Abstract
Background
The infectious and diagnostic form of Entamoeba histolytica (Eh), cause of amebic dysentery and liver abscess, is the quadranucleate cyst. The cyst wall of Entamoeba invadens (Ei), a model for Eh, is composed of chitin fibrils and three sets of chitin-binding lectins that cross-link chitin fibrils (multivalent Jacob lectins), self-aggregate (Jessie lectins), and remodel chitin (chitinase). The goal here was to determine how well the Ei model applies to Entamoeba cysts from humans.
Methods/Results
An Eh Jacob lectin (EhJacob2) has three predicted chitin-binding domains surrounding a large, Ser-rich spacer. Recombinant EhJacob2 made in transfected Eh trophozoites binds to particulate chitin. Sequences of PCR products using primers flanking the highly polymorphic spacer of EhJacob2 may be used to distinguish Entamoeba isolates. Antibodies to the EhJacob2, EhJessie3, and chitinase each recognize cyst walls of clinical isolates of Entamoeba. While numerous sera from patients with amebic intestinal infections and liver abscess recognize recombinant EhJacob1 and EhJessie3 lectins, few of these sera recognize recombinant EhJacob2.
Conclusions/Significance
The EhJacob2 lectin binds chitin and is polymorphic, and Jacob2, Jessie3, and chitinase are present in cyst walls of clinical isolates of Entamoeba. These results suggest there are substantial similarities between cysts of the human pathogen (Eh) and the in vitro model (Ei), even though there are quantitative and qualitative differences in their chitin-binding lectins.
Author Summary
For many years, we and others have used cysts of Entamoeba invadens (Ei), a reptilian parasite, to model the infectious and diagnostic cysts of the human pathogen Entamoeba histolytica (Eh). The Ei cyst wall is composed of chitin fibrils, as well as Jacob and Jessie lectins that have unique chitin-binding domains. Our recent results suggest a “wattle and daub” model of the Ei cyst wall, where the wattle or sticks (chitin fibrils bound by multivalent Jacob lectins) is constructed prior to the addition of the mortar or daub (self-aggregating Jessie3 lectins). Here we “humanize” the Ei model of the cyst wall with four findings. First, a recombinant Eh Jacob2 lectin, which has three predicted chitin-binding domains surrounding a large spacer domain, binds chitin beads. Second, polymorphisms in the spacer domain of EhJacob2 discriminate clinical isolates of Entamoeba. Third, chitinase, Jacob2 lectin, and Jessie3 lectin are present in cyst walls of clinical isolates of Entamoeba. Finally, numerous sera from patients infected with Entamoeba recognize recombinant Eh Jacob1 and Jessie3 lectins.
Introduction
The infectious and diagnostic stage of Entamoeba histolytica (Eh), the cause of amebic dysentery and liver abscess, is a quadranucleate cyst [1], [2]. Eh is morphologically indistinguishable from Entamoeba dispar (Ed), a human commensal that does not cause disease [3]. Because Eh does not readily encyst in axenic culture, we have studied cyst walls formed in vitro by Entamoeba invadens (Ei) that infects reptiles [4], [5].
The Ei cyst wall is composed of chitin (a homopolymer of β-1,4-linked GlcNAc) and three unique sets of chitin-binding lectins called Jacob, Jessie, and chitinase [6], [7]. Ei Jacob lectins contain 3 to 6 tandemly arranged chitin-binding domains (CBDs), each of which contains six Cys residues (see Table S1 for a list of database accession numbers and a brief description of each protein). Spacer regions between CBDs of Ei Jacob lectins contain sites for cleavage by Cys proteases, as well as Ser residues that are modified by O-phosphodiester-linked glycans [7]. Ei Jessie lectins and chitinase each contain an N-terminal CBD, which contains eight Cys residues [8]–[10]. Ei Jessie3 lectins contain a self-aggregating domain that forms the mortar or daub between chitin fibrils [11].
As Eh cysts are difficult to obtain from patient stool in quantity, we have predicted components of the cyst wall from the whole genome sequence of Eh [2], [12]. An Eh Jacob lectin (EhJacob1) that has two CBDs binds chitin when expressed as a recombinant protein in transfected Eh trophozoites (Table S1) [10]. Similarly, the N-terminal CBDs of Eh chitinase, Jessie2, and Jessie3 each bind chitin [10]. The Eh chitinase, chitin synthase, and chitin deacetylases each have the expected activities when expressed as recombinant proteins in bacteria or yeast [8], [13], [14]. Messenger RNAs for chitinases, Jessie lectins, and Jacob lectins are expressed by Eh encysting in xenic culture [15].
A low complexity spacer region between the CBD and enzymatic domain of Eh and Ed chitinases contains a series of heptapeptide repeats that are polymorphic among clinical isolates [16], [17]. Polymorphic tandem repeats have also been observed in the Ser-rich Eh protein (SREHP or K2 antigen) [16]–[19]. While abundant and immunogenic Eh trophozoite proteins such as the Gal/GalNAc lectin and SREHP are immunogenic and are therefore vaccine candidates [20]–[24], little is known about the immunogenicity of Eh cyst wall proteins.
In an effort here to test how well the Ei cyst model fits the human pathogen Eh, we characterized here a second Eh Jacob lectin (EhJacob2: EHI_044500; see Table S1) that contains three predicted CBDs separated by a long, Ser-rich spacer similar to those present in EiJacob6 and EiJacob7) [7]. Questions asked included the following:
Does the EhJacob2 lectin bind chitin?
Is the low complexity spacer of EhJacob2 polymorphic from isolate to isolate?
Are Jacob2, Jessie3, and chitinase present in cyst walls of clinical isolates of Entamoeba?
Do human anti-amebic sera recognize recombinant Eh cyst wall proteins?
Materials and Methods
Ethics statement
Culture and manipulation of Entamoeba, including production of cysts in vitro and handling of cysts from patient samples, has been has been approved by the Boston University Institutional Biosafety Committee (BU IBC). Similarly, recombinant expression of Entamoeba proteins in bacteria has been approved by the BU IBC. Rabbit antibodies were made using approved protocols from the BU IACUC. An exemption has been received from the Boston University IRB for de-identified patient sera and for de-identified stool samples containing Entamoeba cysts. Patient sera, all of which bound to Gal/GalNAc lectin, came from five individuals with amebic liver abscess and five individuals with intestinal amebiasis. All of these serum samples, which were de-identified, were collected prior to the initiation of these studies. The Ethical Review Committee of the International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR,B) and the Human Investigation Committee of the University of Virginia reviewed and approved the design of the previous study under which these samples were obtained.
Identification of Eh and Ed Jacob2 lectins
Eh and Ed Jacob2 lectins were identified in BLASTP searches of the NR database at NCBI or at AmoebaDB using the EhJacob1 sequence (see Table S1 for accession numbers) [10], [12], [25]. N-terminal signals and transmembrane helices were predicted using Phobius [26].
Analysis of Jacob2 gene polymorphisms
Genomic DNA from axenic Eh strains (HM-1:IMSS, HK-9, 200:NIH, and SD157) was isolated using the Wizard Genomic DNA Purification Kit (Promega). DNA from an axenic strain of Ed (SAW760) was a generous gift from Graham Clark. DNAs from numerous clinical isolates of Eh were a generous gift from Egbert Tannich. PCR primers flanking the Ser-rich region between the second and third CBDs of Jacob 2 were designed from sequences that were identical in the Eh and Ed genomic sequences. The sense primer (GCTGATGGATTCTACTGTGTT) encoded the heptapeptide (ADGFYCV). The anti-sense primer (ACAGAAAAGACCATCTTGAGT) was anti-sense to heptapeptide (TQDGLFC). In the Eh genome project strain HM-1:IMSS the predicted product was 1260-nt long [12]. PCR was performed for 35 cycles of 30 sec at 94°C, 30 sec at 50°C, and 3 min at 72°C using the PCR Master Mix system (Promega). Amplified products were analyzed using a 0.8% agarose gel in 1× Tris-acetate-EDTA (TAE) buffer. Selected PCR products were cloned into a TA-vector and sequenced from both ends.
Expression of EhJacob2 in transfected amebae
The entire coding region of the EhJacob2 gene (1722 nt encoding a 574-aa protein) was PCR amplified from HM-1:IMSS strain gDNA using the Expand High Fidelity PCR system (Roche). The sense primer (GCGGTACC ATGAAACAACTTATATTAGCA) began at the start codon (italic) and included a KpnI site (underline). The anti-sense primer (GCGGATCC TTA TAAATCTTCTTCTGAAATTAATTTTTGTTCCTTGTTTTCATTGTTATTATT ) included a BamHI site (single underline) and was anti-sense to the 3′ end of the coding region of EhJacob2 (bold underline). This primer was anti-sense to a c-myc sequence (bold) and to a stop codon (italic). This product was cloned into the pJST4 vector [27] between the 5′ and 3′ untranslated regions of the Eh actin gene, and this construct was used to transfect HM-1:IMSS trophozoites. Transfected Eh trophozoites were lysed by incubation in lysis/wash buffer (20 mM Tris-HCl, pH 8.0; 1 M NaCl, 0.1% Triton X-100) plus 250 µM E64 for 1 hr on ice. The lysate was centrifuged for 1 min at maximum speed in a microcentrifuge, and the supernatant was incubated with chitin beads (New England Biolabs) for 1 hr at room temperature. Unbound material was then removed, and the beads were washed 5 times in lysis/wash buffer. Bound material was removed by boiling the beads for 5 min in SDS buffer (50 mM Tris-HCl, pH 6.8; 2% SDS; 5% 2-mercaptoethanol, 5% glycerol).
Protein samples were analyzed by SDS-PAGE on 4–20% Tris-glycine gels. After electrophoresis, proteins were stained with Coomassie Blue or blotted onto nitrocellulose. EhJacob2 was detected on the blots using an anti-c-myc antibody (Invitrogen) followed by a peroxidase-conjugated goat anti-mouse IgG antibody (Jackson ImmunoResearch). Bound antibodies were detected with the LumiGLO chemiluminescent substrate (KPL).
Expression of Eh cyst wall proteins in bacteria and production of rabbit antibodies
The region of the EhJacob1 gene encoding a 53-aa C-terminal 6-Cys CBD, which begins with VNCTEVKE and ends with the stop codon, was PCR amplified from Eh DNA. The sense primer (CGGGATCCGTCAATTGTACTGAAGTGAAAGAA) had a BamHI site at the 5′-end (underline). The anti-sense primer (CCCAAGCTT TTA GTGGTGGTGGTGGTGGTGATAACATGGATTGTTATAAC ), which had a 5′ HindIII site (underlined), was anti-sense to a stop codon (italics), a polyHis tail (bold), and the 3′ end of the coding region of the EhJacob1 gene (bold underline).
The coding region of the EhJacob2 gene (minus the first 48 nt that encode the N-terminal 16-aa-long signal peptide and minus 24-aa at the C-terminus) was PCR amplified from Eh DNA. The sense primer (GGGTACCTAATGGTATACCCAACTGGATGTAAGAAGAAA) had a Kpn1 site at the 5′-end (underline) and encoded the peptide (VYPTGCKKK) that is C-terminal to the predicted cleavage site in EhJacob2 for the signal peptidase [26]. The anti-sense primer ( GGATCC TTA GTGGTGGTGGTGGTGGTGGTATTGGTAAGGACCTTCTTGT ), which had a BamH1 site (underline), was anti-sense to a stop codon (italics), a polyHis tail (bold), and the 3′ end of the coding region of the EhJacob2 gene (bold underline). The EhJacob1 and EhJacob2 PCR products were cloned into pMAL-p2E (New England Biolabs), using the same methods we used to clone Eh Jessie3 into this vector [11]. Maltose-binding protein (MBP) fusions containing the Eh cyst wall lectins were expressed in E. coli (Bl21-DE3 strain) using IPTG induction and an amylose resin (NEB) for purification. The purity of these recombinant proteins was checked on SDS-PAGE.
The 363-aa long catalytic domain of Eichitinase1, which begins with the peptide (KVVSYYT) was amplified from Ei DNA using a sense primer ( GGATCCATGAAGGTTGTCTCGTATTACACC) that had a 5′ KpnI site (underline). The anti-sense primer ( CTCGAG TTA GCAACCGATCAAGCTCTTTC ) had a XhoI site (underline) and was anti-sense to a stop codon (italic) and the C-terminal peptide (KKELDQC) (bold underline). The Eichitinase1 PCR product was cloned into the pQE30 vector (Qiagen) and expressed in M15 strain of E. coli that contains a lac repressor-expressing plasmid (pREP4). Recombinant Eichitinase1, which contains a C-terminal polyHis tag, was induced with IPTG and purified on Ni-NTA agarose beads.
Mono-specific polyclonal rabbit antibodies to amylose resin-purified MBP-EhJacob1 and MBP-EhJacob2 were made at Strategic Biosolutions, using methods similar to that used previously to make a rabbit anti-EhJessie3 antibody [11]. Prior to their use in microscopy, rabbit antibodies were purified using MBP-EhJacob1 or MBP-EhJacob2 fusion-proteins chemically coupled to agarose. Similar methods were used to raise a mono-specific rabbit antibody to the catalytic domain of Eichitinase1. This antibody cross-reacts with the catalytic domain of Ehchitinase1.
Binding of anti-cyst wall lectin antibodies to Entamoeba cysts in stool samples
Approximately 2 to 3 grams of stool sample from patients infected with Entamoeba (in Kharagpur, India) were emulsified in 10 ml of chilled phosphate-buffered saline (PBS) and then passed through a mesh to remove the larger particles from the materials. Each sample was washed with ice cold PBS by centrifugation at 5000 rpm for 5 min thrice, and the pellet was resuspended in 1 ml PBS. The presence of Entamoeba cyst in sample was confirmed by iodine staining and light microscopy or by calcofluor white staining and epifluorescent microscopy.
To localize the Jacob2, chitinase and Jessie3 in the Eh cyst wall, we fixed stool samples fixed with 2% paraformaldehyde for 15 min at room temperature and washed three times with PBS. Fixed samples were incubated for two hrs with 1∶200 dilutions of rabbit anti-EhJacob2, anti-Eh Jessie3, and anti-chitinase (catalytic domain) antibodies (described above). Samples were washed with PBS and then incubated with TRITC-conjugated goat anti-rabbit antibody (1∶500 dilution) for 1 hr. Secondary antibody alone was used as negative control. Samples were again washed with PBS and examined with a FV1000 confocal microscope (Olympus). Images were captured by FV10-ASW 1.6 viewer and processed with Adobe photoshop CS3.
Finally, xenic cysts of Eh, which were incubated with anti-Jacob2 and anti-Jessie3 antibodies, were a generous gift of Upinder Singh [15].
Binding of anti-amebic sera to Western blots of recombinant cyst wall lectins
For Western blotting, ∼2 µg each of MBP, MBP-EhJacob1, MBP-EhJacob2 and MBP-EhJessie3 were separated in 4–16% gradient SDS-PAGE (Invitrogen, USA) and transferred to a PVDF membrane by semi-dry method. Blotted membranes were incubated with each patient's sera (1∶10 dilution) (Dacca, Bangladesh) on a rocker overnight at 4°C. Membranes were washed three times for 15 min with PBS-Tween 20 and then incubated with HRP-conjugated anti-human antibody (Sigma) (1∶2000 dilution) for 1 hr. Bound antibody was detected using Super Signal West Pico Chemiluminescent kit (Pierce), as per manufacturer's instruction. The strength of bound antibodies was qualitatively scored as no signal (−), barely detectable signal (+/−), weak signal (+), stronger signal (++), and strongest signal (+++).
Results and Discussion
The EhJacob2 lectin has a large Ser-rich spacer
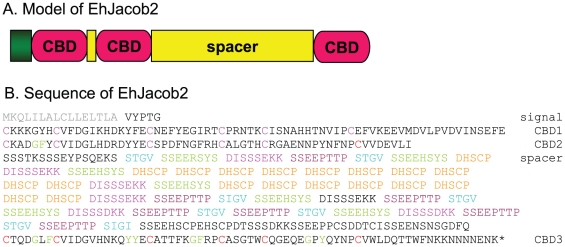
Eh has only two predicted Jacob lectins [2], [10], [12]. EhJacob1, which we previously characterized [10], is present in three nearly identical copies in the genome (see Table S1). EhJacob1, which contains two CBDs, is 151-amino acids long, has a predicted molecular weight of 17377 daltons, and has a predicted pI of 5.2. In contrast, the EhJacob2 lectin, which contains three predicted CBDs, is 574-amino acids long, has a predicted molecular weight of 62862 daltons, and has a predicted pI of 4.65 (Figs. 1A and 1B). The first two predicted CBDs of EhJacob2 are separated from the third CBD by a large spacer domain, which is 30% Ser. Large Ser-rich spacer domains are also present in minor components of the Ei cyst wall (EiJacob6 and EiJacob7) and in chitin-binding proteins of insects (peritrophins) that are present in the wall surrounding the blood meal [7], [28]. Large Ser-rich domains in EhJacob2 suggest the possibility of numerous O-phosphodiester-linked glycans, as demonstrated in Ei Jacob lectins [7]. In contrast, there are no sites for Asn-linked glycosylation in EhJacob2 [29]. Within the spacer domain of EhJacob2 are numerous short repeats that are polymorphic (see next section). These repeats include sequences (e.g. TTPSTGV) that resemble sites for cleavage by Cys proteases in Ei Jacob lectins (TTPVD) [7].
Figure 1. Eh Jacob2 has three chitin-binding domains (CBDs) surrounding a large Ser-rich spacer.
A. EhJacob2 has an N-terminal signal peptide, three CBDs, and a large spacer between the first two CBDs and the last CBD. EhJacob2 has no transmembrane helix or GPI-anchor. B. Sequence of EhJacob2 where signal peptide (grey) and Cys residues (red) within CBDs are highlighted. Also highlighted are short repeats in the spacer, which fall into five families: A (light blue), B (green), C (pink), D (purple), and E (orange). Polymorphisms in these repeat families are further described in Fig. 4. The Ed Jacob2 is shown in Fig. S1.
The predicted Jacob2 from the commensal parasite Ed (EDI_246160) is 743-amino acids long and contains three CBDs that closely resemble those of EhJacob2 (Fig. S1 and Table S1). In contrast, the Ser-rich domain of EdJacob2 contains numerous repeats (marked in bold letters in Fig. S1) that are distinct from those of EhJacob2.
The EhJacob2 lectin binds chitin
To determine if EhJacob2 is a chitin-binding lectin, EhJacob2 was expressed with a myc-tag at its C-terminus in transfected Eh trophozoites [10], [27]. A total lysate from transfected Eh was incubated with chitin beads, and unbound proteins (the vast majority) were removed (Fig. 2A). A single, ∼78-kDa protein binds to the chitin beads. A Western blot using an anti-myc antibody confirmed that this chitin-binding protein is the recombinant EhJacob2 protein (Fig. 2B). In control non-transfected E. histolytica trophozoites, no proteins bind to chitin beads (data not shown).
Figure 2. EhJacob2 is a chitin-binding protein.
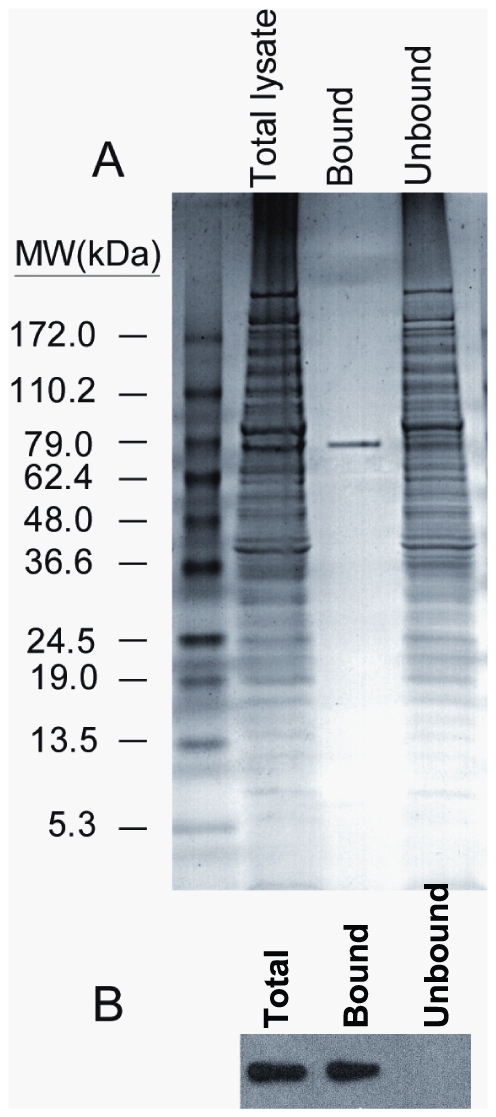
A. Coomassie blue-stained SDS-PAGE gel showing a total lysate of Eh trophozoites transfected with c-myc tagged EhJacob2, the fraction that binds chitin beads, and the fraction that does not bind chitin. B. Western blot confirms the chitin binding of EhJacob2, which is detected with anti-c-myc antibodies and chemiluminescence.
The Ser-rich domain of EhJacob2 is highly polymorphic
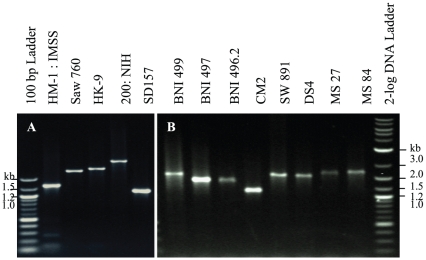
We hypothesized that the Ser-rich region of EhJacob2 might be polymorphic because similar low complexity regions containing internal repeats in Entamoeba SREHP and chitinase are polymorphic [16]–[19]. EhJacob2 PCR products from DNA of axenized strains of Eh (HM-1:IMSS, HK-9, and 200:NIH), one clinical isolate (SD157), and axenized Ed strain (SAW760) range in size from 1.1 kb to 2.3 kb (Fig. 3A). EhJacob2 PCR products also range in size from clinical isolates of Eh (Fig. 3B).
Figure 3. Ser-rich domains of EhJacob2 are polymorphic.
Amplification products were generated using PCR primers flanking the Ser-rich region between the second and third chitin-binding domains of Jacob2. A. Jacob2 PCR products from axenized Eh isolates (HM-1:IMSS, HK-9, 200:NIH, and SD157) and Ed isolate (SAW760) have distinct mobilities on agarose gels. B. Jacob2 PCR products from clinical Eh isolates also have distinct mobilities.
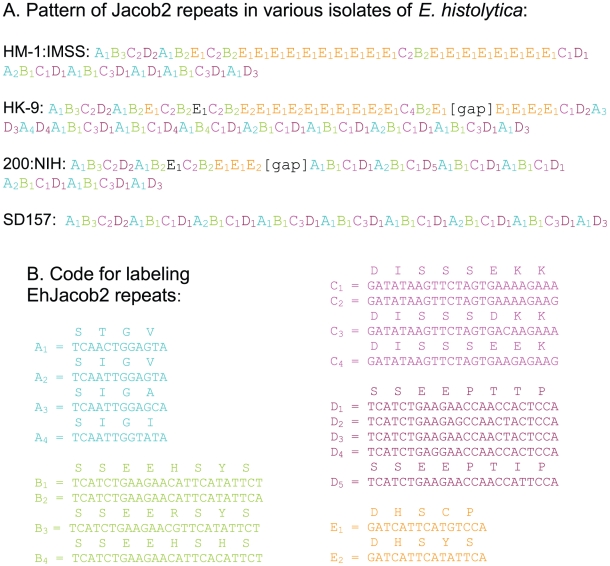
Selected EhJacob2 PCR products were cloned and sequenced at both ends, and five groups of repetitive elements in the Ser-rich spacer were coded (A to E in Fig. 4), using methods to describe Entamoeba chitinase and Ser-rich protein repeats [16]. Nucleotide differences within groups included both silent and non-silent changes. While the repetitive elements differ among the four isolates examined, there are some similarities. For example, the repetitive regions all start with A1B3C2D2A1 and end with D1A1B1C4D1A1D3. Blocks of ABCD are common, and HM-1:IMSS, HK-9, and 200:NIH all have CB followed by variable numbers of E. In contrast, E repeats did not occur in the SD157 sequence.
Figure 4. EhJacob2 PCR products are distinct for each axenized strain.
A. Coded representations of EhJacob2 repeats from PCR products shown in Fig. 3. Complete sequences were obtained for HM-1:IMSS and SD157. Gaps in the middle of sequences in the HK-9 and 200:NIH products are marked. B. Five EhJacob2 repeats are each assigned a letter (A to E) and a color (as described in Fig. 1). The nucleotide sequences coding for each repeat are numbered in the order of their frequency of occurrence in the sequenced products.
Antibodies to the EhJacob2, EhJessie3, and chitinase each recognize cyst walls of clinical isolates of Entamoeba
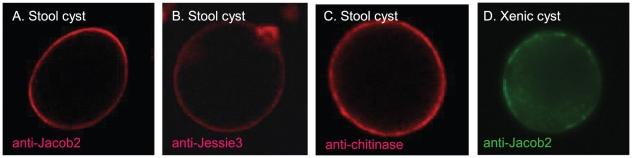
Previously we made polyclonal, mono-specific rabbit antibodies to recombinant EiJacob1 and EhJessie3 and to a heptapeptide repeat present in the spacer domain of the Ei chitinase [11]. Sequential binding of these antibodies to cysts of Ei was used to develop the “wattle and daub” model of Ei cyst wall formation [11]. Here we made polyclonal, mono-specific rabbit antibodies to the entire EhJacob2 and the catalytic domain of Ei chitinase (that is nearly identical to catalytic domains of Eh and Ed chitinases). Antibodies to EhJacob2, EhJessie3, and the catalytic domain of Entamoeba chitinases do not bind to Eh trophozoites but bind to cysts of Entamoeba isolated from patient stools (Fig. 5). Because these cysts were not characterized by molecular methods, we do not know whether they are composed of Eh, Ed, or both. Because the Eh and Ed sequences for Jacob2, Jessie3, and chitinases are so similar (Table S1), we assume but have not proven that antibodies to these cyst wall proteins react with cysts of both Eh and Ed. In addition, anti-EhJacob2 antibodies but not anti-EhJessie3 antibodies bind to Eh cysts made in xenic culture [15].
Figure 5. Antibodies to EhJacob2, Jessie3, and chitinase bind to Entamoeba cysts isolated from patient stools.
A to C. Confocal micrographs of stool cysts detected with rabbit antibodies to Eh Jacob2, Jessie3, or chitinase. D. Confocal micrograph of an Eh cyst from xenic culture labeled with antibodies to EhJessie3. Thanks to Upinder Singh for the micrograph shown in D.
Anti-amebic sera recognize to varying degrees recombinant EhJacob1, EhJacob2, and EhJessie3 lectins
The idea here was to determine whether sera from patients with liver abscess or amebic intestinal infection, each of which recognizes the Gal/GalNAc lectin of Eh trophozoites [22], [23], also recognize recombinant Eh cyst wall proteins on Western blots. MBP alone was used as negative control. While 9 of 10 human anti-amebic sera recognized EhJessie3, 6 of 10 sera recognized EhJacob1 (Table 1). In contrast, just 2 of 10 sera bound to EhJacob2, suggesting EhJacob2 may be less antigenic than the other Eh cyst wall lectins.
Table 1. Binding of human anti-amebic sera to recombinant Eh cyst wall lectins.
| Sl. | Sera Number | Jessie 3* | Jacob1 | Jacob2 |
| 1 | LAI 09 | + | +++ | −− |
| 2 | LAI 12 | + | + | −− |
| 3 | LAI 28 | ++ | −− | −− |
| 4 | LAI 30 | + | +++ | + |
| 5 | LAI 43 | ++ | + | −− |
| 6 | 041 | + | + | + |
| 7 | 163 | + | −− | −− |
| 8 | 1022 | +/− | −− | −− |
| 9 | 1028 | + | ++ | −− |
| 10 | 3040 | −− | −− | −− |
*The strength of bound antibodies was qualitatively scored as no signal (−), barely detectable signal (+/−), weak signal (+), stronger signal (++), and strongest signal (+++).
Major conclusions and unresolved questions
The results here and elsewhere generally support the idea that Ei is a good model for Eh cysts:
Eh Jacob lectins have a similar structure to those described for Ei, and both bind chitin when expressed as recombinant proteins (Figs. 1 and 2) [10]. As an aside, EhJacob2 shows the best expression of any protein we have tried to overexpress in transfected trophozoites. Whether Eh Jacob lectins have post-translational addition of O-phosphodiester-linked glycans to Ser in the spacer domains and cleavage by endogenous Cys proteases, as shown for Ei [7], cannot be determined using the present experimental strategy. Whether differences in the repetitive elements of EhJacob2 and EdJacob2 (Fig. S1) can be exploited for diagnostic purposes also remains to be determined.
The major components of the Ei cyst wall (Jacob lectins, Jessie lectins, and chitinase), all of which contain unique CBDs, are also present in Entamoeba cyst walls of clinical isolates (Fig. 5) [6]–[11]. The finding that Eh cysts from xenic cultures bind anti-Jacob antibodies but not anti-Jessie3 antibodies suggests the possibility that the in vitro cysts may have an incompletely assembled wall [15]. This is because in the Ei model, Jacob lectins are added to cyst walls prior to addition of Jessie lectins [11].
Eh Jacob lectins, Jessie3 lectins, and chitinase are immunogenic in rabbits [6], [11], and it appears that EhJessie3 and EhJacob1 are immunogenic in some persons infected with Entamoeba (Table 1). Whether the immune response to Entamoeba cyst wall lectins inhibits encystation or excystation and so has an effect on transmission of cysts from person to person is interesting but cannot be determined from this data. In contrast, a mucosal IgA anti-lectin antibody response is associated with immune protection against Eh colonization in Bangladeshi children [22], [23].
Differences between Eh and Ed cysts and cysts of Ei include the failure of Eh or Ed to encyst in axenic culture using the conditions that cause Ei to encyst [4], [15]. Ei has seven distinct Jacob lectin genes rather than two present in Eh and Ed (Table S1) [2], [7], [10], [12]. Eh and Ed each has a single chitinase with a C-terminal CBD, while Ei has three chitinases with an N-terminal CBD and two chitinases that have no CBD [8], [9]. Eh and Ed each has a single Jessie3 lectin, while Ei has two Jessie3 lectins [7], [10].
Finally, it appears that EhJacob2 genes are at least as polymorphic as SREHP genes and are more polymorphic than chitinase genes [16]–[19]. These results support the general idea that polymorphisms in surface proteins that contain repetitive elements of Entamoeba, Cryptosporidium (e.g. gp40/15), and Plasmodium (e.g. merozoite and circumsporozoite antigens) may be used to distinguish clinical isolates [30]–[32]. The EhJacob2 polymorphisms may complement other methods such as tRNA gene-linked tandem repeats for finger-printing clinical isolates of Eh [33], [34].
Supporting Information
Ed Jacob2 differs from Eh Jacob2 primarily in the large Ser-rich spacer. Sequence of EdJacob2 (Table S1) where the signal peptide (grey) and Cys residues (red) within CBDs are highlighted (see Fig. 1 for comparison to EhJacob2). Also highlighted are short repeats in the spacer, which fall into five families: A (light blue), B (green), C (pink), D (purple), and E (orange). Differences between the sequence of EdJacob2 and EhJacob2 are marked in bold letters. Because the number and arrangement of these short repeats differs between EdJacob2 and EhJacob2, it was not possible to directly align the two sequences.
(2.68 MB EPS)
Entamoeba proteins with chitin-binding domains (CBDs).
(0.05 MB DOC)
Acknowledgments
Thanks to Graham Clark, Upinder Singh, and Egbert Tannich for Eh cysts or DNA from clinical isolates of Eh.
Footnotes
The authors have declared that no competing interests exist.
This work was supported in part by National Institutes of Health (NIH) grants AI44070 (J.S.) and GM31318 (P.W.R.) and by grants from the Government of India (IMCR to S.K.G. and CSIR to TD). Collection of patient sera was supported in part by an NIH grant to William Petri. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Haque R, Huston CD, Hughes M, Houpt E, Petri WA., Jr Amebiasis. N Engl J Med. 2003;348:1565–1573. doi: 10.1056/NEJMra022710. [DOI] [PubMed] [Google Scholar]
- 2.Clark CG, Alsmark UC, Tazreiter M, Saito-Nakano Y, Ali V, Marion S, et al. Structure and content of the Entamoeba histolytica genome. Adv Parasitol. 2007;65:51–190. doi: 10.1016/S0065-308X(07)65002-7. [DOI] [PubMed] [Google Scholar]
- 3.Diamond LS, Clark CG. A redescription of Entamoeba histolytica Schaudinn, 1903 (emended Walker, 1911) separating it from Entamoeba dispar Brumpt, 1925. J Eukaryot Microbiol. 1993;40:340–344. doi: 10.1111/j.1550-7408.1993.tb04926.x. [DOI] [PubMed] [Google Scholar]
- 4.Eichinger D. Encystation in parasitic protozoa. Curr Opin Microbiol. 2001;4:421–426. doi: 10.1016/s1369-5274(00)00229-0. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Samuelson J, Clark CG, Eichinger D, Paul J, et al. Gene discovery in the Entamoeba invadens genome. Mol Biochem Parasitol. 2003;129:23–31. doi: 10.1016/s0166-6851(03)00073-2. [DOI] [PubMed] [Google Scholar]
- 6.Frisardi M, Ghosh SK, Field J, Van Dellen K, Rogers R, et al. The most abundant glycoprotein of amebic cyst walls (Jacob) is a lectin with five Cys-rich, chitin-binding domains. Infect Immun. 2000;68:4217–4224. doi: 10.1128/iai.68.7.4217-4224.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Dellen KL, Chatterjee A, Ratner DM, Magnelli PE, Cipollo J, et al. Unique posttranslational modifications of chitin-binding lectins of Entamoeba invadens cyst walls. Eukaryotic Cell. 2006;5:836–848. doi: 10.1128/EC.5.5.836-848.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de la Vega H, Specht CA, Semino CE, Robbins PW, Eichinger D, et al. Cloning and expression of chitinases of Entamoebae. Mol Biochem Parasitol. 1997;85:139–147. doi: 10.1016/s0166-6851(96)02817-4. [DOI] [PubMed] [Google Scholar]
- 9.Dey T, Basu R, Ghosh SK. Entamoeba invadens: cloning and molecular characterization of chitinases. Exp Parasitol. 2009;123:244–249. doi: 10.1016/j.exppara.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 10.Van Dellen K, Ghosh SK, Robbins PW, Loftus B, Samuelson J. Entamoeba histolytica lectins contain unique 6-Cys or 8-Cys chitin-binding domains. Infect Immun. 2002;70:3259–3263. doi: 10.1128/IAI.70.6.3259-3263.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterjee A, Ghosh SK, Jang K, Bullitt E, Moore L, et al. Evidence for a “wattle and daub” model of the cyst wall of entamoeba. PLoS Pathog. 2009;5:e1000498. doi: 10.1371/journal.ppat.1000498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 13.Das S, Van Dellen K, Bulik D, Magnelli P, Cui J, et al. The cyst wall of Entamoeba invadens contains chitosan (deacetylated chitin). Mol Biochem Parasitol. 2006;48:86–92. doi: 10.1016/j.molbiopara.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 14.Van Dellen KL, Bulik D, Specht C, Robbins PW, Samuelson J. Heterologous expression of an Entamoeba histolytica chitin synthase in Saccharomyces cerevisiae. Eukaryotic Cell. 2006;5:203–206. doi: 10.1128/EC.5.1.203-206.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehrenkaufer GM, Haque R, Hackney JA, Eichinger DJ, Singh U. Identification of developmentally regulated genes in Entamoeba histolytica: insights into mechanisms of stage conversion in a protozoan parasite. Cell Microbiol. 2007;9:1426–1444. doi: 10.1111/j.1462-5822.2006.00882.x. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh SK, Frisardi M, Ramierez-Avila L, Descoteaux S, Sturm-Ramirez K, et al. Molecular epidemiology of Entamoebae: Evidence of a bottleneck (demographic sweep) and transcontinental spread of diploid parasites. J Clin Microbiol. 2000;38:3815–3821. doi: 10.1128/jcm.38.10.3815-3821.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haghighi A, Kobayashi S, Takeuchi T, Masuda G, Nozaki T. Remarkable genetic polymorphism among Entamoeba histolytica isolates from a limited geographic area. J Clin Microbiol. 2002;40:4081–4090. doi: 10.1128/JCM.40.11.4081-4090.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohler S, Tannich E. A family of transcripts (K2) of Entamoeba histolytica contains polymorphic repetitive regions with highly conserved elements. Mol Biochem Parasitol. 1993;59:49–58. doi: 10.1016/0166-6851(93)90006-j. [DOI] [PubMed] [Google Scholar]
- 19.Clark CG, Diamond LS. Entamoeba histolytica: a method for isolate identification. Exp Parasitol. 1993;77:450–455. doi: 10.1006/expr.1993.1105. [DOI] [PubMed] [Google Scholar]
- 20.Mann BJ, Torian BE, Vedvick TS, Petri WA., Jr Sequence of a cysteine-rich galactose-specific lectin of Entamoeba histolytica. Proc Natl Acad Sci USA. 1991;88:3248–3252. doi: 10.1073/pnas.88.8.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang T, Cieslak PR, Stanley SL., Jr Protection of gerbils from amebic liver abscess by immunization with a recombinant Entamoeba histolytica antigen. Infect Immun. 1994;62:1166–1170. doi: 10.1128/iai.62.4.1166-1170.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haque R, Ali IM, Sack RB, Farr BM, Ramakrishnan G, et al. Amebiasis and mucosal IgA antibody against the Entamoeba histolytica adherence lectin in Bangladeshi children. J Infect Dis. 2001;183:1787–1793. doi: 10.1086/320740. [DOI] [PubMed] [Google Scholar]
- 23.Haque R, Mondal D, Duggal P, Kabir M, Roy S, et al. Entamoeba histolytica infection in children and protection from subsequent amebiasis. Infect Immun. 2006;74:904–909. doi: 10.1128/IAI.74.2.904-909.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaudhry OA, Petri WA., Jr Vaccine prospects for amebiasis. Expert Rev Vaccines. 2005;4:657–668. doi: 10.1586/14760584.4.5.657. [DOI] [PubMed] [Google Scholar]
- 25.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Käll L, Krogh A, Sonnhammer ELL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 27.Ghosh SK, Lohia A, Kumar A, Samuelson J. Overexpression of P-glycoprotein gene 1 by transfected Entamoeba histolytica confers emetine-resistance. Mol Biochem Parasitol. 1996;82:257–260. doi: 10.1016/0166-6851(96)02733-8. [DOI] [PubMed] [Google Scholar]
- 28.Shen Z, Jacobs-Lorena M. Evolution of chitin-binding proteins in invertebrates. J Mol Evol. 1999;48:341–347. doi: 10.1007/pl00006478. [DOI] [PubMed] [Google Scholar]
- 29.Magnelli P, Cipollo JF, Ratner DM, Cui J, Kelleher D, et al. Unique Asn-linked oligosaccharides of the human pathogen Entamoeba histolytica. . J Biol Chem. 2008;283:18355–18364. doi: 10.1074/jbc.M800725200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muthusamy D, Rao SS, Ramani S, Monica B, Banerjee I, et al. Multilocus genotyping of Cryptosporidium sp. isolates from human immunodeficiency virus-infected individuals in South India. J Clin Microbiol. 2006;44:632–634. doi: 10.1128/JCM.44.2.632-634.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anders RF, McColl DJ, Coppel RL. Molecular variation in Plasmodium falciparum: polymorphic antigens of asexual erythrocytic stages. Acta Trop. 1993;53:239–253. doi: 10.1016/0001-706x(93)90032-7. [DOI] [PubMed] [Google Scholar]
- 32.Rich SM, Hudson RR, Ayala FJ. Plasmodium falciparum antigenic diversity: evidence of clonal population structure. Proc Natl Acad Sci USA. 1997;94:13040–13045. doi: 10.1073/pnas.94.24.13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ali IK, Zaki M, Clark CG. Use of PCR amplification of tRNA gene-linked short tandem repeats for genotyping Entamoeba histolytica. J Clin Microbiol. 2005;43:5842–5847. doi: 10.1128/JCM.43.12.5842-5847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samuelson J, Caplivski D, Sturm-Ramirez K, Kretzinger K, Descoteaux S, et al. A proposal for a molecular biologic system for classifying isolates of Entamoeba histolytica and Entamoeba dispar. Arch Med Res. 1997;28:274–275. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ed Jacob2 differs from Eh Jacob2 primarily in the large Ser-rich spacer. Sequence of EdJacob2 (Table S1) where the signal peptide (grey) and Cys residues (red) within CBDs are highlighted (see Fig. 1 for comparison to EhJacob2). Also highlighted are short repeats in the spacer, which fall into five families: A (light blue), B (green), C (pink), D (purple), and E (orange). Differences between the sequence of EdJacob2 and EhJacob2 are marked in bold letters. Because the number and arrangement of these short repeats differs between EdJacob2 and EhJacob2, it was not possible to directly align the two sequences.
(2.68 MB EPS)
Entamoeba proteins with chitin-binding domains (CBDs).
(0.05 MB DOC)