGC GT-3′. The Tmem49 primers spanned intron 4 to prevent any possible amplification of the pri-Mir21 transcript, which starts in intron 10. The standard cycling conditions were 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min; for Sybr Green assays, a dissociation curve was performed to monitor potential nonspecific amplification. Data were collected during the 60°C extension step. For small RNA analysis (i.e., mature Mir21), Rnu6 (U6) was used to normalize data due to an equivalent size. For larger RNA analysis (i.e., pri-Mir21 and Tmem49), Gapdh was used to normalize data. We have previously shown both Gapdh and U6 to be valid for normalization [22]. Each sample was run in triplicate for both the target and the normalizer, and the average Ct was used in subsequent calculations. Each sample was also run as a minus RT control to confirm lack of DNA contamination. The delta-delta Ct method was used to calculate relative fold change values between samples, with one control sample set to 1 and all other samples compared against it. At least three independent replicate experiments were used to calculate means and SEM values [22].
Immunoblotting
Granulosa cell protein lysate concentrations were determined using the Bio-Rad protein assay (Hercules, CA). Each lysate (15 μg) was denatured by diluting the sample 1:2 with sample buffer (2.8 ml of distilled water, 1.0 ml of 0.5 M Tris-HCl [pH 6.8], 0.8 ml of glycerol, 1.6 ml of 10% SDS, 0.4 ml of 2-β-mercaptoethanol, and 0.4 ml of 0.05% [w/v] bromophenol blue) and heating for 5 min at 95°C. Proteins were separated on 12% SDS-PAGE gels in 5× electrode running buffer, pH 8.3 (25 mM Tris base, 192 mM glycine, and 0.1% [w/v] SDS), and transferred to polyvinylidene fluoride membranes (Millipore, Billerica, MA) in transfer buffer (12 mM Tris-HCl, 96 mM glycine, and 20% [v/v] MeOH). Blots were incubated for 1 h at room temperature in a 5% milk solution to block nonspecific binding. Blots were then incubated overnight at 4°C with one of the following antibodies: caspase 3 antibody (Cell Signaling Technology), SPRY2 (Santa Cruz Biotechnology, Santa Cruz, CA), tropomyosin 1 (Santa Cruz Biotechnology), PDCD4 (ProSci, Poway, CA), PTEN (Sigma), and ACTB (Santa Cruz Biotechnology). After washing and incubation with the appropriate secondary antibody, protein-antibody complexes were visualized using West Pico Chemiluminescent Substrate (Pierce) following the manufacturer's protocol.
Statistical Analysis
Data are displayed as means ± SEM of replicate samples. Statistical analysis of data was performed using GraphPad Prism (version 4). When two groups were compared (i.e., LNA-NS vs. LNA-21), a Student t-test was used to detect differences between treatments. For multiple-group comparisons (i.e., in vivo and in vitro time courses), one-way ANOVA followed by a student Newman-Keuls posthoc test was used to determine differences among the means. In cases where two variables were examined (i.e., +/− cAMP time course and the LNA-NS/21 concentration experiment), two-way ANOVA was used to determine differences among the means. A value of P < 0.05 was considered significant.
RESULTS
Mir21 Expression Increases after In Vivo eCG/Follicle-Stimulating Hormone and hCG/LH Administration
To determine whether transcription of Mir21 increased in response to the eCG, the expression of pri-Mir21 was examined by quantitative RT-PCR. Mural granulosa cells collected from preovulatory follicles (i.e., 0–48 h after eCG and before hCG) exhibited a 4-fold increased expression of pri-Mir21 transcript 12 h after eCG, returned to basal levels at 24 h, then increased to an intermediate level at 36 and 48 h after eCG (Fig. 1A). Mature Mir21 followed pri-Mir21 expression, with levels increasing 6.4-fold at 24 h after eCG before returning to basal concentrations at 36 h (Fig. 1B). Despite having its own promoter, a recent study indicated that Mir21 might be coexpressed with the gene Tmem49, in which it resides [35]. Expression of Tmem49 within granulosa cells showed no change after in vivo eCG administration (Fig. 1C).
FIG. 1.
Induction of miRNA 21 by eCG and hCG. Quantitative RT-PCR of pri-Mir21 (A and D), mature Mir21 (B and E), or Tmem49 (C and F) in murine granulosa cells with eCG alone 0, 12, 24, 36, or 48 h (A–C) or after eCG (48 h) plus hCG for 0, 1, 2, 4, 6, 8, or 12 h (D–F). All quantitative RT-PCR data (n = 3) were normalized to Gapdh; one-way ANOVA was used to determine statistical differences between time points. Means ± SEM with different letters above the bars are different (P < 0.05).
Mural granulosa cells collected from periovulatory follicles (i.e., after hCG) exhibited increased pri-Mir21 expression 2 h after hCG, reaching a maximal 30-fold induction 4 h after hCG, and then remained elevated through 12 h after hCG (Fig. 1D). In vivo expression of mature Mir21 trailed that of pri-Mir21 by several hours, with mature Mir21 reaching its maximum 5.8-fold induction at 6 h after hCG; mature Mir21 remained elevated through 12 h after hCG (Fig. 1E). Expression of Tmem49 within granulosa cells showed no change after in vivo hCG administration (Fig. 1F).
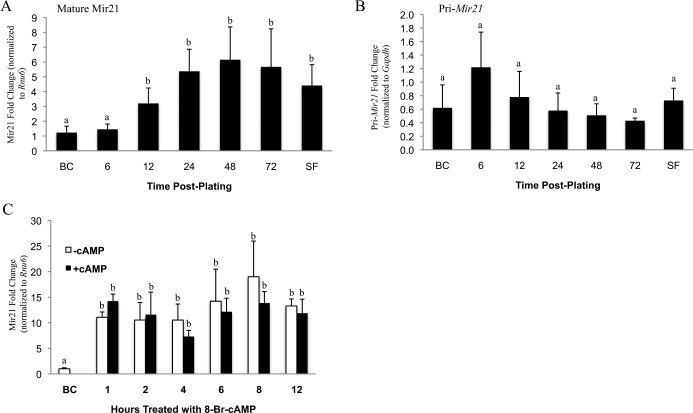
In contrast to in vivo Mir21 expression, which demonstrated a clear transcriptional response, mature Mir21 in cultured granulosa cells increased 5-fold upon plating, with no corresponding increase in pri-Mir21 detection (Fig. 2, A and B). Granulosa cell expression of mature Mir21 increased (P < 0.05) within 12 h of plating, reaching its apex at 48 h (Fig. 2A). To determine whether serum withdrawal affected Mir21 expression, serum was removed after 48 h of culture, and no significant effect on basal mature Mir21 expression was observed (Fig. 2A). Granulosa cells grown (72 h) in the complete absence of serum exhibited increased Mir21 expression that was similar to those with serum (data not shown), and addition/withdrawal of serum was eliminated as a source of involvement in LNA-21 action in similarly serum-free-grown granulosa cells (data not shown). Subsequent cAMP treatments and transfection experiments were done in granulosa cells removed from serum-containing media and placed in serum-free media at 48 h after plating. We observed that cultured granulosa cell expression of Mir21 did not change in response to treatment of cells with the cell-permeable 8-bromo-cAMP (Fig. 2C) or after treatment with hCG (data not shown).
FIG. 2.
MicroRNA 21 increases in response to plating and does not change in response to cAMP. Quantitative RT-PCR of mature Mir21 (A) and pri-Mir21 (B) before culture/plating (BC), and 6, 12, 24, 48, or 72 h, or SF, which represents cells 72 h after plating, with the last 24 h in serum-free media. Mature Mir21 in granulosa cells before culture (BC) and after 72 h of plating (minus serum for the last 24 h) and then treated with 8-bromo-cAMP for 1, 2, 4, 6, 8, or 12 h of 8-bromo-cAMP treatment are shown in C. Data (n = 3) were normalized to Gapdh; one-way (A and B) and two-way ANOVA (C) were used to determine statistical significance; Means ± SEM with different letters above the bars are different (P < 0.05).
Differential Knockdown of Mir21 by LNA and 2′-O-Methyl Oligonucleotides Leads to Functional Differences
To analyze the function of Mir21, granulosa cells were treated with either a 2′-O-methyl oligonucleotide complementary to Mir21 (anti-21) or a control scrambled 2′-O-methyl oligonucleotide (anti-NS). Anti-21-treated cells had one ninth of the expression of mature Mir21 compared with anti-NS-treated cells (Fig. 3A). The specificity of this anti-21 treatment was confirmed because detection of two other miRNAs, Mir132 and Mir212, were not affected (data not shown). Similarly, knockdown of Mir132 and Mir212 in cultured granulosa cells had no effect on Mir21 expression [22]. LNA oligonucleotides have been shown to block miRNA expression and function to a greater degree than 2′-O-methyl oligonucleotides [40, 41]. Consistent with these observations, Mir21 in granulosa cells treated with LNA-21 was one twenty-seventh of that in granulosa cells treated with the scrambled LNA (LNA-NS; Fig. 3B). The specificity of LNA-21 was confirmed because Mir132 and Mir212 expression was not affected (data not shown). Additionally, there was no change in Mir21 in granulosa cells that were mock transfected (i.e., lipofectamine alone added) or nontransfected (i.e., no LNA or lipofectamine added; data not shown).
FIG. 3.
Knockdown of Mir21 by 2′-O-methyl oligonucleotides or LNA oligonucleotides in cultured granulosa cells. Quantitative RT-PCR of mature Mir21 in cultured granulosa cells after transfection with 2′-O-methyl anti-21 or anti-NS oligonucleotides (A) or with LNA-21 or LNA-NS (i.e., control) oligonucleotides (B). Data (n = 3) were normalized to Gapdh; t-tests were used to determine statistical significance within the oligonucleotide treatment groups. *Means ± SEM are different (P < 0.05). Two-way ANOVA comparison of relative values across treatment groups indicated a 3-fold decline (P < 0.05) in Mir21 between anti-21 and LNA-21 treatment groups, with no difference between anti-NS and LNA-NS-treated cells.
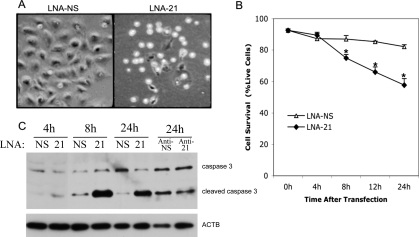
The 3-fold difference in Mir21 knockdown between the different Mir21 inhibitors was associated with a marked difference in cell morphology because only the LNA-21-treated cells rounded up and pulled off of the dish, a well-described “late step” in the process of apoptosis (Fig. 4A) [42]. To confirm apoptosis, cells were stained with annexin A5 and live/dead violet stain, fixed, and then analyzed by flow cytometry. Annexin A5 labeling indicated that granulosa cells began undergoing apoptosis within 8 h of LNA-21 treatment, and by 24 h of treatment 42% of the cells had undergone apoptosis (Fig. 4B). No change in apoptosis was observed in the control LNA-NS-treated cells (Fig. 4B). Flow cytometry also indicated that granulosa cells transfected with anti-21 or precursor-Mir21 (which increases Mir21) had no effect on apoptosis at 24 and 48 h after transfection (data not shown). To further confirm the increase of apoptosis in LNA-21-treated cells, immunoblotting was performed for cleaved caspase 3, an active mediator of apoptosis [43]. Detection of cleaved caspase 3 increased in LNA-21-treated cells within 8 h after transfection but was not increased in anti-21-treated cells (Fig. 4C). To confirm that serum withdrawal was not having an impact on the ability of LNA-21 to induce apoptosis, cells were transfected in the presence of serum or with serum replaced 4 h after transfection, and in both cases cleaved caspase 3 increased after transfection (data not shown). Dose-response analysis using decreasing concentrations of LNA-21 (20, 5, and 1.25 nM; Fig. 5A) displayed decreasing amounts of cleaved caspase 3 (Fig. 5B). Although the 1.25 nM dose of LNA-21 showed a similar knockdown to anti-21 (one ninth of basal expression), there was still evidence of increased cleaved caspase 3 in these cells (Fig. 5).
FIG. 4.
Induction of apoptosis upon LNA-21 treatment of cultured murine granulosa cells. Bright field image of rounded up cells after 24 h of treatment with LNA-21 (A). Original magnification ×10. LNA-NS-transfected and LNA-21-transfected cells were double stained with annexin A5 and live/dead violet stain and were analyzed by fluorescence-activated cell sorting 0–24 h after transfection (B). Line graph summarizes the percentage of live cells 0–24 h after transfection, and data (n = 3) were analyzed by t-test; *Means ± SEM are different (P < 0.05) between the LNA-21 and LNA-NS at that time point. C) Representative Western blot (n = 3) of caspase 3 and cleaved (active) caspase 3 in LNA-NS-transfected and LNA-21-transfected granulosa cells 4, 8, and 24 h after transfection, or 24 h after transfection with anti-NS or anti-21.
FIG. 5.
Reduced concentrations of LNA result in a decrease in the amount of caspase 3 cleavage. Quantitative RT-PCR of mature Mir21 in cultured granulosa cells with 1.25, 5, or 20 nM LNA-NS or LNA-21 (A). Cells transfected with 1.25 nM LNA-21 had comparable fold reductions in mature Mir21 to those transfected with 42 nM anti-21. Data (n = 3) were normalized to Gapdh; two-way ANOVA was used to determine statistical significance. Means ± SEM with different letters above the bar are different (P < 0.05). B) Representative Western blot (n = 3) of caspase 3, cleaved (active) caspase 3, and ACTB in granulosa cells transfected with 1.25, 5, or 20 nM LNA-NS or LNA-21.
Known Apoptotic Mir21 Target Transcripts Were Unaffected by LNA-21 Treatment of Granulosa Cells
MicroRNA 21 has been implicated in the posttranscriptional regulation of eight apoptotic genes in other cell systems [30, 33–37]. Western blot analyses examined the expression of four of these proteins at 4, 8, and 24 h after transfection with the LNA-21 oligonucleotide (Fig. 6). Two others, RECK and SERPINB5, were also analyzed by Western blot analysis, but expression of both appeared extremely low in granulosa cells, as evidenced by the almost nonexistent signal and lack of differences between LNA-NS and LNA-21 samples (data not shown). With the exception of PTEN, which exhibited a slight nonsignificant increase 4 h after treatment with LNA-21, none of the other proteins exhibited any change in protein expression (Fig. 6). To confirm whether PTEN might be involved in mediating Mir21 action, additional studies knocking down Pten and using blocking anti-miRs specific to the Pten 3′ untranslated region were performed; no effect was observed on granulosa cell apoptosis (see Supplemental Methods and Supplemental Fig. S1, A–C; all supplemental data are available online at www.biolreprod.org).
FIG. 6.
Granulosa cell expression of known Mir21 targets is not affected by LNA-21 knockdown. Representative Western blots (n = 3) for PDCD4, PTEN, TPM1, SPRY2, and control ACTB after 4, 8, or 24 h of transfection with LNA-NS or LNA-21, or 8 h after transfection with anti-NS or anti-21. Densitometry indicated no significant differences for any of the targets.
Caspase 3 Cleavage Decreases in Granulosa Cells after Administration of an Ovulatory Surge of LH/hCG
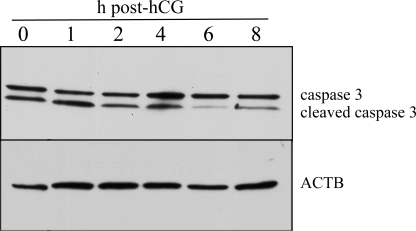
To determine the incidence of apoptosis in follicular granulosa cells after the LH/hCG surge, we examined cleaved caspase 3 by Western blot analysis. Granulosa cell uncleaved caspase 3 levels were unchanged after hCG, except for a slight increase at 4 h after hCG (Fig. 7), whereas cleaved caspase 3 decreased 6 h after in vivo administration of hCG, where it then remained at 8 h after hCG (Fig. 7), at a time coincident with elevated Mir21 (Fig. 1).
FIG. 7.
Cleaved caspase 3 is decreased in vivo after LH/hCG. Representative Western blots (n = 3) of caspase and cleaved caspase 3 and ACTB 0, 1, 2, 4, 6, and 8 h after hCG administration.
In Vivo Knockdown of Mir21 Causes an Increase in Apoptosis
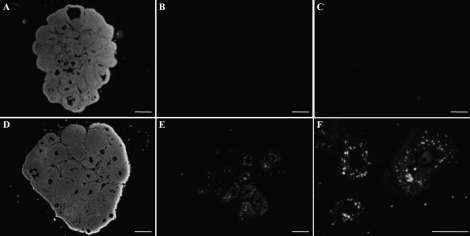
To determine the in vivo effect of Mir21 on ovarian apoptosis, Mir21 function was knocked down in vivo. After follicular stimulation (i.e., 24 h of eCG), an LNA-21ps oligonucleotide targeting Mir21 was injected into one ovarian bursa, whereas the other ovary received vehicle or the fluorescent-tagged nonspecific LNA-NSps. Twenty-two hours after surgery, animals were injected with hCG to stimulate ovulation and luteinization. Ovaries were collected 16 h after hCG and were analyzed for apoptosis (Fig. 8). Apoptosis was significantly increased in LNA-21-treated ovaries compared with saline-treated ovaries (Fig. 8). Transfection efficiency was measured by distribution of the fluorescent-labeled LNA-NS (Supplemental Fig. S2). Although labeling was found throughout the ovary, there were punctate regions of increased fluorescence (Supplemental Fig. S2). When tested for apoptosis, there was no increase in TUNEL fluorescence between the LNA-NS negative control and the TUNEL-labeled LNA-NS samples (Supplemental Fig. S2).
FIG. 8.
Ovarian bursal injections of LNA-21ps cause an increase in apoptosis. Representative fluorescence images (n = 4) of mouse ovaries exposed to saline (A) or LNA-21ps (D) stained with the DNA-binding dye Hoechst. TUNEL assay to detect apoptotic cells in the saline-treated (B) and LNA-21ps-treated (E; F is a 10× magnification of E) ovaries. C) Negative control TUNEL assay (nonworking enzyme) of the LNA-21ps-injected mouse ovary in E. Bar = 500 μm.
In Vivo Knockdown of Mir21 Causes a Reduction in Ovulation Rates
To examine whether the increase in ovarian apoptosis affects ovulation, the number of cumulus-oocyte complexes recovered from the oviduct was recorded as the “ovulation rate.” Ovaries treated with LNA-21 displayed decreased ovulation rates compared with LNA-NS or saline-treated controls (Fig. 9).
FIG. 9.
Ovulation rate is decreased in LNA-21-treated ovaries. Number of cumulus-oocyte complexes (COCs) within the oviduct of LNA-NS/saline-treated ovaries (n = 4) or LNA-21-treated ovaries (n = 4). *Significance (P < 0.05).
DISCUSSION
During the LH-induced transition of a granulosa cell to a luteal cell, apoptosis must be inhibited to allow the formation of a functional corpus luteum. The cleavage of caspase 3 irrevocably commits the cell to apoptosis, and thus measurement of cleaved caspase 3 provides a measurement of the amount of cell death occurring. We examined the amount of cleaved caspase 3 in granulosa cells after the LH/hCG surge and found that cleaved caspase 3 decreases within 6 h of hCG, concurrent with the highest expression of mature Mir21. To examine the in vivo effect of Mir21 in granulosa cells, ovarian bursa were injected with LNA-21ps, and apoptosis was examined. Apoptosis was significantly increased in LNA-21ps-treated ovaries. Here, knockdown of Mir21 may prevent its antiapoptotic action, thereby increasing cleavage of caspase 3 and promotion of apoptosis. In caspase 3 knockout mice, atretic follicles fail to undergo atresia [44]. During atresia, granulosa cells undergo apoptosis and apoptotic bodies appear in the antral space, and our TUNEL assay does show staining within the granulosa and antrum of antral follicles (Fig. 8F) [44]. These follicles may be undergoing atresia due to Mir21 knockdown. Further studies are needed to examine the expression of Mir21 in healthy and atretic granulosa cells to determine its importance in atresia. Unexpectedly, there was a relatively low amount of TUNEL staining within corpora lutea of the LNA-21ps-injected ovaries. This may be because the apoptotic cells have already been cleared, and nonapoptotic cells (perhaps those that were not effectively transfected) remain.
Our in vivo expression studies indicate that Mir21 gene expression in ovarian granulosa cells is regulated predominantly by increased transcription of the pri-Mir21 transcript. Indeed, this is the first study to examine the expression of Mir21 in a physiologic system; all other studies have examined Mir21 expression in cultured cells. The Mir21 promoter is well conserved across a number of species and lies within the 10th intron of a protein-coding gene, Tmem49 [29]. Although at least one study has suggested that Tmem49 and Mir21 are coexpressed [35], we show here, in agreement with two others, that Tmem49 and Mir21 are expressed independently [29, 45]. Our results also indicate that in vivo, Mir21 is regulated by the endocrine hormones, FSH and LH, in the preovulatory and periovulatory follicular granulosa cells, respectively. Within the first 300 bases of the Mir21 transcription start site, a number of known transcription regulatory element-binding sites are located [29]. Promoter analysis studies indicate that at least five regulatory elements (i.e., AP-1, Ets/IPU.1, NF1, Stat3, CCAAT/enhancer binding protein) have an impact on MIR21 expression in HL-60 cells and multiple myeloma cells [29, 46]. Of these transcription factors, LH is known to upregulate the expression of three of these transcription factors in mural granulosa cells [47–49]. Indeed, both FOS and JUN, which interact with AP-1 response elements, were shown to be upregulated within 1 h of LH/hCG administration [47, 48]. Distinct AP-1-binding factors, different from those upregulated by LH, are induced in response to FSH (2–8 h after FSH) and may account for FSH-induced upregulation of pri-Mir21 at 12 h after eCG [48]. Similarly, CCAAT/enhancer binding protein (C/EBP) beta upregulation in ovarian granulosa cells is critical for ovulation and occurs almost simultaneously with the increase in MIR21 expression [49], suggesting that it too might play a role in miRNA 21 regulation. Ongoing studies will define the factors involved in LH/hCG induction of pri-Mir21 expression; however, it should be noted that these experiments may require in vivo models because granulosa cells removed from their natural environment and placed into culture exhibit spontaneous expression of Mir21. Moreover, although expression of mature Mir21 followed that of the pri-Mir21 in vivo after the LH/hCG surge (a clear transcriptional response), our in vitro studies are suggestive that Mir21 expression can also be regulated by much more complex mechanisms (i.e., posttranscriptional).
Quantitative RT-PCR analysis of mature Mir21 in cultured granulosa cells failed to show an increase in response to 8-Br-cAMP treatment. This was unexpected, because the other two LH-induced miRNAs, Mir132 and Mir212, exhibited increased expression in cultured granulosa cells after 8-Br-cAMP treatment [22]. Further examination of in vitro granulosa cell pri-Mir21 and mature Mir21 expression indicated that mature Mir21 increased in the absence of a change in transcription (i.e., pri-Mir21). Recently, a similar observation in human smooth muscle cells was observed [50]. In these cells, members of the transforming growth factor superfamily (BMP4, BMP2, and TGFB) increased expression of the precursor and mature MIR21 transcripts without affecting pri-MIR21 expression [50]. The mechanism of action indicated that the ligands activate the signal transducer SMAD1, which in turn recruits DDX5, an RNA helicase p68 subunit of the Drosha microprocessor complex, to the pri-MIR21, thus stimulating the processing of the abundantly present pri-MIR21 transcripts [50]. The disconnection of LH activity and granulosa cell function in cultured granulosa cells is well known. A large body of literature in rodents from the early 1970s describes a phenomenon known as “spontaneous luteinization,” whereby granulosa cells when removed from their follicular environment take on the attributes of luteal cells over a few days in culture [51, 52]. Interestingly, plating of the granulosa cells initiates some of the LH-mediated molecular events occurring in the follicle after LH, such as increases in transcription factors involved in cell survival and cell signaling [53, 54]. The phenomenon of spontaneous luteinization has never fully been explained but is thought to result from a withdrawal of inhibitory signaling molecules once cells are dispersed and placed into culture. In these studies, we show that amounts of mature Mir21 increased in the absence of increased transcription. It is interesting to speculate that Mir21 may be one of several factors that contribute to the phenotypic changes occurring in luteinizing granulosa cells. The fact that most cancer cell lines and tumors also exhibit increased MIR21 expression may be indicative of their disconnection with factors that normally prevent excessive MIR21 accumulation, which is then able to block apoptosis and promote oncogenesis.
Recent in vitro studies in cultured cancer cells, cancer cell lines, and embryonic stem cells suggest that Mir21 expression is important for differentiation and blockade of apoptosis [15, 31, 32, 55, 56]. Here, we show for the first time that Mir21 also plays a role in apoptosis in an endocrine-responsive tissue. During LH-induced terminal differentiation of the granulosa cell, it is necessary to block apoptosis to allow for the formation of a functional corpus luteum [3–5]. We show here that the loss of ovarian Mir21 activity after ovarian bursal injection of a Mir21-blocking LNA oligonucleotide was able to increase apoptosis within ovarian follicles and thus may be important for the prevention of apoptosis during luteinization. Further experiments will be necessary to determine whether LNA-21 is having effects at sites other than the granulosa cell within the ovary and the mechanism that causes the accumulation of the fluorescent nonspecific LNA in the granulosa cells of the follicle. Notably, the loss of Mir21 in the ovary resulted in a reduced ovulation rate (Fig. 9).
Although we were unable to mimic the transcriptional regulation of Mir21 gene expression in cultured granulosa cells, we were able to functionally demonstrate that granulosa cells exhibit apoptosis in vitro after loss of Mir21 action, similar to that seen after in vivo blockade of Mir21 action. To examine the function of Mir21 in cultured granulosa cells, Mir21 action was knocked down in cultured granulosa cells using 2′-O-methyl or LNA oligonucleotides. Consistent with other investigations using these two different types of inhibitors, we observed a greater effect (3-fold) with the LNA oligonucleotide [36, 40, 41]. The increased effectiveness in knockdown was associated with the appearance of the apoptotic phenotypic response associated with loss of Mir21 in other cell types [31, 32]. One possible explanation for this may be that basal Mir21 expression is much higher in granulosa cells compared with cell lines examined by other investigators, and the reduction induced by 2′-O-methyl oligonucleotides was insufficient to knock down Mir21 expression to levels that cause apoptosis. Results similar to ours were recently described in glioblastoma cells, in which only the LNA-21 oligonucleotides caused significant amounts of apoptosis, whereas the 2′-O-methyl oligonucleotides had little effect [36]. However, when we tested lower doses of LNA-21 at a dose that reached an equivalent (i.e., one ninth of basal Mir21 expression) to the 2′-O-methyl, we still detected apoptosis (cleaved caspase 3: Fig. 5). It presently is unclear why similar levels of knockdown of Mir21 would not generate similar phenotypical responses; this is an interesting phenomenon and suggests that blocking miRNA action with different types of oligonucleotides can have dramatic differences in biological responses, a finding with important implications to the therapeutic potential of miRNA blockers.
MicroRNA 21 has been shown to block apoptosis in a number of cell types, and several targets have been identified as directly regulated by Mir21 to mediate its antiapoptotic activity [30, 33–37]. Tropomyosin 1 (TPM1) was the first identified direct MIR21 target transcript [33]. Posttranscriptional regulation of TPM1 by MIR21 caused a 2-fold increase in TPM1 protein in breast cancer cells after MIR21 knockdown; no effect on TPM1 mRNA expression was detected [33]. TPM1, an actin-binding protein, promotes the specialized type of apoptosis known as anoikis by suppressing anchorage-independent growth [57]. PTEN is another proapoptotic protein, which inhibits the AKT and MTOR kinase pathways to cause apoptosis [58]. Knockdown of MIR21 in human hepatocytes caused a 2- to 3-fold increase in PTEN protein but no change on PTEN mRNA [37]. We observed a slight increase in PTEN protein at 4 h after transfection; however, specific tests of Pten regulation by blockade of Pten expression, or masking potential Mir21 sites failed to prevent apoptosis. Programmed cell death 4 (PDCD4) inhibits global cell translation by inhibiting the translation initiation factor 4A [59]. Knockdown of MIR21 in human breast cancer cells causes an increase in PDCD4 mRNA and protein [34] but failed to show any difference in our granulosa cells. Recently, two factors involved in p53 tumor suppression, the p53-activator heterogeneous nuclear ribonucleoprotein K (HNRNPK) and the p53 homologue TRP63, were identified as direct MIR21 targets in glioblastoma cells [36]. However, knockdown of these proteins in combination with MIR21 knockdown failed to rescue the apoptotic phenotype, suggesting that other MIR21 targets are involved in glioblastoma apoptosis [36]. SERPINB5, RECK, and SPRY1 have also been indicated as playing roles in apoptosis, and all have been identified as direct MIR21 targets [30, 33]. Recent findings by a number of laboratories have shown that target transcripts for miRNA are very cell and tissue specific [34, 36].
In this work, we examined the protein levels of six of these known targets (i.e., those we could identify reliable antibodies for) in response to Mir21 knockdown in murine granulosa cells, and none showed any change (Fig. 6). Our studies were unable to demonstrate that any of the known Mir21 target transcripts were factors in mediating apoptosis in granulosa cells. Recently, MIR21 was knocked down in MCF-7 breast cancer cells [60], and knockdown resulted in changes in target proteins, without corresponding changes in mRNA expression. Furthermore, Yang et al. [60] identified 58 putative MIR21 targets in their proteomic analysis. Examination of genes expressed in murine granulosa cells at 0 and 1 h after hCG (microarray analysis, see Carletti and Christenson [47]) indicated that 22 of these 58 genes are expressed in periovulatory granulosa cells. Future studies examining the proteins and gene expression profiles of these miRNA 21 target transcripts will be needed to determine whether any are involved in apoptosis in ovarian granulosa cells.
In conclusion, this study has demonstrated that Mir21 is transcriptionally upregulated in vivo in response to LH, and we show that this upregulation occurs synchronously with a decrease in cleaved caspase 3. In addition, we have provided evidence that both in vivo and in vitro Mir21 knockdown cause an increase in granulosa cell apoptosis. These results suggest that miRNA 21 plays an important role in posttranscriptionally regulating transcripts that are involved in preventing apoptosis in LH-induced terminally differentiating granulosa cells.
Footnotes
Supported by National Institutes of Health (NIH) grants HD051870 and P20 RR016475 from the National Center for Research Resources, a component of the NIH, to L.K.C., and The Madison and Lila Self Graduate Fellowship to M.Z.C.
REFERENCES
- Oonk RB, Krasnow JS, Beattie WG, Richards JS.Cyclic AMP-dependent and -independent regulation of cholesterol side chain cleavage cytochrome P-450 (P-450scc) in rat ovarian granulosa cells and corpora lutea. cDNA and deduced amino acid sequence of rat P-450scc. J Biol Chem 1989; 264: 21934–21942. [PubMed] [Google Scholar]
- Chaffin CL, Stouffer RL.Local role of progesterone in the ovary during the periovulatory interval. Rev Endocr Metab Disord 2002; 3: 65–72. [DOI] [PubMed] [Google Scholar]
- Robker RL, Richards JS.Hormone-induced proliferation and differentiation of granulosa cells: a coordinated balance of the cell cycle regulators cyclin D2 and p27Kip1. Mol Endocrinol 1998; 12: 924–940. [DOI] [PubMed] [Google Scholar]
- Robker RL, Richards JS.Hormonal control of the cell cycle in ovarian cells: proliferation versus differentiation. Biol Reprod 1998; 59: 476–482. [DOI] [PubMed] [Google Scholar]
- Chaffin CL, Schwinof KM, Stouffer RL.Gonadotropin and steroid control of granulosa cell proliferation during the periovulatory interval in rhesus monkeys. Biol Reprod 2001; 65: 755–762. [DOI] [PubMed] [Google Scholar]
- Shao R, Rung E, Weijdegard B, Billig H.Induction of apoptosis increases SUMO-1 protein expression and conjugation in mouse periovulatory granulosa cells in vitro. Mol Reprod Dev 2006; 73: 50–60. [DOI] [PubMed] [Google Scholar]
- Lee J, Park HJ, Choi HS, Kwon HB, Arimura A, Lee BJ, Choi WS, Chun SY.Gonadotropin stimulation of pituitary adenylate cyclase-activating polypeptide (PACAP) messenger ribonucleic acid in the rat ovary and the role of PACAP as a follicle survival factor. Endocrinology 1999; 140: 818–826. [DOI] [PubMed] [Google Scholar]
- Dineva J, Wojtowicz AK, Augustowska K, Vangelov I, Gregoraszczuk EL, Ivanova MD.Expression of atrial natriuretic peptide, progesterone, apoptosis-related proteins and caspase-3 in in vitro luteinized and leptin-treated porcine granulosa cells. Endocr Regul 2007; 41: 11–18. [PubMed] [Google Scholar]
- Ademokun A, Turner M.Regulation of B-cell differentiation by microRNAs and RNA-binding proteins. Biochem Soc Trans 2008; 36: 1191–1193. [DOI] [PubMed] [Google Scholar]
- Gangaraju VK, Lin H.MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol 2009; 10: 116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno MJ, de Castro IP, Malumbres M.Control of cell proliferation pathways by microRNAs. Cell Cycle 2008; 7: 3143–3148. [DOI] [PubMed] [Google Scholar]
- Chivukula RR, Mendell JT.Circular reasoning: microRNAs and cell-cycle control. Trends Biochem Sci 2008; 33: 474–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, He L, Hannon GJ.The guardian's little helper: microRNAs in the p53 tumor suppressor network. Cancer Res 2007; 67: 11099–11101. [DOI] [PubMed] [Google Scholar]
- Jovanovic M, Hengartner MO.miRNAs and apoptosis: RNAs to die for. Oncogene 2006; 25: 6176–6187. [DOI] [PubMed] [Google Scholar]
- Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S.REST maintains self-renewal and pluripotency of embryonic stem cells. Nature 2008; 453: 223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N.miRNA profiling of naive, effector and memory CD8 T cells. PLoS ONE 2007; 2: e1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimi ST, Fulcher JA, Chang MH, Gov L, Wang S, Lee B.MicroRNA profiling identifies miR-34a and miR-21 and their target genes JAG1 and WNT1 in the co-ordinate regulation of dendritic cell differentiation. Blood 2009; 114: 404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF.Semiconserved regulation of mesendoderm differentiation by microRNAs. Dev Cell 2009; 16: 487–488. [DOI] [PubMed] [Google Scholar]
- Yang GH, Wang F, Yu J, Wang XS, Yuan JY, Zhang JW.MicroRNAs are involved in erythroid differentiation control. J Cell Biochem 2009; 107: 548–556. [DOI] [PubMed] [Google Scholar]
- Hong X, Luense LJ, McGinnis LK, Nothnick WB, Christenson LK.Dicer1 is essential for female fertility and normal development of the female reproductive system. Endocrinology 2008; 149: 6207–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja AK, Andreu-Vieyra C, Franco HL, Ma L, Chen R, Han DY, Zhu H, Agno JE, Gunaratne PH, DeMayo FJ, Matzuk MM.Deletion of Dicer in somatic cells of the female reproductive tract causes sterility. Mol Endocrinol 2008; 22: 2336–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler SD, Carletti MZ, Hong X, Christenson LK.Hormonal regulation of microRNA expression in periovulatory mouse mural granulosa cells. Biol Reprod 2008; 79: 1030–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho WC.OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer 2007; 6: 60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese ET, Hanby AM, Speirs V, Hughes TA.Small is beautiful: microRNAs and breast cancer-where are we now? J Pathol 2008; 215: 214–221. [DOI] [PubMed] [Google Scholar]
- Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 2008; 299: 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M.MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg 2008; 12: 2171–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feber A, Xi L, Luketich JD, Pennathur A, Landreneau RJ, Wu M, Swanson SJ, Godfrey TE, Litle VR.MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg 2008; 135: 255–260;.discussion 260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A.SMAD proteins control DROSHA-mediated microRNA maturation. Nature 2008; 454: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, Iba H.miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol 2008; 378: 492–504. [DOI] [PubMed] [Google Scholar]
- Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008; 456: 980–984. [DOI] [PubMed] [Google Scholar]
- Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY.miR-21-mediated tumor growth. Oncogene 2007; 26: 2799–2803. [DOI] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS.MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 2005; 65: 6029–6033. [DOI] [PubMed] [Google Scholar]
- Zhu S, Si ML, Wu H, Mo YY.MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem 2007; 282: 14328–14336. [DOI] [PubMed] [Google Scholar]
- Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH.Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem 2008; 283: 1026–1033. [DOI] [PubMed] [Google Scholar]
- Hu SJ, Ren G, Liu JL, Zhao ZA, Yu YS, Su RW, Ma XH, Ni H, Lei W, Yang ZM.MicroRNA expression and regulation in mouse uterus during embryo implantation. J Biol Chem 2008; 283: 23473–23484. [DOI] [PubMed] [Google Scholar]
- Papagiannakopoulos T, Shapiro A, Kosik KS.MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res 2008; 68: 8164–8172. [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, Patel T.Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology 2006; 130: 2113–2129. [DOI] [PubMed] [Google Scholar]
- Campbell KL.Ovarian granulosa cells isolated with EGTA and hypertonic sucrose: cellular integrity and function. Biol Reprod 1979; 21: 773–786. [DOI] [PubMed] [Google Scholar]
- Otsuka M, Zheng M, Hayashi M, Lee JD, Yoshino O, Lin S, Han J.Impaired microRNA processing causes corpus luteum insufficiency and infertility in mice. J Clin Invest 2008; 118: 1944–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Kauppinen S, Lund AH.LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene 2006; 372: 137–141. [DOI] [PubMed] [Google Scholar]
- Marquez RT, McCaffrey AP.Advances in microRNAs: implications for gene therapists. Hum Gene Ther 2008; 19: 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcieri E, Gobbi P, Zamai L, Vitale M.Ultrastructural features of apoptosis. Scanning Microsc 1994; 8: 653–665. [PubMed] [Google Scholar]
- Earnshaw WC, Martins LM, Kaufmann SH.Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem 1999; 68: 383–424. [DOI] [PubMed] [Google Scholar]
- Matikainen T, Perez GI, Zheng TS, Kluzak TR, Rueda BR, Flavell RA, Tilly JL.Caspase-3 gene knockout defines cell lineage specificity for programmed cell death signaling in the ovary. Endocrinology 2001; 142: 2468–2480. [DOI] [PubMed] [Google Scholar]
- Wickramasinghe NS, Manavalan TT, Dougherty SM, Riggs KA, Li Y, Klinge CM.Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res 2009; 37: 2584–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK, Burger R, Gramatzki M, Blumert C, Bauer K, Cvijic H, Ullmann AK, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood 2007; 110: 1330–1333. [DOI] [PubMed] [Google Scholar]
- Carletti MZ, Christenson LK.Rapid effects of LH on gene expression in the mural granulosa cells of mouse periovulatory follicles. Reproduction 2009; 137: 843–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SC, Richards JS.Regulation of AP1 (Jun/Fos) factor expression and activation in ovarian granulosa cells. Relation of JunD and Fra2 to terminal differentiation. J Biol Chem 2000; 275: 33718–33728. [DOI] [PubMed] [Google Scholar]
- Sterneck E, Tessarollo L, Johnson PF.An essential role for C/EBPbeta in female reproduction. Genes Dev 1997; 11: 2153–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A.SMAD proteins control DROSHA-mediated microRNA maturation. Nature 2008; 454: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channing CP.Influences of the in vivo and in vitro hormonal environment upon luteinization of granulosa cells in tissue culture. Recent Prog Horm Res 1970; 26: 589–622. [DOI] [PubMed] [Google Scholar]
- Channing CP, Ledwitz-Rigby F.Methods for assessing hormone-mediated differentiation of ovarian cells in culture and in short-term incubations. Methods Enzymol 1975; 39: 183–230. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Robayna IJ, Alliston TN, Buse P, Firestone GL, Richards JS.Functional and subcellular changes in the A-kinase-signaling pathway: relation to aromatase and Sgk expression during the transition of granulosa cells to luteal cells. Mol Endocrinol 1999; 13: 1318–1337. [DOI] [PubMed] [Google Scholar]
- Murphy BD.Models of luteinization. Biol Reprod 2000; 63: 2–11. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, Xiao S, Lu H.miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest 2008; 88: 1358–1366. [DOI] [PubMed] [Google Scholar]
- Chen A, Luo M, Yuan G, Yu J, Deng T, Zhang L, Zhou Y, Mitchelson K, Cheng J.Complementary analysis of microRNA and mRNA expression during phorbol 12-myristate 13-acetate (TPA)-induced differentiation of HL-60 cells. Biotechnol Lett 2008; 30: 2045–2052. [DOI] [PubMed] [Google Scholar]
- Raval GN, Bharadwaj S, Levine EA, Willingham MC, Geary RL, Kute T, Prasad GL.Loss of expression of tropomyosin-1, a novel class II tumor suppressor that induces anoikis, in primary breast tumors. Oncogene 2003; 22: 6194–6203. [DOI] [PubMed] [Google Scholar]
- Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada KM.Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science 1998; 280: 1614–1617. [DOI] [PubMed] [Google Scholar]
- Yang HS, Jansen AP, Komar AA, Zheng X, Merrick WC, Costes S, Lockett SJ, Sonenberg N, Colburn NH.The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol 2003; 23: 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Chaerkady R, Beer MA, Mendell JT, Pandey A.Identification of miR-21 targets in breast cancer cells using a quantitative proteomic approach. Proteomics 2009; 9: 1374–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]