Abstract
A spectral analysis approach was used to estimate kinetic model parameters of the -[1-11C]leucine positron emission tomography (PET) method and regional rates of cerebral protein synthesis (rCPS) in predefined regions of interest (ROIs). Unlike analyses based on the assumption that tissue ROIs are kinetically homogeneous, spectral analysis allows for heterogeneity within a region. To improve estimation performance, a new approach was developed—spectral analysis with iterative filter (SAIF). In simulation SAIF produced low bias, low variance estimates of the influx rate constant for leucine (K1), blood volume fraction (Vb), fraction of unlabeled leucine in the tissue precursor pool for protein synthesis derived from arterial plasma (λ), and rCPS. Simulation of normal count rate studies showed that SAIF applied to ROI time-activity curves (TACs) performed comparably to the basis function method (BFM) applied to voxel TACs when voxelwise estimates were averaged over all voxels in the ROI. At low count rates, however, SAIF performed better. In measured -[1-11C]leucine PET data, there was good agreement between ROI-based SAIF estimates and average voxelwise BFM estimates of K1, Vb, λ, and rCPS. We conclude that SAIF sufficiently addresses the problem of tissue heterogeneity in ROI data and provides a valid tool for estimation of rCPS, even in low count rate studies.
Keywords: brain, kinetic analysis, leucine, positron emission tomography
Introduction
The -[1-11C]leucine positron emission tomography (PET) method was developed and validated to make possible fully quantitative measurement of regional rates of cerebral protein synthesis (rCPS) in human subjects (Schmidt et al, 2005; Smith et al, 2005). Biosynthesis of proteins is a fundamental process necessary for physiological maintenance and functioning of organisms. In the central nervous system, de novo protein synthesis is critical for adaptive responses such as long-term memory formation. Animal studies indicate that rCPS are altered in models of certain clinical disorders (Qin et al, 2005; Smith and Kang, 2000; Widmann et al, 1991, 1992; Collins et al, 1980) and in some physiologic states such as slow-wave sleep (Nakanishi et al, 1997). rCPS also changes during brain development (Sun et al, 1995) and normal aging (Ingvar et al, 1985). We have shown that measurements of rCPS with the -[1-11C]leucine PET method in awake, healthy young men are reproducible, have low variability (Bishu et al, 2008), and are unaffected by propofol anesthesia (Bishu et al, 2009). The latter point is particularly important for the study of subjects who are unable to tolerate the PET scanning procedure while awake, such as subjects with neurodegenerative and neurodevelopmental disorders.
In the first fully quantitative -[1-11C]leucine PET studies in human subjects, (Sundaram et al, 2006; Bishu et al, 2008, 2009) rCPS was determined at the region of interest (ROI) level. Because the analysis was based on a homogeneous tissue kinetic model for leucine, heterogeneity within a region was not taken into account. Tomasi et al (2009) recently developed a basis function method (BFM) to estimate kinetic model parameters and rCPS on a voxel-by-voxel basis. By averaging the estimated parameters and rCPS over all voxels within a ROI, this method produces estimates for the region as a whole that takes into account differences in kinetics among the region's voxels. In simulation studies, estimates derived from the voxelwise approach were shown to be more accurate than those derived by fitting ROI time-activity curves (TACs) to a homogeneous tissue kinetic model (Tomasi et al, 2009).
In this study, we sought an approach to analysis of ROI TACs that is not based on the assumption of tissue homogeneity. The spectral analysis method (Cunningham and Jones, 1993; Turkheimer et al, 1994) applies to both homogeneous and heterogeneous tissues, and no previous hypotheses are required regarding the number of compartments in the tissue. Application of previously reported spectral analysis approaches to analysis of -[1-11C]leucine PET data, however, did not consistently lead to reliable estimates of the kinetic parameters for leucine or rCPS, principally because of inadequate discrimination of components with very slow kinetics from trapping of tracer, and of rapidly equilibrating components from activity in blood in brain. We developed a new approach that uses spectral analysis together with an iterative numerical filter to improve component discrimination and thereby improve parameter estimation. The method was tested in simulated ROI TACs and shown to produce parameter estimates with low bias and variance. Voxel-level simulations were also performed and the average of the voxelwise BFM estimates was compared with estimates derived by applying spectral analysis with iterative filter (SAIF) to the corresponding ROI TACs. The two methods performed comparably at normal count rates, but at low count rates SAIF performance was better. SAIF was then used to analyze ROI TACs from previously acquired -[1-11C]leucine PET studies in nine awake healthy young men. The results were in good agreement with those determined by the voxelwise estimation method of Tomasi et al (2009).
Materials and methods
Spectral Analysis
Spectral analysis uses a time-invariant single input, single output model to identify kinetic components of tissue tracer activity (Cunningham and Jones, 1993; Turkheimer et al, 1994). The system output is described by the convolution of the input function with an exponential transfer function that has real-valued, nonpositive exponents. This corresponds to using a generic first-order compartmental model without fixing a priori the number of compartments in the system. Once the spectrum of kinetic components has been estimated, knowledge of the tracer's biochemical and/or physiololgical properties can be used to relate parameters of the identified spectrum to measures of the process of interest. Spectral analysis has been used with a number of PET tracers to study brain (see, e.g., Cunningham and Jones, 1993; Tadokoro et al, 1993; Turkheimer et al, 1994; Fujiwara et al, 1996; Richardson et al, 1996; Bertoldo et al, 1998; Hammers et al, 2007; Hinz et al, 2008; Brooks et al, 2008) and heart (Bertoldo et al, 1998).
In spectral analysis, the concentration of radioactivity in tissue at time t, Ctiss*(t), is modeled as a convolution of the plasma TAC, Cp*(t), with the sum of M+1 distinct exponential terms as
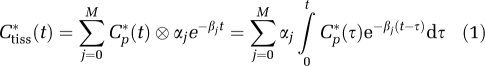 |
where αj and βj are assumed to be real-valued and nonnegative. Not all compartmental models satisfy these assumptions, but they are met by the kinetic models used with many PET tracers (Schmidt, 1999); we show in the next section that models describing the kinetics of leucine in homogeneous and heterogeneous tissues satisfy the conditions for application of spectral analysis. In equation (1), the maximum number of terms to be included in the model, M+1, is predefined and set to an arbitrary large number, for example, 100. Possible values of βj are fixed so that they cover an appropriate spectral range, and values of αj are estimated from blood and tissue TACs by a nonnegative least squares (NNLS) estimator. In practice, only a few components with αj>0 are detected. From equation (1) one observes that ‘high-frequency' components, that is, components with βj very large, become proportional to Cp*(t), whereas ‘low-frequency' components, that is, those with βj=0 or near zero, become proportional to ∫Cp*(t)dt and account for trapping of tracer. Components with intermediate values of βj reflect tissue compartments that exchange material directly or indirectly with plasma. Equation (1) is often written to explicitly show trapping of tracer as
 |
where βj>0, j=1,2…M.
Leucine Homogeneous Tissue and Heterogeneous Tissue Kinetic Models
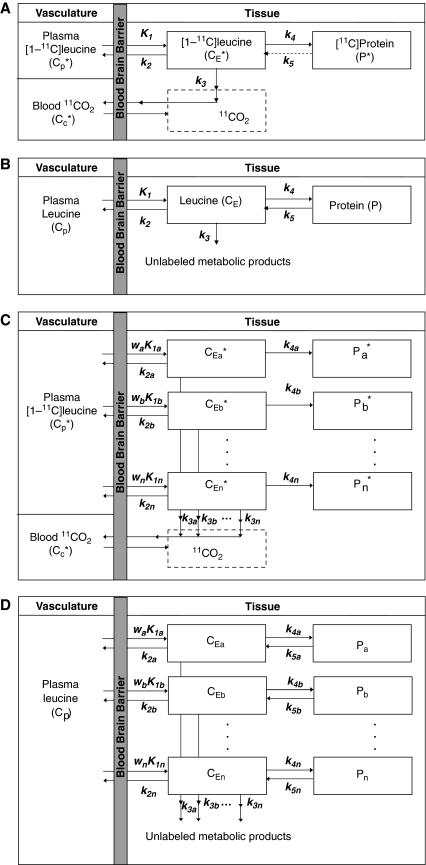
Figure 1A illustrates the model used to describe the behavior of -[1-11C]leucine in brain (Schmidt et al, 2005). Total concentration of 11C in the field of view of the PET camera (CT*) includes free -[1-11C]leucine (CE*), -[1-11C]leucine incorporated into tissue protein (P*), and activity in blood (VbCb*, where Vb is the fraction of the volume occupied by blood and Cb* is the total 11C concentration in whole blood). CT* also includes 11CO2, the predominant labeled product of -[1-11C]leucine metabolism. The simplified model shown assumes that (1) there is negligible fixation of 11CO2 during the experimental period (Siesjo and Thompson, 1965) and (2) diffusible 11CO2 in brain rapidly equilibrates with arterial blood (Buxton et al, 1987), that is, its concentration can be approximated by VDCc*, where Cc* is the 11CO2 concentration in whole blood and VD is the brain–blood equilibrium distribution volume of 11CO2, which was fixed at 0.41 (Smith et al, 2005; Brooks et al, 1984). Therefore,
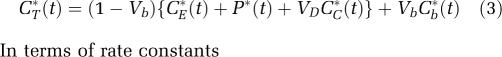 |
In terms of rate constants
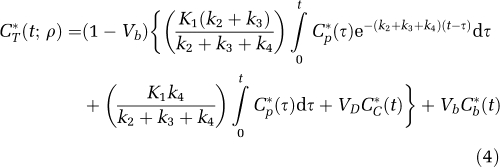 |
where K1 and k2 are the rate constants for transport of leucine from plasma to tissue and back, respectively; k3 is the rate constant for the first two steps in leucine catabolism, transamination, and decarboxylation, which yield 11CO2; k4 is the rate constant for leucine incorporation into protein; and ρ is the parameter vector (K1, k2+k3, k4, Vb). Because of the long average half-life of protein in brain breakdown of labeled protein (k5P*, where k5 is the rate constant for release of free leucine from proteolysis) is assumed to be negligible during the experimental interval (Lajtha et al, 1976). The kinetic model assumes that the tissue is homogeneous with respect to concentrations of amino acids, blood flow, rates of transport and metabolism of amino acids, and rates of incorporation into protein (Schmidt et al, 2005).
Figure 1.
(A) Simplified homogeneous tissue model for -[1-11C]leucine. K1 and k2 are the rate constants for transport of leucine from plasma to tissue and back, respectively. k3 is the rate constant for the first two steps in leucine catabolism, transamination, and decarboxylation, which yield 11CO2. k4 and k5 are the rate constants for leucine incorporation into protein and for the release of free leucine from proteolysis, respectively. Because of the long half-life of protein in brain (Lajtha et al, 1976), it is assumed that there is no significant breakdown of labeled product (P*) during the experimental interval, that is, k5P*∼0. Under the assumptions of negligible fixation of 11CO2 during the experimental period (Siesjo and Thompson, 1965) and rapid equilibration of 11CO2 between brain and blood (Buxton et al, 1987), the model reduces to two tissue compartments (free -[1-11C]leucine, CE*, and -[1-11C]leucine incorporated into tissue protein, P*) plus the 11CO2 compartment in which the concentration is assumed known. The model assumes that the tissue is homogeneous with respect to concentrations of amino acids, blood flow, rates of transport and metabolism of amino acids, and rates of incorporation into protein. (B) Homogeneous tissue model for unlabeled leucine. The model used to describe labeled leucine also holds for unlabeled leucine. Assuming no isotope effect, the rate constants for labeled and unlabeled leucine are identical. In each homogeneous tissue the unlabeled leucine (CE) and protein (P) are assumed to be in steady state, and the steady-state breakdown unlabeled protein (k5P) is greater than zero. (C) Heterogeneous tissue model for -[1-11C]leucine. The heterogeneous tissue is composed of n homogeneous subregions; each subregion is described by the model shown in (A). The constants wa, wb, …, wn represent the relative tissue weights of the subregions, where wa+wb+…+wn=1. (D) Heterogeneous tissue model for unlabeled leucine. Model for leucine in heterogeneous tissue is composed of n homogeneous subregions; each subregion is described by the model shown in (B).
The model used to describe labeled leucine holds also for unlabeled leucine (Figure 1B), except that unlabeled leucine and protein are in steady state. CE and P represent, respectively, free unlabeled leucine and unlabeled leucine incorporated into tissue protein. Assuming no isotope effect, rate constants for unlabeled and labeled leucine are identical. The steady-state breakdown of unlabeled protein (k5P) is greater than zero.
On the basis of this kinetic model, rCPS can be expressed as a function of the measured concentration of unlabeled leucine in arterial plasma, Cp, and the rate constants K1, k2+k3, and k4 as
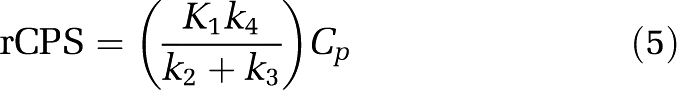 |
(Schmidt et al, 2005). The fraction of unlabeled leucine in the precursor pool for protein synthesis derived from arterial plasma, λ, can be expressed as
 |
the remainder (1−λ) derives from protein breakdown in tissue.
The assumption of tissue homogeneity may not always be a good approximation for a given region. Rates of each step in the protein synthesis pathway vary regionally throughout the brain. Owing to the limited spatial resolution of even a high-resolution PET scanner, the thinness of the cerebral cortex (∼3 to 4 mm), and white matter tracts in subcortical areas, it is likely that the activity measured in most ROIs derives from a heterogeneous mixture of kinetically distinct tissues. A model for -[1-11C]leucine that explicitly takes tissue heterogeneity into account is shown in Figure 1C; it assumes that each tissue is composed of n kinetically homogenous subregions. Activity in the tissue as a whole can be expressed as a convex combination of activity in its subregions (a,b,…,n) so that total activity in the volume is given by
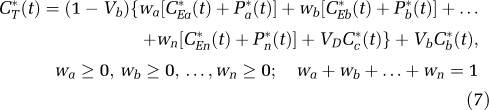 |
If subregion i is characterized by the parameter set {K1i, (k2i+k3i), k4i,} then
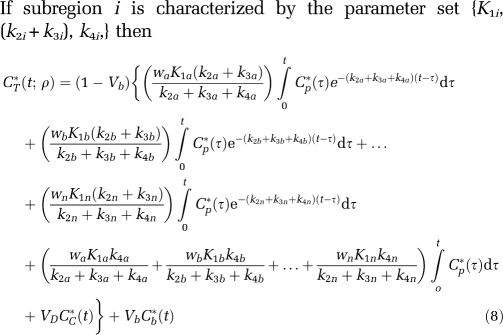 |
For this model the parameter vector ρ=(waK1a, wbK1b,…,wnK1n, k2a+k3a, k2b+k3b,…, k2n+k3n, k4a, k4b,…,k4n, Vb). (Recall that VD is assumed known a priori.) Thus, there are a total of 3n+1 parameters. If we assume that the i values of k2i+k3i+k4i are distinct, then 2n+2 parameters can be estimated: n+1 integral coefficients, n exponents, and Vb. Therefore, a total of n−1 constraints are necessary to identify all model parameters. Motivated by the observation that regional variation in λ is small (Bishu et al, 2008), the n−1 constraints λa=λb=…=λn were chosen.
The corresponding heterogeneous tissue model for unlabeled leucine is shown in Figure 1D. In the heterogeneous tissue, weighted average rCPS can be calculated as
and λ as
The weighted average influx rate constant for the mixed tissue, K1, can also be determined as
Weighted averages of k2+k3 and k4 in the mixed tissue are not identifiable from estimated parameters since relative tissue weights wi are not known.
Although equation (8) provides a good description of -[1-11C]leucine kinetics in heterogeneous tissue, it does not lend itself well to nonlinear least squares parameter estimation for two reasons: (1) the number of subregions is not known, and (2) the large number of parameters to be estimated, even for two subregions, leads to poor convergence. Similarities between equation (2) and equation (8), however, suggest that spectral analysis could be used to estimate parameters. In fact, the -[1-11C]leucine heterogeneous tissue kinetic model is equivalent to a first-order compartmental model with a trapping component, n equilibrating components, one metabolite compartment of known concentration, and one blood component, that is,
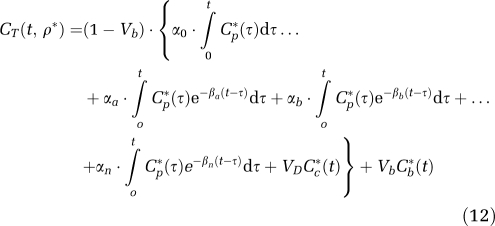 |
where
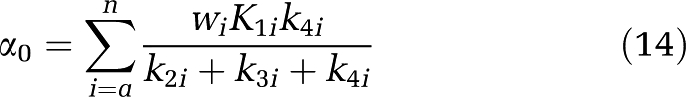 |
Furthermore, because all rate constants and weights are real-valued and nonnegative, α0, αi and βi (i=a,b,…,n) are real-valued nonnegative numbers, so the conditions necessary for application of spectral analysis are met. For this model, the parameter vector is ρ*=(α0,αa,αb,…,αn,βa,βb, …,βn,Vb). From equations (9), (10), (11), (13), (14) and (15), the relationship between rate constants of the heterogeneous tissue model for leucine and the spectral analysis components can be defined. The weighted average influx rate constant for the mixed tissue, K1, is given by
 |
Lambda can be computed as
 |
and
 |
Implementation of Spectral Analysis
Implementation of spectral analysis requires defining the grid of values of βj, selecting an algorithm and determining weighting factors for NNLS estimation, addressing artifacts that arise from the non-continuous nature of the grid, and determining standard errors in parameter estimates. In addition, numerical filters can be used to lessen the effect of noise in the data on estimated spectra and on parameters of interest.
In this study, a logarithmic distribution of βj, j=1,2,…,M was chosen (DiStefano, 1981; Turkheimer et al, 1994), with a lower limit defined as β1=1/(3Tend), where Tend is the end time of the scan, and upper limit βM=(3/Tin), where Tin is the duration of the first frame (Turkheimer et al, 1994). One hundred points were included (M=100). Therefore,
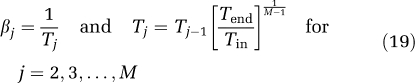 |
The M+1 unknown values αj were estimated with lsqnonneg, the NNLS algorithm implemented in Matlab (The MathWorks, Natik, MA, USA). Weights were inversely proportional to the variance of decay-corrected measured activity in the region, which was modeled assuming Poisson statistics as
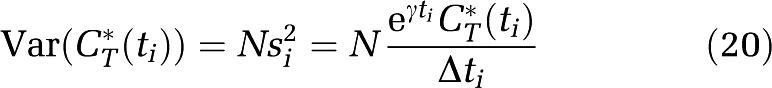 |
(Wu and Carson, 2002) where γ is the decay constant for 11C, Δti is the length of Frame i, and N is a proportionality coefficient reflecting the noise level in the data. In PET data, N is not known a priori, but its value does not affect parameter estimates. Weights were based on whole brain activity instead of activity in the ROI itself to facilitate comparison between parameter estimates obtained with spectral and voxelwise analyses. Under the assumption that the βj were fixed rather than estimated parameters, a lower bound on the standard deviation of the estimated αj was determined from the inverse of the Fisher Information matrix; N (equation (20)) was estimated a posteriori from the weighted residual sum of squares (WRSS) (Landaw and DiStefano, 1984). The difference between tracer arrival time in brain and arterial sampling site was estimated by shifting blood curves 0 to 20 secs, fitting the whole brain TAC by either nonlinear least squares and a homogeneous tissue model or by spectral analysis, and selecting the delay that produced the smallest WRSS. Tracer arrival delays estimated by the two methods did not differ by more than 1 sec; therefore, values estimated with the former method were used for consistency with the voxelwise analysis method (Tomasi et al, 2009). Because tracer appearance times in various parts of the brain differ from the mean of the brain as a whole by ±2 secs (Iida et al, 1988) whole brain tracer arrival delay was used for all ROIs in each study.
Owing to the discrete nature of the grid of βs, the algorithm cannot always place a component in the exact position that would yield the best fit of the data; it is constrained to use only βs included in the grid. When a suitable approximation of the optimal β is not on the grid, the algorithm chooses instead two consecutive values of β that bracket the optimal value. Under the assumption that consecutive components result from this phenomenon, we replaced each consecutive component pair (βj; αj), (βj+1; αj+1) with a single component (βNEW, αNEW) calculated from the amplitude-weighted average as follows:
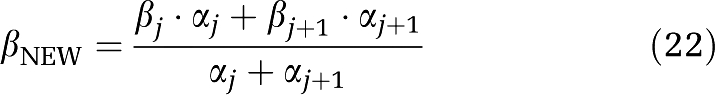 |
Numerical Filtering
Noise in the data greatly influences the accuracy with which trapping (very low-frequency) and high-frequency components can be detected, but numerical filters can improve the quality of the estimated spectrum. Turkheimer et al (1994) suggested a high-pass filter for equilibrating components to improve estimates of α0, which was used for determining regional cerebral metabolic rate for glucose in [18F]fluorodeoxyglucose PET studies. The filter assumes that equilibrating components in the tissue have βj≥some cutoff frequency βcutoff, and that components detected with 0<βj<βcutoff result from noise in the data shifting trapping components from their true position at βj=0. The filter was effected by subtracting from the measured ROI TAC all components with βj≥βcutoff; the remainder, which was assumed to equal the integrated component α0∫Cp*(t)dt, was used to re-estimate α0. Use of this filter produced better estimates of α0 and hence glucose metabolic rate than use of the unfiltered spectrum. Initial application of this filter to -[1-11C]leucine PET data, however, did not produce reliable estimates of λ and rCPS.
A New Filter: Spectral Analysis Iterative Filter
Determination of rCPS with spectral analysis requires good estimates of both α0 and 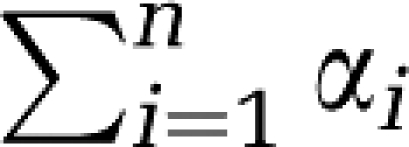 (see equations (16), (17) and (18)). It is, therefore, not only necessary to separate low-frequency equilibrating components from trapping, but also to separate high-frequency equilibrating components from blood. If the latter is not performed successfully,
(see equations (16), (17) and (18)). It is, therefore, not only necessary to separate low-frequency equilibrating components from trapping, but also to separate high-frequency equilibrating components from blood. If the latter is not performed successfully,  will be substantially overestimated.
will be substantially overestimated.
The starting point for definition of a new filter was the filter of Turkheimer et al (1994). Instead of considering the components with 0<βj<βcutoff as resulting from trapping components and noise in the data, the new filter allows that these components may also be due, at least in part, to slowly equilibrating components present in the system. Therefore, the effect on remaining equilibrating components of removing components with 0<βj<βcutoff must be considered.
The second aspect of the new filter was setting an upper cutoff frequency above which equilibrating components are assumed to be indistinguishable from blood in brain. Removal of these components from the spectrum affects estimates of remaining equilibrating components as well as blood volume.
The filtering includes two steps that are repeated until stabilization of WRSS is reached. Because of the iterative cycle we have called this the ‘SAIF'. The procedure is as follows:
Define the grid for βj, j=1,2,…,M and perform spectral analysis without filtering.
Set up Bandpass filter: Define an equilibrating component passband [βL,βU] such that all components with β∈[βL,βU] are considered to be real equilibrating components of the system.
New estimates of trapping and blood volume: Assume all components with 0<βj<βL result from noise having shifted some or all of the trapping components from their true position in the spectrum, and all components with βj>βU are shifted blood components. Subtract components inside the passband from measured activity (CT*(t)) and estimate new values of the coefficient of the trapping compartment (α0) and the blood volume fraction (Vb).
New estimates of equilibrating components: Recognizing that components outside the passband may have at least partially resulted from real equilibrating components that were shifted because of noise, re-estimate equilibrating components as follows. Using the new values of α0 and Vb from Step 3, subtract the contribution of the trapping and blood components from the measured data, and re-estimate the set of equilibrating components using NNLS with the grid restricted to β∈[βL,βU].
Stop criterion: Calculate the difference between WRSS after Step 3 and after Step 4; if it is less than ɛ=0.001 stop (In our data, WRSS ranged from 2 to 30, depending on the region. The algorithm stopped, therefore, when the change in WRSS was <0.05% to 0.003%.). Otherwise restart at Step 3.
Using change in WRSS as the stop criterion allows the data to determine the number of iterations. When all equilibrating components initially identified with spectral analysis (before filtering) are inside the passband, only one iteration is required.
Simulation studies were used to determine the optimal equilibrating component passband for -[1-11C]leucine data and for comparing SAIF with previously published spectral analysis techniques as well as with the BFM voxelwise estimation method. SAIF was also used for estimation of K1, Vb, λ, and rCPS in a group of healthy young adult males who had previously undergone -[1-11C]leucine PET studies.
Simulation Studies
As with the filter of Turkheimer et al (1994), the choice of upper and lower cutoff frequencies βL and βU for SAIF represents incorporation of prior information regarding the data into the algorithm. The particular choice of interval influences the final estimated spectrum, and therefore also estimates of the parameters of interest. To choose an interval, we simulated a mixed tissue with a trapping component (α0=0.01 mL g−1 min−1; β0=0 min−1) and two equilibrating components, one slow (αslow=0.01 mL g−1 min−1; βslow=0.05 min−1) and one fast (αfast=0.04 mL g−1 min−1; βfast=0.20 min−1). Vb was set to 0.05. These data correspond to cortical and white matter tissues in which the rate constants k2+k3 and k4 are four times higher in cortex. To allow for a larger, more conservative interval of βs to be included in the passband, the fast tissue value of k2+k3+k4 was set somewhat higher and the slow tissue value lower than mean values determined by voxelwise estimation in cortex and white matter, respectively (Tomasi et al, 2009). Arterial input functions measured in one subject were used. Noise was generated from a Gaussian distribution, with zero mean and variance given by equation (20). The coefficient of proportionality N was fixed consistent with mid-level noise in ROI data. Five hundred noisy TACs were generated and fitted with SAIF. Different passbands for SAIF were used (βL=0.005 to 0.06 min−1; βU=0.1 to 0.6 min−1) and results compared. To select the best interval, performance indices percent bias (Bias%) and percent root mean square error (RMSE%) in rCPS were computed as
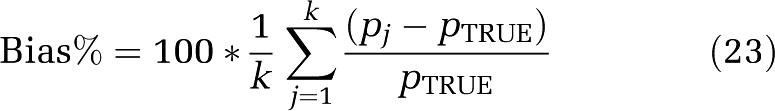 |
and
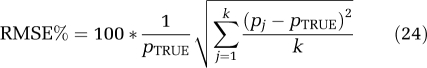 |
where pj is the SAIF estimate of rCPS at the jth noise realization, pTRUE is the simulated value of rCPS, and k is the number of noise repetitions.
Once the choice of [βL,βU] was made, whole brain and three ROIs showing low, moderate, and high noise levels (frontal cortex, corona radiata, and hippocampus, respectively) were simulated to examine the effect of noise on estimated parameters of interest. For each region, the components and noise coefficient estimated in one subject with nonlinear least squares and a two-tissue heterogeneous tissue model (equation (8)) were the ‘true' values for the simulation. One thousand noisy TACs for each ROI were generated, and parameters were estimated with unfiltered spectral analysis, spectral analysis with the equilibrating component high-pass filter (βcutoff=1/Tend) (Turkheimer et al, 1994), spectral analysis with the grid restricted to the bandpass interval, and SAIF. Bias% and RMSE% (equations (23) and (24)) in K1, Vb, λ, and rCPS were computed for each analysis method.
A set of multiple-voxel ROI simulations was also performed to compare analysis of ROI TACs with voxelwise estimation using BFM, as described in Tomasi et al (2009). Briefly, parameters and noise levels in all voxels of a cortical, subcortical, and white matter ROI, that is, the frontal cortex (central slices), thalamus, and corona radiata, respectively, in one subject were estimated with BFM and became the ‘true' values for the simulation. Noisy voxel TACs were generated from the ‘true' parameters of the voxel, the measured plasma and blood curves, and added Gaussian noise (equation (20)). A noisy ROI TAC was computed as the average of the noisy voxel TACs. Parameters were then estimated in two ways: (1) using BFM at the voxel level and averaging estimates over all voxels in the ROI, and (2) using SAIF to fit the ROI TAC. One hundred repetitions were made, and average Bias% (equation (23)) and RMSE% (equation (24)) were computed. To simulate the effects of a low count rate study, the simulations were repeated with the same set of ‘true' parameters, but with the measured plasma and blood curves reduced to 25% of their original values.
PET Studies
Data from nine healthy awake male subjects (age 20 to 24) from our previous study (Bishu et al, 2008) were used. The criteria for subject inclusion and the procedure for -[1-11C]leucine PET studies are described in detail in Bishu et al (2008). All studies were performed on the High-Resolution Research Tomograph (HRRT) (CPS Innovations, Knoxville, TN, USA), which has spatial resolution of ∼2.6 mm full width at half maximum (Wienhard et al, 2002). The 90-min emission scan was initiated coincident with a 2-min intravenous infusion of 20 to 30 mCi of -[1-11C]leucine (one subject received a 1-min infusion). Estimated leucine-specific activity was 3 mCi/nmol. Images were reconstructed using motion-compensated three-dimensional ordinary Poisson ordered subset expectation maximization (Carson et al, 2003) as 42 frames (16 × 15, 4 × 30, 4 × 60, 4 × 150, 14 × 300 secs); voxel size was 1.21 × 1.21 × 1.23 mm. Arterial blood sampling was performed; concentrations of unlabeled and labeled leucine in plasma and total 11C and 11CO2 activities in whole blood were measured according to methods detailed in Bishu et al (2008). All subjects underwent a T1-weighted magnetic resonance imaging of the brain for ROI placement. A total of 18 regions and whole brain were evaluated in all subjects except one in which the hypothalamus could not be drawn. Estimates of K1, Vb, λ, and rCPS determined in each ROI by use of SAIF with the passband determined in the simulation study were compared with the mean value over all voxels in the ROI of K1, Vb, λ, and rCPS estimated voxelwise with the BFM of Tomasi et al (2009).
Results
SAIF Passband
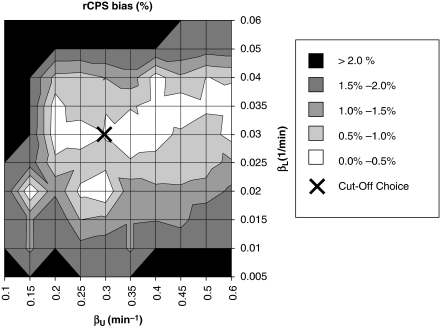
In simulations to observe the best passband, bias in rCPS ranged from 0.03% to 7% (Figure 2), depending on the cutoff frequencies βL and βU. RMSE% ranged from 2.4% to 21.5%. More than one pair of cutoff frequencies provided rCPS bias <1% cutoff values of 0.03 and 0.3 min−1 for βL and βU, respectively, were chosen as a good compromise between precision and accuracy. For this interval, bias and RMSE in rCPS were small (0.5% and 5.7%, respectively). This interval also provided good estimates of λ (bias 0.2%, RMSE 1.2%).
Figure 2.
Bias of regional rates of cerebral protein synthesis (rCPS) for different passbands. The map illustrates the bias of rCPS estimated in simulation studies for different lower and upper beta cutoff frequencies, βL and βU, respectively. Darker areas of the grayscale indicate higher rCPS biases. The cutoff frequencies were fixed in the range from 0.005 to 0.06 min−1 for βL and from 0.1 to 0.6 min−1 for βU. The cross (located at βL=0.03 min−1, βU=0.3 min−1) indicates the best choice for the endpoints of the passband; this choice represents a compromise between lower biases in rCPS (equation (23)) and lower root mean square error (RMSE) (equation (24)).
Performance of SAIF in Simulation
Table 1 provides estimates of K1, Vb, λ, and rCPS determined by SAIF with the chosen passband in 1000 simulated TACs for whole brain and 3 ROIs. Parameter estimates determined by spectral analysis without filtering, spectral analysis with high-pass filter, and spectral analysis with the grid restricted to include only β0 and βj∈[βL,βU] are also shown. There was a substantial overestimation of K1 and λ, and underestimation of rCPS, with unfiltered spectral analysis, primarily because of shifting of the trapping component away from β0. Vb was underestimated. Use of the high-pass filter tended to overcompensate for the shift away from β0, that is, the coefficient of the trapping component was overestimated. As a result, K1 and λ were underestimated and rCPS overestimated. In this case, Vb was also overestimated. Simply restricting the grid to the passband improved results, even in the simulated frontal cortex, which included a slow component with βj<βL. SAIF produced the lowest biases in all parameters of interest; biases remained <4.8% in absolute value. In whole brain, frontal cortex, and corona radiata SAIF also produces the lowest RMSE (<5.7%), but in hippocampus, the smallest region with the highest noise level, spectral analysis with the grid restricted to the passband produced the smallest RMSE.
Table 1. Performance of spectral analysis algorithms in simulation dataa.
| Whole brainb | K1 (mL g−1 min−1) | Vb | λ | rCPS (nmol g−1 min−1) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ‘True' value | 0.050 | 0.068 | 0.764 | 1.69 | ||||||||
| Estimated value (mean±s.d.) | Bias (%) | RMSE (%) | Estimated value (mean±s.d.) | Bias (%) | RMSE (%) | Estimated value (mean±s.d.) | Bias (%) | RMSE (%) | Estimated value (mean±s.d.) | Bias (%) | RMSE (%) | |
| Unfiltered | 0.081±0.047 | 61.4% | 94.5% | 0.065±0.005 | −5.0% | 7.3% | 0.851±0.073 | 11.3% | 9.6% | 1.27±0.44 | −24.5% | 25.9% |
| Highpass EC filter | 0.049±0.003 | −2.4% | 6.0% | 0.072±0.006 | 5.2% | 8.8% | 0.746±0.037 | −2.4% | 4.8% | 1.80±0.25 | 6.9% | 14.6% |
| With restricted grid | 0.051±0.001 | 1.8% | 2.0% | 0.068±0.001 | −0.6% | 1.5% | 0.773±0.006 | 1.1% | 0.8% | 1.64±0.08 | −3.0% | 4.6% |
| SAIF | 0.051±0.001 | 0.9% | 1.9% | 0.068±0.001 | −0.5% | 1.9% | 0.762±0.006 | −0.3% | 0.8% | 1.72±0.05 | 2.0% | 2.7% |
| Frontal cortexc | K1 (mL g−1 min−1) | Vb | λ | rCPS (nmol g−1 min−1) | ||||||||
| ‘True' value | 0.051 | 0.071 | 0.757 | 1.79 | ||||||||
| Estimated value (mean±s.d.) | Bias (%) | RMSE (%) | Estimated value (mean±s.d.) | Bias (%) | RMSE (%) | Estimated value (mean±s.d.) | Bias (%) | RMSE (%) | Estimated value (mean±s.d.) | Bias (%) | RMSE (%) | |
| Unfiltered | 0.087±0.060 | 70.1% | 118.1% | 0.067±0.006 | −6.1% | 8.5% | 0.869±0.083 | 14.8% | 11.0% | 1.14±0.59 | −36.0% | 33.2% |
| Highpass EC filter | 0.050±0.002 | −2.2% | 3.9% | 0.075±0.005 | 5.1% | 7.0% | 0.721±0.038 | −4.8% | 5.0% | 2.11±0.31 | 18.0% | 17.6% |
| With restricted grid | 0.052±0.001 | 1.7% | 2.0% | 0.071±0.002 | −0.5% | 2.8% | 0.750±0.008 | −0.9% | 1.1% | 1.88±0.06 | 5.3% | 3.2% |
| SAIF | 0.052±0.001 | 1.4% | 2.1% | 0.071±0.001 | −0.7% | 1.4% | 0.764±0.008 | 0.9% | 1.0% | 1.75±0.06 | −2.3% | 3.3% |
| Corona radiatad | K1 (mL g−1 min−1) | Vb | λ | rCPS (nmol g−1 min−1) | ||||||||
| ‘True' value | 0.031 | 0.032 | 0.792 | 0.89 | ||||||||
| Estimated value (mean±s.d.) | Bias (%) | RMSE (%) | Estimated value (mean±s.d.) | Bias (%) | RMSE (%) | Estimated value (mean±s.d.) | Bias (%) | RMSE (%) | Estimated value (mean±s.d.) | Bias (%) | RMSE (%) | |
| Unfiltered | 0.074±0.064 | 137.5% | 210.1% | 0.027±0.007 | −16.8% | 22.2% | 0.887±0.075 | 11.9% | 9.5% | 0.66±0.27 | −26.2% | 30.3% |
| Highpass EC filter | 0.029±0.003 | −6.9% | 9.9% | 0.036±0.005 | 11.0% | 15.8% | 0.762±0.049 | −3.8% | 6.2% | 0.99±0.18 | 11.2% | 20.2% |
| With restricted grid | 0.032±0.001 | 2.7% | 3.3% | 0.032±0.002 | −1.4% | 6.2% | 0.799±0.012 | 0.8% | 1.5% | 0.87±0.06 | −2.5% | 6.2% |
| SAIF | 0.032±0.001 | 1.5% | 3.9% | 0.032±0.002 | −1.7% | 5.7% | 0.793±0.012 | 0.1% | 1.5% | 0.90±0.05 | 1.1% | 5.4% |
| Hippocampuse | K1 (mL g−1 min−1) | Vb | λ | rCPS (nmol g−1 min−1) | ||||||||
| ‘True' value | 0.036 | 0.082 | 0.674 | 1.91 | ||||||||
| Estimated value (mean±s.d.) | Bias (%) | RMSE (%) | Estimated value (mean±s.d.) | Bias (%) | RMSE (%) | Estimated value (mean±s.d.) | Bias (%) | RMSE (%) | Estimated value (mean±s.d.) | Bias (%) | RMSE (%) | |
| Unfiltered | 0.165±0.197 | 356.2% | 649.7% | 0.067±0.021 | −18.7% | 31.6% | 0.838±0.137 | 24.4% | 31.8% | 1.34±0.76 | −29.8% | 49.8% |
| Highpass EC filter | 0.033±0.003 | −8.4% | 12.0% | 0.089±0.007 | 8.1% | 12.0% | 0.578±0.086 | −14.3% | 19.1% | 2.71±0.75 | 42.1% | 57.6% |
| With restricted grid | 0.037±0.002 | 1.6% | 0.5% | 0.081±0.005 | −1.7% | 0.2% | 0.659±0.032 | −2.2% | 0.1% | 2.07±0.21 | 8.5% | 0.1% |
| SAIF | 0.037±0.002 | 1.9% | 6.7% | 0.080±0.005 | −1.9% | 6.0% | 0.668±0.032 | −0.9% | 4.8% | 2.00±0.20 | 4.8% | 11.7% |
rCPS, regional rates of cerebral protein synthesis; RMSE, root mean square error; SAIF, spectral analysis with iterative filter.
Number of noise realizations=1000.
Simulation parameters: waK1a=0.042 mL g−1 min−1; wbK1b=0.0082 mL g−1 min−1; k2a+k3a=0.15 min−1; k2b+k3b=0.029 min−1; k4a=0.047 min−1; k4b=0.0089 min−1; Vb=0.068; noise level N=0.20 nCi min/mL.
Simulation parameters: waK1a=0.044 mL g−1 min−1; wbK1b=0.0075 mL g−1 min−1; k2a+k3a=0.13 min−1; k2b+k3b=0.018 min−1; k4a=0.042 min−1; k4b=0.0059 min−1; Vb=0.071; noise level N=0.25 nCi min/mL.
Simulation parameters: waK1a=0.021 mL g−1 min−1; wbK1b=0.0102 mL g−1 min−1; k2a+k3a=0.15 min−1; k2b+k3b=0.041 min−1; k4a=0.040 min−1; k4b=0.0107 min−1; Vb=0.032; noise level N=0.36 nCi min/mL.
Simulation parameters: waK1a=0.020 mL g−1 min−1; wbK1b=0.016 mL g−1 min−1; k2a+k3a=0.115 min−1; k2b+k3b=0.022 min−1; k4a=0.055 min−1; k4b=0.010 min−1; Vb=0.082; noise level N=0.70 nCi min/mL.
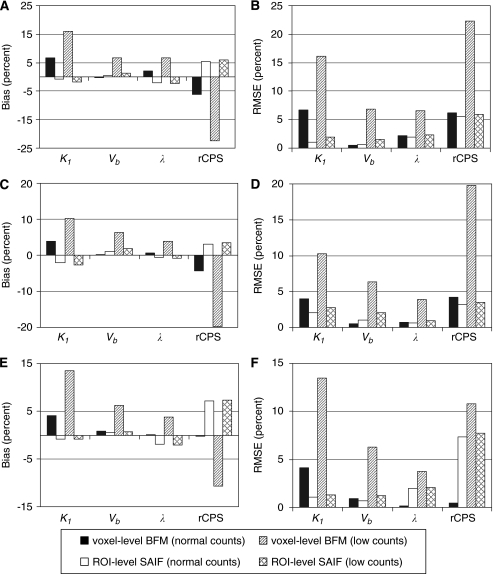
Bias% and RMSE% for multiple-voxel ROI simulations are shown in Figure 3. For each of the regions simulated, the mean of the values of K1, Vb, λ, and rCPS estimated in each voxel with BFM was compared with the corresponding parameter estimated from the ROI TACs with SAIF. At normal count rates, K1 and λ tended to be somewhat overestimated with BFM and underestimated with SAIF, whereas rCPS was slightly underestimated with BFM and slightly overestimated with SAIF; Vb was well estimated with both methods. RMSE% was below 8% for all parameters in all ROIs with both methods. At simulated low count rates, however, the performance of BFM worsened while that of SAIF was very little affected; consequently, SAIF analysis of the ROI TACs provided better estimates of all parameters.
Figure 3.
Bias% and root mean square error (RMSE)% for multiple-voxel region of interest (ROI) simulation. Shown are the simulated central slices of the frontal cortex (A and B), thalamus (C and D), and corona radiata (E and F). Parameter estimates were computed in two ways: using basis function method (BFM) at the simulated voxel level and averaging the parameter estimates over all voxels within the ROI (voxel-level BFM), and using spectral analysis with iterative filter (SAIF) to fit the simulated ROI TAC (ROI-level SAIF). Normal count rate simulations were based on data measured in one subject administered 25.9 mCi [11C]leucine (peak scanner count rate 3.4 × 106 counts per second). Low count rate simulations used blood input functions 25% of the measured values. At normal count rates, performance of the two analysis methods was similar. At low count rates, performance of the voxelwise estimation method worsened while changes in ROI-level SAIF performance were minimal.
SAIF Application to Measured PET Data
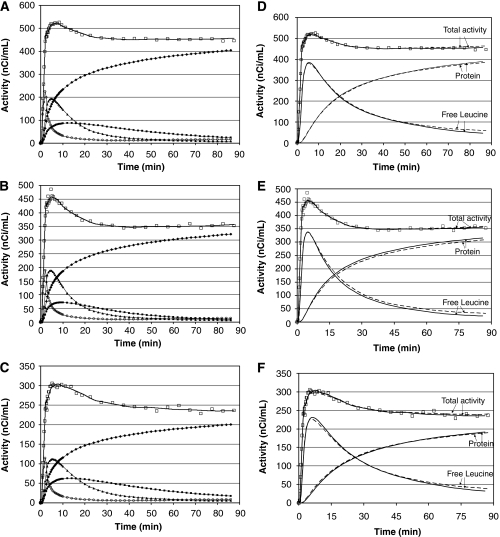
SAIF produced good fits of ROI TACs (Figures 4A–4C). In all ROIs one trapping component, one blood component, and at least two equilibrating components were detected. Components detected in frontal cortex, thalamus, and corona radiata in one subject are illustrated in Figures 4A–4C. Of the 170 ROIs analyzed (19 in 8 subjects and 18 in 1 subject) 3 equilibrating components were necessary to describe total activity in 24 of the ROIs (14%). It was not consistent among subjects, however, which ROIs required the larger number of components. Figures 4D–4F compare the best-fit of measured total activity determined with SAIF to that obtained with voxelwise analysis in one subject. In general, total activity estimated with SAIF fits measured data as well as or slightly better than the mean of voxelwise-estimated activities. The two methods also provided comparable trends for predicted time courses of labeled free leucine and protein in tissue. After the first approximately 15 to 20 mins, labeled free leucine tended to be slightly lower, and labeled protein slightly higher, when estimated with SAIF compared with voxelwise analysis. In the initial portion of the curves, particularly in corona radiata, these trends were reversed. Overall, similarities between estimation methods in predicted time courses of labeled free leucine and protein are reflected in similarities in estimated rCPS in this subject: the percent differences between SAIF and voxelwise estimates were 1.5%, 1.9%, and 3.1% for frontal cortex, thalamus and corona radiata, respectively.
Figure 4.
(A–C) Time courses of total activity and activity in tissue components estimated with spectral analysis with iterative filter (SAIF). Total activity was measured in the frontal cortex (A), thalamus (B), and corona radiata (C) in one subject. Open squares (□) and solid lines represent the measured and fitted total activity in the regions of interest (ROIs). Open circles (○) represent activity of blood in the brain. Solid diamonds (⧫), solid squares (▪), and solid triangles (▴) represent trapping, slow, and fast components, respectively, detected by SAIF. Concentration of 11CO2 in brain is not shown. (D–F) SAIF and voxelwise fits. The graphs show the time courses of total activity, labeled protein and labeled free leucine in the frontal cortex (D), thalamus (E), and corona radiata (F) of one subject. Open squares (□) indicate measured total activity in the ROI, and solid lines represent activity estimated by fitting the ROI TAC using SAIF. The average time course of the activities determined in each voxel from the parameter estimates for that voxel is shown by the dashed lines. Free leucine and protein time courses estimated by the two methods are similar in the very early portion of the curve; they gradually separate with increasing time. In the last 15 to 30 mins, one sees slightly lower total activity and free leucine estimated by SAIF, and slightly higher concentrations of labeled protein.
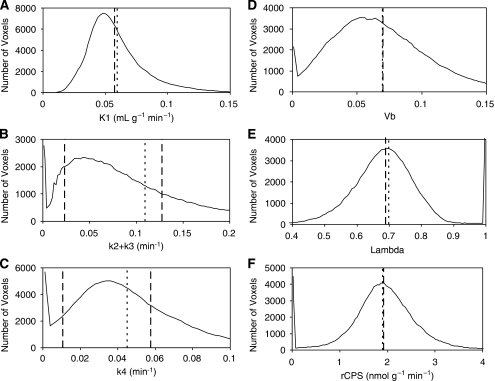
Figures 5A–5E compare values of K1, k2+k3, k4, Vb, λ and rCPS estimated from the frontal cortex TAC by SAIF with values determined voxelwise. The distribution of BFM-estimated parameters in all voxels within the ROI is shown (solid line) along with the mean value over all voxels in the ROI (dotted lines). SAIF estimates (dashed lines) represent values of the parameters calculated from the spectra identified from the ROI TAC. For the parameters K1, Vb, λ, and rCPS (Figures 5A, 5D–5F) dashed and dotted lines are very close to each other; this indicates good agreement between estimates provided by SAIF and the mean of the voxelwise estimates. In Figure 5B (k2+k3) and 5C (k4) there are two dashed lines corresponding to fast and slow components detected with SAIF; this indicates that the ROI is heterogeneous and is comprised of two homogeneous subregions. Because relative weights of the two subregions cannot be independently identified, no weighted average for k2+k3 or k4 can be evaluated.
Figure 5.
Distribution of voxelwise estimates of K1 (A), k2+k3 (B), k4 (C), Vb (D), λ (E), and regional rates of cerebral protein synthesis (rCPS) (F) in all voxels of the frontal cortex of one subject. Solid lines indicate the number of voxels with the basis function method (BFM)-estimated parameter value indicated on the abscissa. Dotted vertical lines represent the mean of the BFM-estimated parameters values, averaged over all voxels in the region. Dashed vertical lines represent the values of the estimates provided by applying the spectral analysis with iterative filter (SAIF) method to the region of interest (ROI) TAC. Note the good agreement between the mean voxelwise estimates of K1, Vb, λ, and rCPS and the corresponding estimates provided by SAIF. SAIF provides one estimate of k2+k3 for each of the two subregions detected by the method (B). Similarly, there are two estimates for k4 (C). The weighted average values of k2+k3 and k4 cannot be computed, however, since the relative weights of the two subregions in the tissue are unknown (see text). The small fraction of the region's 160,000 voxels having BFM estimates of k2+k3, k4, and rCPS near zero, and λ near one, may be due to non-brain voxels included in the ROI, for example, voxels located in the sulci.
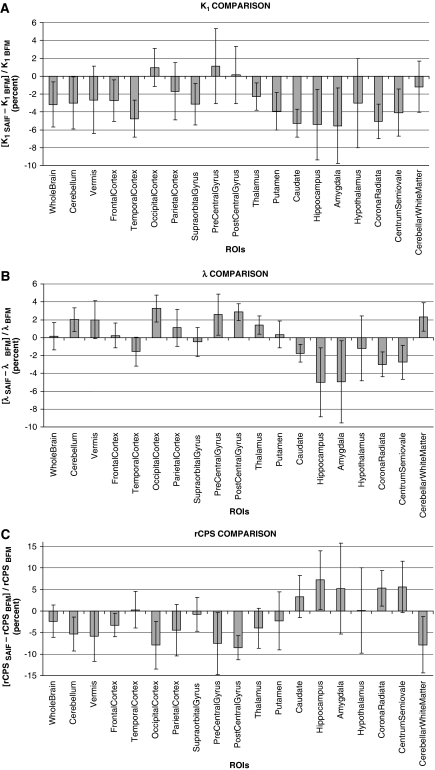
Figures 6A–6C illustrate comparisons of SAIF estimates with regional mean values of voxelwise estimates of K1, λ, and rCPS in individual subjects. The percent difference between SAIF and voxelwise estimates for each parameter was computed for each subject. Graphs represent intersubject means and s.d. of the percent differences between analysis methods. In general, differences between methods were small, but intersubject variability tended to be greater in smaller and therefore noisier regions. Across all regions and subjects, the percent difference in K1 ranged from a minimum of −12% (in the hippocampus of one subject) to a maximum of 6% (precentral gyrus); intersubject mean values ranged from −5% to 1% (Figure 6A). The percent difference in λ, across all regions and subjects, ranged from −14% (hippocampus) to 6% (hypothalamus); intersubject mean value range: −5% to 3% (Figure 6B). The range of percent differences in rCPS, across all regions and subjects, was −18% (occipital cortex of one subject) to 23% (hippocampus); intersubject mean values ranged from −9% to 7% (Figure 6C). The range of percent differences in Vb, across all regions and subjects, was −14% (hypothalamus) to 6% (corona radiata); intersubject mean values ranged from −5% to 1% (data not shown).
Figure 6.
Percent difference between spectral analysis with iterative filter (SAIF) and voxelwise estimates of K1 (A), λ (B), and regional rates of cerebral protein synthesis (rCPS) (C). For each of the nine subjects, for each region of interest (ROI), and for each parameter, the percent difference was computed as 100*(pSAIF−pvoxelwise)/pvoxelwise, where pSAIF is the SAIF estimate and pvoxelwise is the mean of the voxelwise estimates of the parameter. For each ROI indicated, the bar represents the intersubject mean of the percent difference in the parameter estimates. The intersubject s.d. is indicated by the error bar. In general, there is good agreement between the estimates provided by the two methods. Some of the greatest differences in the estimates between the two methods, as well as the largest intersubject variabilities, are found in the smallest regions (hippocampus, amygdala, hypothalamus, pre- and postcentral gyrus).
Table 2 summarizes values of K1, λ, and rCPS estimated by SAIF and those estimated voxelwise. Means and s.d. for 9 subjects are reported for 18 ROIs and whole brain. There is good agreement between the mean values of the two methods in most regions, but intersubject variability was somewhat higher in λ and rCPS estimated with SAIF than with the voxelwise method. In most regions, there was also good agreement between the methods in estimates of Vb. SAIF and mean voxelwise estimates of Vb in whole brain were 0.062±0.009 and 0.062±0.008, respectively. In all regions examined, the intersubject mean of the SAIF estimates of Vb was 95 to 102% the corresponding mean of the voxelwise estimates.
Table 2. Regional estimates of K1, λ, and rCPSa.
| ROI TAC-based estimation with SAIFb | Voxelwise estimation with BFMc | |||||
|---|---|---|---|---|---|---|
| Region (volume, mean±s.d.) | K1 (mL g−1 min−1) | Lambda | rCPS (nmol g−1 min−1) | K1 (mL g−1 min−1) | Lambda | rCPS (nmol g−1 min−1) |
| Whole brain (1397±133 cm3) | 0.047±0.004 | 0.77±0.03 | 1.57±0.13 | 0.049±0.004 | 0.77±0.02 | 1.61±0.08 |
| Cerebellum (115±15 cm3) | 0.067±0.007 | 0.80±0.02 | 1.85±0.14 | 0.070±0.007 | 0.78±0.02 | 1.96±0.10 |
| Vermis (6±1 cm3) | 0.064±0.007 | 0.79±0.04 | 1.85±0.16 | 0.066±0.008 | 0.77±0.03 | 1.96±0.09 |
| Cortical | ||||||
| Frontal cortex (135±61 cm3) | 0.051±0.005 | 0.75±0.03 | 1.83±0.11 | 0.052±0.005 | 0.75±0.02 | 1.89±0.09 |
| Temporal cortex (58±12 cm3) | 0.044±0.004 | 0.73±0.03 | 1.74±0.15 | 0.046±0.004 | 0.75±0.02 | 1.73±0.10 |
| Occipical cortex (29±12 cm3) | 0.064±0.006 | 0.78±0.03 | 1.93±0.21 | 0.063±0.006 | 0.76±0.03 | 2.09±0.17 |
| Parietal cortex (17±5 cm3) | 0.053±0.004 | 0.76±0.04 | 1.78±0.28 | 0.054±0.004 | 0.76±0.02 | 1.86±0.20 |
| Supraorbital gyrus (4±1 cm3) | 0.047±0.006 | 0.74±0.03 | 1.76±0.14 | 0.048±0.005 | 0.75±0.03 | 1.77±0.10 |
| Precentral gyrus (3±0 cm3) | 0.057±0.005 | 0.78±0.04 | 1.75±0.29 | 0.056±0.006 | 0.76±0.02 | 1.88±0.22 |
| Postcentral gyrus (2±0 cm3) | 0.059±0.006 | 0.79±0.03 | 1.71±0.16 | 0.059±0.006 | 0.77±0.02 | 1.87±0.18 |
| Subcortical | ||||||
| Thalamus (15±6 cm3) | 0.050±0.004 | 0.79±0.02 | 1.47±0.07 | 0.051±0.003 | 0.78±0.02 | 1.54±0.10 |
| Caudate (8±1 cm3) | 0.033±0.004 | 0.77±0.02 | 1.04±0.10 | 0.034±0.005 | 0.79±0.02 | 1.01±0.07 |
| Putamen (9±1 cm3) | 0.048±0.005 | 0.80±0.03 | 1.30±0.13 | 0.051±0.006 | 0.80±0.02 | 1.33±0.08 |
| Hippocampus (4±1 cm3) | 0.035±0.004 | 0.71±0.04 | 1.58±0.20 | 0.037±0.004 | 0.74±0.02 | 1.47±0.15 |
| Amygdala (5±1 cm3) | 0.031±0.003 | 0.71±0.05 | 1.42±0.18 | 0.033±0.003 | 0.74±0.03 | 1.35±0.11 |
| Hypothalamusd (0.6±0.2 cm3) | 0.040±0.007 | 0.78±0.05 | 1.25±0.34 | 0.041±0.005 | 0.79±0.03 | 1.24±0.24 |
| White matter | ||||||
| Corona radiata (46±5 cm3) | 0.026±0.004 | 0.76±0.02 | 0.87±0.08 | 0.027±0.004 | 0.79±0.02 | 0.83±0.06 |
| Centrum semiovale (35±11 cm3) | 0.028±0.003 | 0.76±0.03 | 0.97±0.12 | 0.030±0.004 | 0.78±0.02 | 0.91±0.08 |
| Cerebellar peduncles (14±2 cm3) | 0.043±0.004 | 0.81±0.03 | 1.07±0.13 | 0.044±0.004 | 0.80±0.02 | 1.16±0.08 |
BFM, basis function method; rCPS, regional rates of cerebral protein synthesis; ROI, region of interest; SAIF, spectral analysis with iterative filter.
Values are mean±s.d. for nine subjects, except where indicated.
SAIF estimates based on the ROI time activity curves and optimal cut-off interval choice; weights based on whole brain TAC;
Voxelwise BFM estimates based on the homogeneous tissue model and voxel time activity curves; weights based on whole brain TAC; parameters averaged over volume of interest.
Values are mean±s.d. for eight subjects.
Discussion
Before this study, estimation of rCPS and kinetic model parameters for leucine from regional time-activity data were based on a compartmental model that assumed each tissue ROI was kinetically homogeneous. Because this model does not account for heterogeneity within a ROI, fits of ROI TACs were poor and estimates of rCPS were biased (Tomasi et al, 2009). To ameriolate, at least partially, effects of heterogeneity on estimation bias, Tomasi et al (2009) developed an approach to estimate parameters of the homogeneous tissue kinetic model for leucine for each voxel in the image; voxelwise estimates were then averaged over all voxels within a ROI to obtain a mean ROI value. This approach reduced substantially estimation bias, although differences between measured and fitted TACs suggested that effects of heterogeneity were not completely eliminated.
In this study, we sought to develop an approach for ROI-based analysis that takes kinetic heterogeneity in the tissue into account. Indeed, voxel-based analyses may not always be feasible for a specific study, for example, when available computational time is severely limited or when images are of poor statistical quality. The latter may be the case, for example, when injected activity is low. In the presence of very high noise levels, voxelwise estimates of rCPS become increasingly negatively biased (Tomasi et al, 2009) and ROI-based analysis may be the only viable alternative. Previous attempts to take kinetic heterogeneity into account using a fixed compartmental model (which describes a heterogeneous tissue as comprised of two homogeneous subregions) required nonlinear least squares estimation of six parameters and suffered frequent problems of convergence of the fitting algorithm and a high degree of uncertainty in the estimates (Tomasi et al, 2009). Spectral analysis, however, uses a set of basis functions together with a NNLS algorithm that is guaranteed to converge in a finite number of iterations to the minimum WRSS over the given basis function set (Lawson and Hanson, 1974). In this study, simulations showed that spectral analysis with iterative filter provided estimates of comparable quality to those derived by averaging voxelwise estimates when injected doses of tracer were in the normal range for our studies. When the count rates were significantly reduced, however, performance of voxelwise estimation deteriorated while that of SAIF applied at the ROI level was effectively unchanged. Consequently, at low count rates SAIF is the preferred alternative. Although it was not surprising to observe that a ROI-based analysis performed better under high noise conditions, it was critical that the ROI-based analysis took tissue heterogeneity into account, which SAIF does by allowing multiple components in the tissue region. By contrast, ROI-based analyses that are based on homogeneity assumptions have distinctly poorer performance than the voxelwise alternative, whether at normal count rates (Tomasi et al, 2009) or low count rates (data not shown).
Spectral analysis is based on a general compartmental model and uses as basis functions a set of exponentials convolved with the plasma input function. Unlike fixed compartmental models, spectral analysis allows the data to determine the number of components in the tissue. It is, however, sensitive to the choice of the range of exponents, and noise in the data can shift components from their true position in the spectrum. These effects impact various parameters of interest differently. For example, the rate constant for influx of tracer from plasma to tissue is determined as the sum of the magnitudes of all components in the spectrum and is little affected by shifts in positions of the components. The net rate constant for trapping of tracer, by contrast, is extremely sensitive to even small shifts of near-zero components. For determination of rCPS, it is necessary to obtain good estimates of both the net rate constant for trapping and the sum of the coefficients of all other non-blood components in the system. We found that previously published spectral analysis approaches, either without or with filtering, did not consistently provide reliable estimates of both factors and thus did not perform well for estimating rCPS.
To improve estimation of rCPS with spectral analysis, we introduced a numerical filter, SAIF, which attempts to sharpen the distinction between trapping of the tracer and slowly equilibrating components while at the same time distinguishing blood from rapidly equilibrating components. The filter is applied iteratively and terminates when an iteration provides no appreciable improvement in fit of the data. In simulation studies at noise levels typical of ROI data, SAIF was found to provide estimates of rCPS that had low bias and low RMSE.
Essential to performance of SAIF is selection of appropriate upper and lower bounds of the passband. The passband interval can be thought of as prior information on the data; it is defined as the interval in which true equilibrating components in the system are expected to be found. We chose passband limits based on results of a simulation study, but if the true kinetics of leucine in the tissue differ appreciably from those used in the simulation, the optimal passband may differ from the one we selected. Other approaches to selection of passband endpoints may lead to further improvements in SAIF performance and should be explored.
An important assumption that was made to identify all parameters of the heterogeneous tissue kinetic model from the estimated spectrum was that the fraction of unlabeled leucine in the precursor pool for protein synthesis in subregion i derived from arterial plasma (λi) is the same for all subregions comprising the mixed tissue (see equation (10)). Examination of voxelwise estimates of λ, however, suggests that values may be slightly higher in white matter (Tomasi et al, 2009). Sensitivity of rCPS determined with SAIF to variations in λ was analyzed by recomputing rCPS under the assumption that λ in the tissue subregion with slower kinetics, presumably white matter, was higher than in the tissue with faster kinetics, for example, cortex. rCPS under this assumption was neglibibly higher, by <2% when λslow was assumed to be 15% higher than λfast, and by <5% when λslow was 25% higher than λfast (data not shown).
The most remarkable outcome is the good agreement between SAIF estimates derived from ROI TACs and estimates derived from voxelwise analyses of PET data acquired on the High-Resolution Research Tomograph after injection of 20 to 30 mCi -[1-11C]leucine. In voxelwise analyses, parameters were estimated based on a homogeneous tissue kinetic model, and parameter estimates were averaged over all voxels in the ROI. Differences between intersubject means of estimates provided with SAIF and voxelwise analysis were small (<9% in absolute value) in all ROIs and in all parameters of interest. Close agreement between methods can also be observed in the time courses of the model fit of total activity in the ROI as well as in time courses of labeled free leucine and protein predicted by the two methods.
Intersubject variability with SAIF was somewhat higher than found in the voxelwise analyses. This may be due to the larger number of parameters estimated for the multiple subregions in the ROI. The variability with SAIF was, however, lower than found with nonlinear least squares estimation with a fixed heterogeneous tissue kinetic model (data not shown), even when the number of compartments in the two approaches was the same. In the case of SAIF, the guaranteed convergence of the NNLS algorithm to the global minimum over the set of basis functions, in comparison with the generally poor convergence of nonlinear least squares when the number of parameters is large, is a distinct advantage. Incorporating stronger prior information may be a means to further reduce variability with SAIF.
The capacity of spectral analysis to take into account heterogeneity in the tissue ROI allows it to provide a good description of the data, much better than was possible with a homogenous tissue kinetic model of the system. Application of the iterative filter, implemented in SAIF, resolved problems of classic spectral analysis by sharpening separation of near-zero components from trapping and rapidly equilibrating components from blood, and thereby provided good estimates for leucine kinetic parameters and rCPS. Moreover, SAIF estimates in this study were in good agreement with those based on voxelwise analyses. SAIF can therefore be considered as a valid tool for estimation of rCPS from -[1-11C]leucine PET data at ROI level.
The authors declare no conflict of interest.
References
- Bertoldo A, Vicini P, Sambuceti G, Lammertsma AA, Parodi O, Cobelli C. Evaluation of compartmental and spectral analysis models of [18F]FDG kinetics for heart and brain studies with PET. IEEE Trans Biomed Eng. 1998;45:1429–1448. doi: 10.1109/10.730437. [DOI] [PubMed] [Google Scholar]
- Bishu S, Schmidt KC, Burlin T, Channing M, Conant S, Huang T, Liu Z-H, Qin M, Vuong B, Unterman A, Xia Z, Zametkin A, Herscovitch P, Smith CB. Regional rates of cerebral protein synthesis measured with L-[1-11C]leucine and PET in conscious, young adult men: normal values, variability, and reproducibility. J Cereb Blood Flow Metab. 2008;28:1502–1513. doi: 10.1038/jcbfm.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishu S, Schmidt KC, Burlin T, Channing M, Horowitz L, Huang T, Liu Z-H, Qin M, Vuong B, Unterman A, Xia Z, Zametkin A, Herscovitch P, Quezado Z, Smith CB. Propofol anaesthesia does not alter regional rates of cerebral protein synthesis measured with L-[1-11C]leucine and PET in healthy malesubjects. J Cereb Blood Flow Metab. 2009;29:1035–1047. doi: 10.1038/jcbfm.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DJ, Doder M, Osman S, Luthra SK, Hirani E, Hume S, Kase H, Kilborn J, Martindill S, Mori A. Positron emission tomography analysis of [11C]KW-6002 binding to human and rat adenosine A2A receptors in the brain. Synapse. 2008;62:671–681. doi: 10.1002/syn.20539. [DOI] [PubMed] [Google Scholar]
- Brooks DJ, Lammertsma AA, Beaney RP, Leenders KL, Buckingham PD, Marshall J, Jones T. Measurement of regional cerebral pH in human subjects using continuous inhalation of 11CO2 and positron emission tomography. J Cereb Blood Flow Metab. 1984;4:458–465. doi: 10.1038/jcbfm.1984.65. [DOI] [PubMed] [Google Scholar]
- Buxton RB, Alpert NM, Babikian V, Weise S, Correia JA, Ackerman RH. Evaluation of the 11CO2 positron emission tomographic method for measuring brain pH. I. pH changes measured in states of altered PCO2. J Cereb Blood Flow Metab. 1987;7:709–719. doi: 10.1038/jcbfm.1987.125. [DOI] [PubMed] [Google Scholar]
- Carson R, Barker W, Liow J-S, Johnson C. Design of a motion-compensation OSEM list-mode algorithm for resolution-recovery reconstruction for the HRRT. IEEE Trans Nucl Sci. 2003;5:3281–3285. [Google Scholar]
- Collins RC, Nandi N, Smith CB, Sokoloff L. Focal seizures inhibit brain protein synthesis. Trans Am Neurol Assoc. 1980;105:43–46. [PubMed] [Google Scholar]
- Cunningham VJ, Jones T. Spectral analysis of dynamic PET studies. J Cereb Blood Flow Metab. 1993;13:15–23. doi: 10.1038/jcbfm.1993.5. [DOI] [PubMed] [Google Scholar]
- DiStefano JJ. Optimized blood sampling protocols and sequential design of kinetic experiments. Am J Physiol. 1981;240:R259–R265. doi: 10.1152/ajpregu.1981.240.5.R259. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Mejia M, Itoh M, Yanai K, Meguro K, Sasaki H, Ono S, Itoh H, Fukuda H, Iwata R, Ido T, Watabe H, Cunningham VJ, Ashburner J, Jones T.1996Quantitative imaging of [11C]benztropine in the human brain with graphical analysis and spectral analysis Quantification of brain function using PET(Myers R, Cunningham V, Bailey D, Jones T, eds),San Diego, CA: Academic Press; 317–320. [Google Scholar]
- Hammers A, Asselin M-C, Turkheimer FE, Hinz R, Osman S, Hotton G, Brooks DJ, Duncan JS, Koepp MJ. Balancing bias, reliability, noise properties and the need for parametric maps in quantitative ligand PET: [11C]diprenorphine test-retest data. Neuroimage. 2007;38:82–94. doi: 10.1016/j.neuroimage.2007.06.035. [DOI] [PubMed] [Google Scholar]
- Hinz R, Selvaraj SS, Venkatesha Murthy N, Bhagwagar Z, Taylor M, Cowen PJ, Grasby PM. Effects of citalopram infusion on the serotonin transporter binding of [11C]DASB in healthy controls. J Cereb Blood Flow Metab. 2008;28:1478–1490. doi: 10.1038/jcbfm.2008.41. [DOI] [PubMed] [Google Scholar]
- Iida H, Higano S, Tomura N, Shishido F, Kanno I, Miura S, Murakami M, Takahashi K, Sasaki H, Uemura K. Evaluation of regional differences of tracer appearance time in cerebral tissues using [15O]water and dynamic positron emission tomography. J Cereb Blood Flow Metab. 1988;8:285–288. doi: 10.1038/jcbfm.1988.60. [DOI] [PubMed] [Google Scholar]
- Ingvar MC, Maeder P, Sokoloff L, Smith CB. Effects of ageing on local rates of cerebral protein synthesis in Sprague-Dawley rats. Brain. 1985;108 (Pt 1:155–170. doi: 10.1093/brain/108.1.155. [DOI] [PubMed] [Google Scholar]
- Lajtha A, Latzkovits L, Toth J. Comparison of turnover rates of proteins of the brain, liver and kidney in mouse in vivo followinglong term labeling. Biochim Biophys Acta. 1976;425:511–520. doi: 10.1016/0005-2787(76)90015-0. [DOI] [PubMed] [Google Scholar]
- Landaw EM, DiStefano JJ. Multiexponential, multicompartmental, and noncompartmental modeling. II. Data analysis and statistical considerations. Am J Physiol. 1984;246:R665–R677. doi: 10.1152/ajpregu.1984.246.5.R665. [DOI] [PubMed] [Google Scholar]
- Lawson CL, Hanson RJ. Solving least squares problems. Englewood Cliffs, NJ: Prentice-Hall; 1974. pp. 158–167. [Google Scholar]
- Nakanishi H, Sun Y, Nakamura RK, Mori K, Ito M, Suda S, Namba H, Storch FI, Dang TP, Mendelson W, Mishkin M, Kennedy C, Gillin JC, Smith CB, Sokoloff L. Positive correlations between cerebral protein synthesis rates and deep sleep in Macaca mulatta. Eur J Neurosci. 1997;9:271–279. doi: 10.1111/j.1460-9568.1997.tb01397.x. [DOI] [PubMed] [Google Scholar]
- Qin M, Kang J, Burlin TV, Jiang C, Smith CB. Postadolescent changes in regional cerebral protein synthesis: an in vivo study in the FMR1 null mouse. J Neurosci. 2005;25:5087–5095. doi: 10.1523/JNEUROSCI.0093-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson MP, Koepp MJ, Brooks DJ, Fish DR, Duncan JS. Benzodiazepine receptors in focal epilepsy with cortical dysgenesis: an [11C]flumazenil PET study. Ann Neurol. 1996;40:188–198. doi: 10.1002/ana.410400210. [DOI] [PubMed] [Google Scholar]
- Schmidt KC. linear compartmental systems can be analyzed by spectral analysis of PET output data summed overall compartments. J Cereb Blood Flow Metab. 1999;19:560–569. doi: 10.1097/00004647-199905000-00010. [DOI] [PubMed] [Google Scholar]
- Schmidt KC, Cook MP, Qin M, Kang J, Burlin TV, Smith CB. Measurement of regional rates of cerebral protein synthesis with L-[1-11C]leucine and PET with correction for recycling of tissue amino acids: I. Kinetic modeling approach. J Cereb Blood Flow Metab. 2005;25:617–628. doi: 10.1038/sj.jcbfm.9600067. [DOI] [PubMed] [Google Scholar]
- Siesjo BK, Thompson WO. The rate of incorporation of gaseous 14CO2 into brain tissue constituents. Experientia. 1965;20:98–99. doi: 10.1007/BF02151265. [DOI] [PubMed] [Google Scholar]
- Smith CB, Kang J. Cerebral protein synthesis in a genetic mouse model of phenylketonuria. Proc Natl Acad Sci USA. 2000;97:11014–11019. doi: 10.1073/pnas.97.20.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CB, Schmidt KC, Qin M, Burlin TV, Cook MP, Kang J, Saunders RC, Bacher JD, Carson RE, Channing MA, Eckelman WC, Herscovitch P, Laverman P, Vuong BK. Measurement of regional rates of cerebral protein synthesis with L-[1-11C]leucine and PET with correction for recycling of tissue amino acids: II. Validation in rhesus monkeys. J Cereb Blood Flow Metab. 2005;25:629–640. doi: 10.1038/sj.jcbfm.9600066. [DOI] [PubMed] [Google Scholar]
- Sun Y, Deibler GE, Jehle J, Macedonia J, Dumont I, Dang T, Smith CB. Rates of local cerebral protein synthesis in the rat during normalpostnatal development. Am J Physiol. 1995;268:R549–R561. doi: 10.1152/ajpregu.1995.268.2.R549. [DOI] [PubMed] [Google Scholar]
- Sundaram SK, Muzik O, Chugani DC, Mu F, Mangner TJ, Chugani HT. Quantification of protein synthesis in the human brain using L-[1-11C]-leucine PET: incorporation of factors for large neutral amino acids in plasma and for amino acids recycled from tissue. J Nucl Med. 2006;47:1787–1795. [PubMed] [Google Scholar]
- Tadokoro M, Jones AKP, Cunningham VJ, Sashin D, Grootoonk S, Ashburner J, Jones T.1993Parametric images of [11C]diprenorphine binding using spectral analysis of dynamic PET images acquired in 3D Quantification of brain function. Tracer kinetics and image analysis in brain PET(Uemura K, Lassen NA, Jones T, Kanno I, eds),Amsterdam: Elsevier Science Publishers; 289–294. [Google Scholar]
- Tomasi G, Bertoldo A, Bishu S, Unterman A, Smith CB, Schmidt K. Voxel-based estimation of kinetic model parameters of the L-[1-11C]leucine PET method for determination of regional rates of cerebral protein synthesis: validation and comparison with region-of-interest-based methods. J Cereb Blood Flow Metab. 2009;29:1317–1331. doi: 10.1038/jcbfm.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkheimer F, Moresco RM, Lucignani G, Sokoloff L, Fazio F, Schmidt K. The use of spectral analysis to determine regional cerebral glucose utilization with positron emission tomography and [18F]fluorodeoxyglucose: theory, implementation, and optimization procedures. J Cereb Blood Flow Metab. 1994;14:406–422. doi: 10.1038/jcbfm.1994.52. [DOI] [PubMed] [Google Scholar]
- Widmann R, Kuroiwa T, Bonnekoh P, Hossmann KA. [14C]leucine incorporation into brain proteins in gerbils after transient ischemia: relationship to selective vulnerability of hippocampus. J Neurochem. 1991;56:789–796. doi: 10.1111/j.1471-4159.1991.tb01993.x. [DOI] [PubMed] [Google Scholar]
- Widmann R, Kocher M, Ernestus RI, Hossmann KA. Biochemical and autoradiographical determination of protein synthesis in experimental brain tumors of rats. J Neurochem. 1992;59:18–25. doi: 10.1111/j.1471-4159.1992.tb08870.x. [DOI] [PubMed] [Google Scholar]
- Wienhard K, Schmand M, Casey ME, Baker K, Bao J, Eriksson L, Jones WF, Knoess C, Lenox M, Lercher M, Luk P, Michel C, Reed JH, Richerzhagen N, Treffert J, Vollmar S, Young JW, Heiss WD, Nutt R. The ECAT HRRT: performance and first clinical application of the new high resolution research tomograph. IEEE Trans Nucl Sci. 2002;49:104–110. [Google Scholar]
- Wu Y, Carson R. Noise reduction in the simplified reference tissue model for neuroreceptor functional imaging. J Cereb Blood Flow Metab. 2002;22:1440–1452. doi: 10.1097/01.WCB.0000033967.83623.34. [DOI] [PubMed] [Google Scholar]