Summary
Aspartyl-phosphate phosphatases underlie the rapid responses of bacterial chemotaxis. One such phosphatase, CheZ, was originally proposed to be restricted to proteobacter, suggesting only a small subset of microbes relied on this protein. A putative CheZ phosphatase was identified genetically in the epsilon proteobacter Helicobacter pylori (Terry et al., 2006, Mol Micro 61:187). H. pylori utilizes a chemotaxis system consisting of CheAY, three CheVs, CheW, CheY and the putative CheZ to colonize the host stomach. Here we investigate whether this CheZ has phosphatase activity. We phosphorylated potential targets in vitro using either a phosphodonor or the CheAY kinase and [γ-32P] ATP, and found that H. pylori CheZ (CheZHP) efficiently dephosphorylates CheYHP and CheAY and has additional weak activity on CheV2. We detected no phosphatase activity toward CheV1 or CheV3. Mutations corresponding to E. coli CheZ active site residues or deletion of the C-terminal region inactivate CheZHP phosphatase activity, suggesting the two CheZ's function similarly. Bioinformatics analysis suggests that CheZ phosphatases are found in all proteobacteria classes, as well as classes Aquificae, Deferribacteres, Nitrospira, and Sphingobacteria, demonstrating that CheZ phosphatases are broadly distributed within Gram-negative bacteria.
Introduction
Chemotaxis is a mechanism that allows microorganisms to sense their chemical environment and respond by moving away from harmful agents or toward attractive ones. Canonical chemotaxis signal transduction involves sensing a chemical signal by a chemoreceptor protein, whose interaction with the coupler protein CheW sends a signal to histidine kinase CheA to undergo ATP-dependent autophosphorylation (Wadhams & Armitage, 2004). The phosphoryl group is then transferred to the response regulator CheY, which binds to the flagellar motor. The level of phosphorylated CheY (CheY-P) effects the rotational direction of the flagellar motor causing the flagella to turn clockwise or counter clockwise. For a microorganism to respond quickly to the external chemical environment, it is necessary for CheY-P to rapidly return to its non-phosphorylated form. CheY-P has an intrinsic dephosphorylation activity that is accelerated by a variety of mechanisms. The first discovered was the CheY phosphatase CheZ, which accelerates the dephosphorylation of CheY-P (Hess et al., 1988, Zhao et al., 2002, Silversmith, 2010). However, CheZ homologs have not been reported in all microorganisms with chemotaxis systems (Szurmant & Ordal, 2004). Many microorganisms have another family of CheY-P phosphatases, the CheC/FliY/CheX phosphatases, whose mechanism is similar to CheZ (Muff et al., 2007, Wuichet et al., 2007, Pazy et al., 2010, Silversmith, 2010). Another way to regulate the cellular level of CheY-P is to decrease the flow of phosphoryl groups to CheY using additional CheY-homologous proteins termed phosphate sinks. In the prototype example of this pathway in Sinorhizobium meliloti, the phosphate sink is an alternative CheY that accepts phosphoryl groups from CheA, but does not interact with the flagellar switch (Sourjik & Schmitt, 1998, Szurmant & Ordal, 2004). By directing phosphate flow to the sink protein, the signaling CheY is able to autodephosphorylate and the signal is terminated.
Helicobacter pylori is a chemotactic ε-proteobacteria that infects 50% of the world’s population (Covacci et al., 1999). Infection with H. pylori can lead to ulcers or gastric cancer (Kandulski et al., 2008). H. pylori relies on chemotaxis to successfully colonize the gastric mucosa of the host (Foynes et al., 2000, Terry et al., 2005). The chemotaxis system in H. pylori deviates from the E. coli model. The major chemotaxis proteins CheAY, CheW, and CheYHP are present. CheAY is a modified CheA with a response regulator receiver domain (REC) fused at its C-terminus (Tomb et al., 1997, Foynes et al., 2000). There are three CheV proteins called CheV1, CheV2, and CheV3, which are hybrids of CheW and a REC domain. Neither CheZ nor any of the CheC/FliY/CheX phosphatases were annotated in the original genome sequencing of H. pylori (Tomb et al., 1997). Jiménez-Pearson et al. found that in vitro CheAY and CheA’ (CheAY without its REC domain) transferred phosphoryl groups to both CheYHP and the CheAY REC domain. Because the REC domain of CheAY would not be expected to interact with the flagellar motor, it was suggested that this phosphorylation event might be akin to a phosphate sink and thus that H. pylori might employ this method for dephosphorylating CheYHP-P (Jiménez-Pearson et al., 2005). The lack of known phosphatases and the presence of three CheV proteins also suggested that CheV proteins might act as phosphate sinks in the chemotaxis system of H. pylori, but the lower affinity of CheA’ for CheV made this an unlikely scenario (Pittman et al., 2001, Jiménez-Pearson et al., 2005).
Previous work from our lab reported the identification of a remote CheZ homologue, HP0170 (Terry et al., 2006). It was identified as a chemotactic (Che+) suppressor of a non-chemotactic cheW mutant. Extended incubation of non-chemotactic cheW mutants resulted in recovered migration ability in soft-agar media. Proteomic analysis of this Che+ suppressor mutant revealed that it had lost expression of HP0170. Analysis of additional suppressor mutants showed various mutations in the HP0170 gene, including single insertions, multiple base pair insertions, deletions, and missense mutations, that all resulted in loss or mutation of the C-terminal domain of the protein. Using the SAM-T02 program that identifies very remote protein homologs, HP0170 was identified as a remote homologue of CheZ.
Here we investigate the potential phosphatase activity of HP0170 on all H. pylori chemotaxis proteins bearing a REC domain: CheAY, the three CheVs, and CheYHP. H. pylori is different from the well-studied E. coli in this respect of having more than two potential phosphatase targets. We show here that HP0170 is a phosphatase for phosphoryl CheYHP (CheYHP-P) and phosphoryl CheAY (CheAY-P), as well as a weak phosphatase for phosphoryl CheV2 (CheV2-P). Mutations at residues corresponding to the E. coli CheZ active site inactivate HP0170’s dephosphorylation activity. The deletion of 12 amino acids at the C-terminus also inactivates HP0170 activity. These findings show that HP0170 functions as a CheZ for H. pylori, and thus we refer to it as CheZHP. Using PSI-BLAST to search for CheZHP orthologs, we found that CheZ is present across several classes of Gram negative bacteria.
Results
HP0170 is a CheYHP-P phosphatase
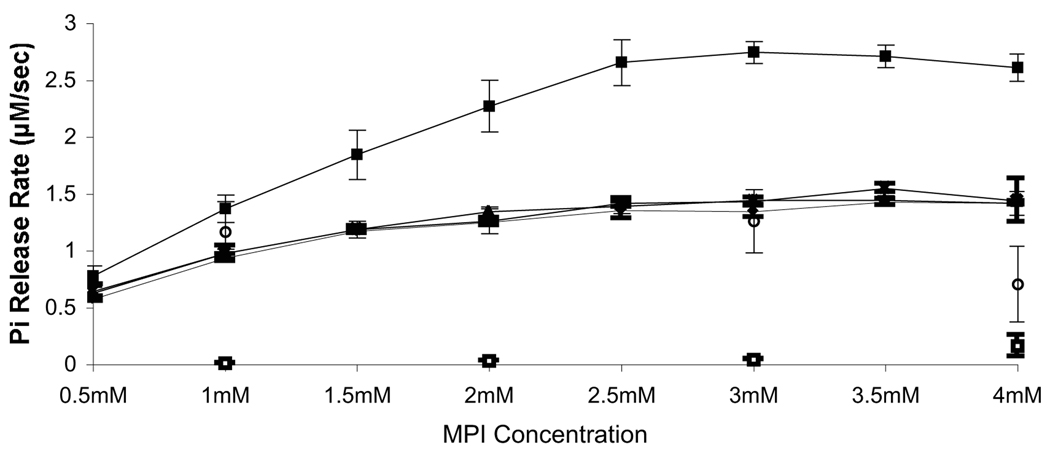
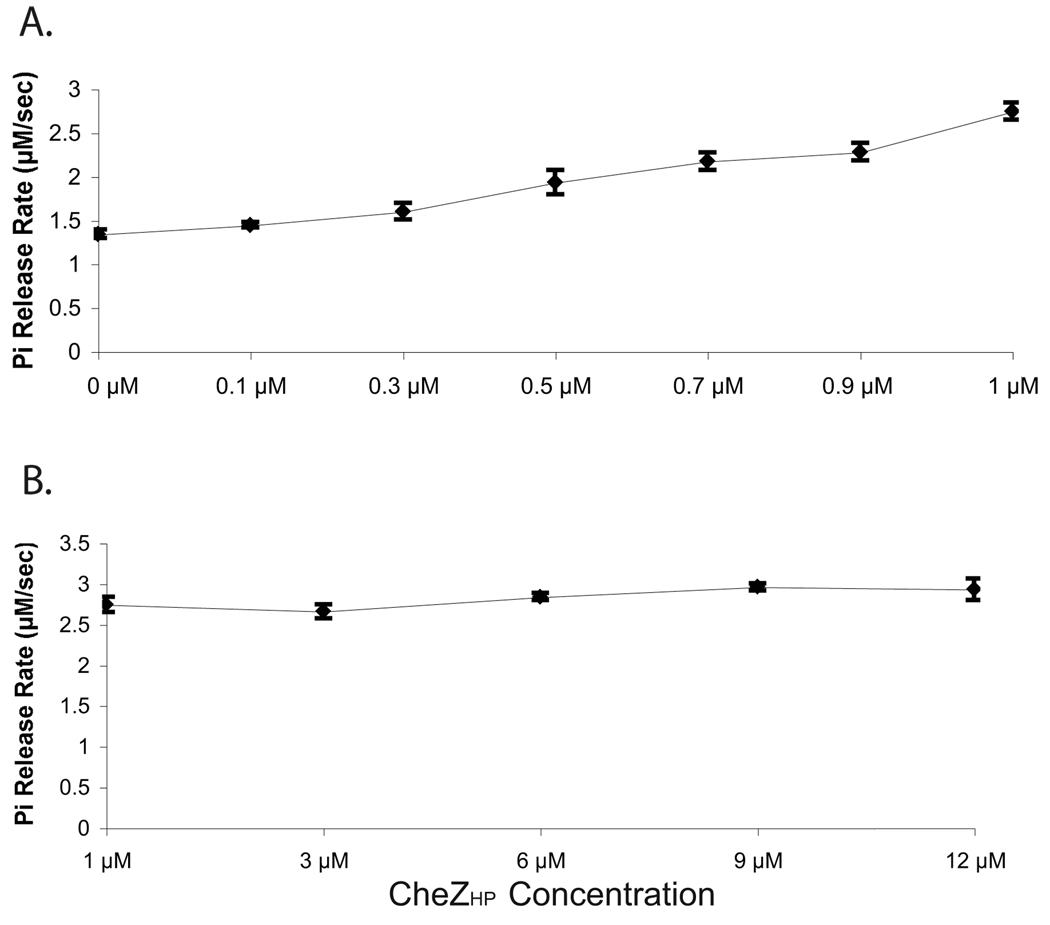
Previous work identified HP0170 as a very remote homolog of the E. coli CheZ phosphatase (Terry et al., 2006) but it was not known whether this protein retained phosphatase activity. To determine whether HP0170 has phosphatase activity, we first employed the EnzChek ® Phosphate Assay Kit as developed by Silversmith et al. 2001 for use with E. coli CheY and CheZ. This method uses the small-molecule phosphodonor monophosphoimidazole (MPI) to phosphorylate CheY, and then measures the steady-state phosphate release due to CheY dephosphorylation. The phosphate release thus depends on both the rates of phosphorylation and dephosphorylation. In the case of E. coli CheZ and CheY, CheY’s autodephosphorylation activity releases a basal amount of phosphate that is enhanced by CheZ. For our assay, we employed recombinant purified H. pylori proteins. We first tested whether H. pylori CheY (CheYHP) would be phosphorylated by MPI, and varied the concentration of the MPI substrate to determine saturating conditions. Similar to E. coli CheY, we found that CheYHP releases phosphate and that this release reaches a saturation point at 2.5 mM MPI, using 5 µM CheYHP (Fig. 1). This behavior suggests that CheYHP can be phosphorylated by MPI and that at concentrations above 2.5 mM MPI, autodephosphorylation is the rate-limiting step because increasing the concentration of phosphodonor does not increase the phosphate release rate. The expected phosphorylation site of CheYHP, aspartate 53, is required for the phosphorylation by MPI as a CheYHP D53A mutant does not show any phosphate release in this phosphate release assay (data not show). We also determined that Mg2+ is the optimal divalent metal ion for CheYHP phosphorylation/dephosphorylation, similar to other CheY proteins (data not shown). The addition of HP0170 to a reaction containing CheYHP and MPI significantly increases the rate of phosphate release indicating that HP0170 is a CheYHP-P phosphatase (Fig. 1). Dephosphorylation of CheYHP-P by HP0170 eventually reaches saturation suggesting that this is the maximum dephosphorylation activity by HP0170 under these experimental conditions. These results demonstrate that HP0170 is a CheYHP-P phosphatase in H. pylori and can be appropriately called CheZHP. Additionally, we expanded the concentrations of CheZHP tested ranging from 0.1 to 12 µM, while keeping the amount of CheYHP constant at 5 µM. The phosphate release rate increases more-or-less linearly as a function of CheZHP concentration, with CheZHP reaching its maximum activity at 1 µM (Fig. 2). This finding is likely due to the autophosphorylation of CheY being the rate-limiting step, such that additional phosphatase cannot increase the release rate. Of note, the presence of CheZHP alone without CheYHP did not result significant phosphate release (Fig. 1).
Figure 1. CheYHP-P autodephosphorylation and dephosphorylation in the presence of CheZHP and its variants.
Shown is the amount of phosphate released using MPI as a phosphodonor. Lines are marked as follows: ♦, CheY alone; ■, CheYHP plus CheZHP; ▲, CheYHP plus CheZHP D189N; x, CheYHP plus CheZHP Q193R; ○, CheYHP plus CheZHPΔC12; □, CheZHP alone. Each reaction contains 5 µM CheY and 1 µM CheZHP and the indicated concentration of MPI. The data represents an average of two to four experiments and error bars show standard error. The lines overlap for CheY alone, CheYHP plus CheZHP D189N, CheYHP plus CheZHP Q193R and CheYHP plus CheZHPΔC12.
Figure 2. CheYHP–P dephosphorylation saturates at 1µM CheZHP.
Shown is the amount of phosphate released using MPI as a phosphodonor. The reaction includes 5 µM CheYHP, 3 mM MPI, and CheZHP at varying concentrations. (A) CheZHP at lower concentration ranging from 0 −1 µM. (B) CheZHP concentration ranging from 1–12 µM. The data represents the average of three experiments, and error bars show standard error.
Using [γ-32P] ATP to phosphorylate CheYHP via CheAY confirms the phosphatase activity of CheZHP
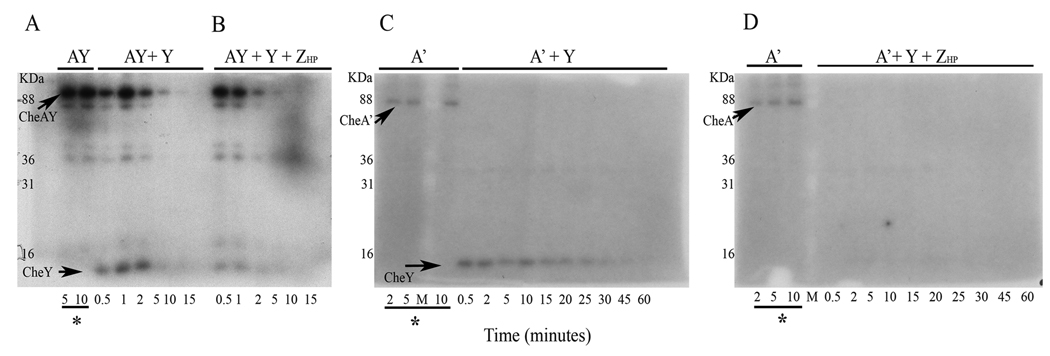
To confirm that CheZHP is a CheYHP-P phosphatase, we employed a traditional method of phosphorylation using the cognate histidine kinase with radioactive ATP. For this assay, H. pylori CheAY was phosphorylated with [γ-32P] ATP which in turn phosphorylated CheYHP. We used full length CheAY, as well as a truncated variant with its REC domain removed called CheA’ (Jiménez-Pearson et al., 2005). We first phosphorylated CheYHP with either CheAY-P or CheA’-P, under multiple turnover conditions in which ATP was supplied throughout. CheYHP was phosphorylated by both CheAY and CheA’ as evident by the appearance of CheY-sized radioactive bands after CheYHP was added to the reaction (Fig. 3A and 3C). This finding is identical to what was found previously by Jiménez-Pearson et al. 2005. CheAY-P bands are visible for up to 10 minutes in a reaction containing CheYHP (Fig. 3A). In the presence of CheZHP, CheY-P and CheAY-P bands are visible for much shorter times, presumably as a result of transfer of phosphoryl group to CheYHP and accelerated dephosphorylation of CheYHP-P by CheZHP (Fig. 3B); as discussed in the next section, however, CheZ also targets CheAY-P but does so with less efficiency as compared to CheY-P. Thus multiple reactions are occurring, with CheZHP targeting CheAY-P and CheY-P, and a further remote possibility that CheZHP has some contaminating phosphatase that directly targets ATP. We thus simplified this assay by using the truncated variant CheA'. CheA'-P dephosphorylated as soon as CheYHP was added but the phosphorylated CheYHP-P remained visible for an extended period of time (Fig. 3C). This rapid movement of phosphate from CheA’-P to CheYHP is presumably due to the absence of the REC domain in CheA’ such that the protein now rapidly transfers phosphoryl groups to CheYHP. In this assay, CheA' autophosphorylation and phosphoryl group transfer is continuous. The observation of stable CheYHP-P indicates that the rate of phosphoryl transfer from CheA' to CheYHP exceeds the rate of CheYHP autodephosphorylation under these conditions. The addition of CheZHP quickly dephosphorylates CheYHP-P generated by CheA’, as CheYHP-P bands are barely visible compared to the reaction that lacks CheZHP (Fig. 3 C and D). We attempted to determine the half-life of CheYHP-P by first phosphorylating CheA’-P, removing excess ATP using gel filtration, and then adding this CheA'-P to CheYHP. Under these single-turnover conditions, we could not visualize CheYHP-P even after only five seconds (data not shown), supporting that CheYHP rapidly dephosphorylates without continuous phosphorylation by CheA'. We estimated the CheYHP autodephosphorylation rate constant, kdephosph, by dividing the saturating dephosphorylation rate from Figure 1 (1.42 µM /sec) by the concentration of CheYHP (5 µM) (Silversmith et al., 2001). This rate of H. pylori CheY dephosphorylation is 0.28 s−1, approximately 8 times higher than the reported kdephosph of E. coli CheY, 0.035 s−1 (Silversmith et al., 2001); this calculation indicates that CheYHP-P has a half-life of approximately 2.5 seconds. This short half-life is consistent with our inability to detect CheYHP-P without repeat phosphorylation. Taken together, these assays results strongly support that CheZHP is a CheYHP-P phosphatase.
Figure 3. Phosphorylation assay of CheYHP using CheAY-P and CheA’-P.
The amount of phosphor-CheY, phosphor-CheAY, and phosphor-CheA’ were monitored using autoradiography of SDS-PAGE gels. CheAY and CheA’ were phosphorylated in vitro for 10 minutes before the addition of CheY with or without CheZHP. 2 µM of CheAY, 12 µM CheA’, 4 µM of CheYHP and 1 µM of CheZHP were used unless specified. (A) CheAY phosphorylated with 43 µM ATP with addition of CheYHP. (B) Same set up as (A) with the addition of CheZHP. (C) CheA’ phosphorylated with 114 µM ATP with the addition of 12 µM CheYHP. (D) Same as (C) with the addition of 3 µM CheZHP. Underlined time points marked with asterisks indicate phosphorylation of CheAY or CheA' before addition of CheYHP and CheZHP. kDa indicates the molecular weight marker lane, with values indicating kilodaltons. The data is a representative of three experiments. The extra bands in are likely to be partially degraded CheAY-P that was also observed shortly after purification of CheAY.
CheZHP has phosphatase activity on CheAY but not on CheA’
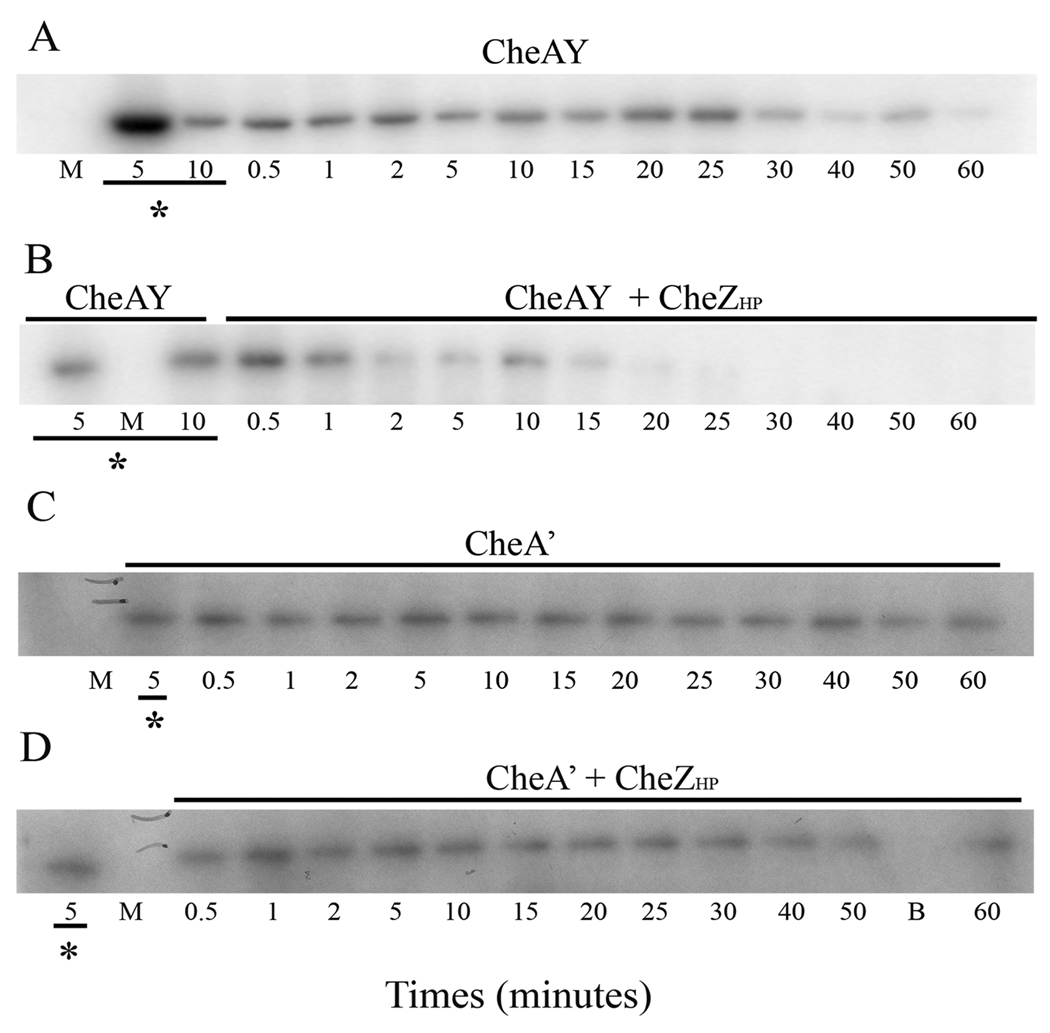
We also examined whether CheZHP has a phosphatase activity on H. pylori CheAY and CheA’. We phosphorylated CheAY and CheA’ with [γ-32P] ATP, followed by addition of excess cold ATP to follow the dephosphorylation of these proteins. CheZHP dephosphorylates CheAY-P as seen in Figure 4A and B. The radioactive bands of CheAY-P start to disappear at 25 minutes after the chase. We quantified the band intensity across this gel, and used that information to calculate the half-life of CheAY-P to be approximately 30 minutes, or about 10 times longer than found previously (Jiménez-Pearson et al., 2005). We are not sure of the reason for this discrepancy. In the presence of CheZHP, the radioactive bands disappear by 15 minutes indicating that CheZHP affects the phosphorylation state of CheAY-P. The calculated half-life of CheAY-P chased with CheZHP is approximately 12 minutes. CheA’-P, however, was not affected by CheZHP even with an extended incubation time (Fig. 4C and D). This finding suggests that CheZHP targets the REC domain of CheAY; however the relatively weak phosphorylation of CheA' as compared to CheAY could indicate that the truncated protein has lost some function. Overall, this set of experiments confirms that CheZHP is a phosphatase whose activity is not restricted to CheYHP-P but also extends to CheAY-P.
Figure 4. CheZHP has phosphatase activity on CheAY but not on CheA’.
Pulse-chase phosphorylation assays were done as described in the text, using CheAY and CheA’ phosphorylated with 203 µM ATP as described in the methods, followed by addition of 50-fold excess unlabelled ATP and CheZHP. Aliquots were removed at the indicated time points. (A) 2 µM CheAY was phosphorylated for 10 minutes followed by addition of 10 mM of unlabelled ATP. (B) Same as (A) with the addition of 1 µM CheZHP at the same time as the unlabelled ATP. (C) 12 µM of purified CheA’-P was chased with 10mM unlabeled ATP. (D) Same as (C) with 6 µM CheZHP added along with the unlabeled ATP. Underlined time points with asterisks indicate the absence of unlabeled ATP and CheZHP. The bands show phosphorylated CheAY in (A) and (B), and phosphorylated CheA’ in (C) and (D). This data is a representative of two experiments. M and B indicated marker lane and blank lane, respectively. Gels were exposed to phosphoroimager cassette for 3 hours to overnight, or to X-ray film for 24–48 hours. CheA’ bands always appear weaker likely due to some loss of activity associated with this truncation.
CheZHP uses the same active site residues as does E. coli CheZ
To further analyze the activity of CheZHP, we generated and analyzed variants of CheZHP that changed aspartate 189 to asparagine (CheZHP D189N), glutamine 193 to arginine (CheZHP Q193R) and deleted the 12 C-terminal amino acids (CheZHP ΔC12). These residues and sequences are critical for E. coli CheZ function. In E. coli, mutation of either D143 or Q147 (equivalent to D189 and Q193 in H. pylori strain 26695 and D177 and Q181 in H. pylori strain SS1, respectively, in CheZHP; see Figure 6) results in proteins that do not dephosphorylate CheY-P (Boesch et al., 2000, Zhao et al., 2002). The C-terminus of E. coli CheZ binds to CheY-P to bring it to the CheZ active site (Blat & Eisenbach, 1996, Zhao et al., 2002, Silversmith, 2005). Additionally, one of our original HP0170 loss-of-function mutants had a point mutation that changed aspartate 189 to asparagine (Terry et al., 2006). Phosphate release assays showed that these point mutations render CheZHP inactive as a phosphatase of CheYHP-P, indicating that D189 and Q193 are important for CheZHP dephosphorylation of CheYHP-P (Fig. 1). The absence of the last 12 amino acids at the C-terminus displays a similar defect to the point mutants (Fig. 1). However, the C-terminus is located approximately 50 amino acids downstream of the point mutations suggesting that the C-terminus might perform the same function as the C-terminus of E. coli CheZ, binding CheY-P, rather than being a part of the active site. Of note, all of these CheZHP mutant proteins were stable and maintained their length (data not shown), suggesting the mutations did not grossly affect protein folding and stability.
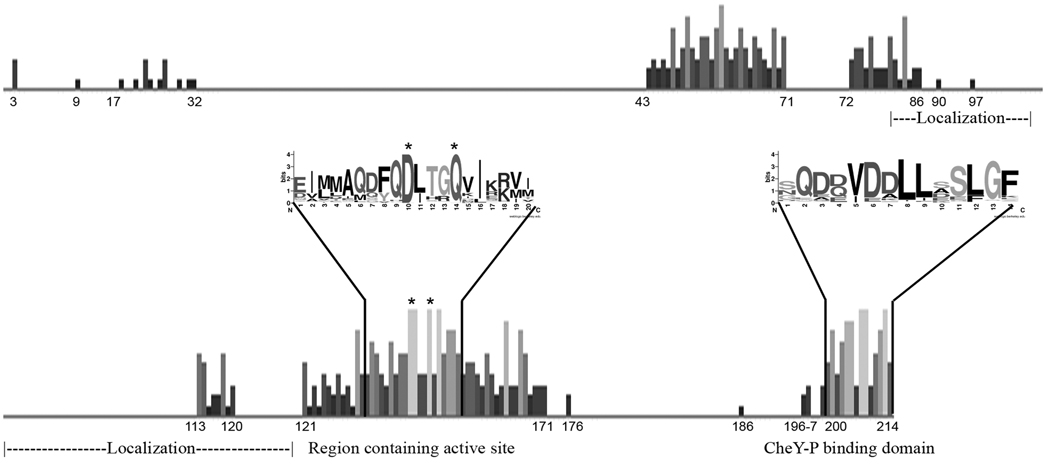
Figure 6. Amino acids conservation among CheZHP orthologs.
Shown is the output of a multiple sequence alignment displayed with degree of conservation as height of the bar at each position. Inset sequence logo format shows sequences of highly conserved regions. The numbering corresponds to E. coli CheZ sequence. The conserved Asp 143 and Gln 147 in E. coli CheZ are indicated by asterisks.
CheZHP dephosphorylates CheV2 but not CheV1 or CheV3
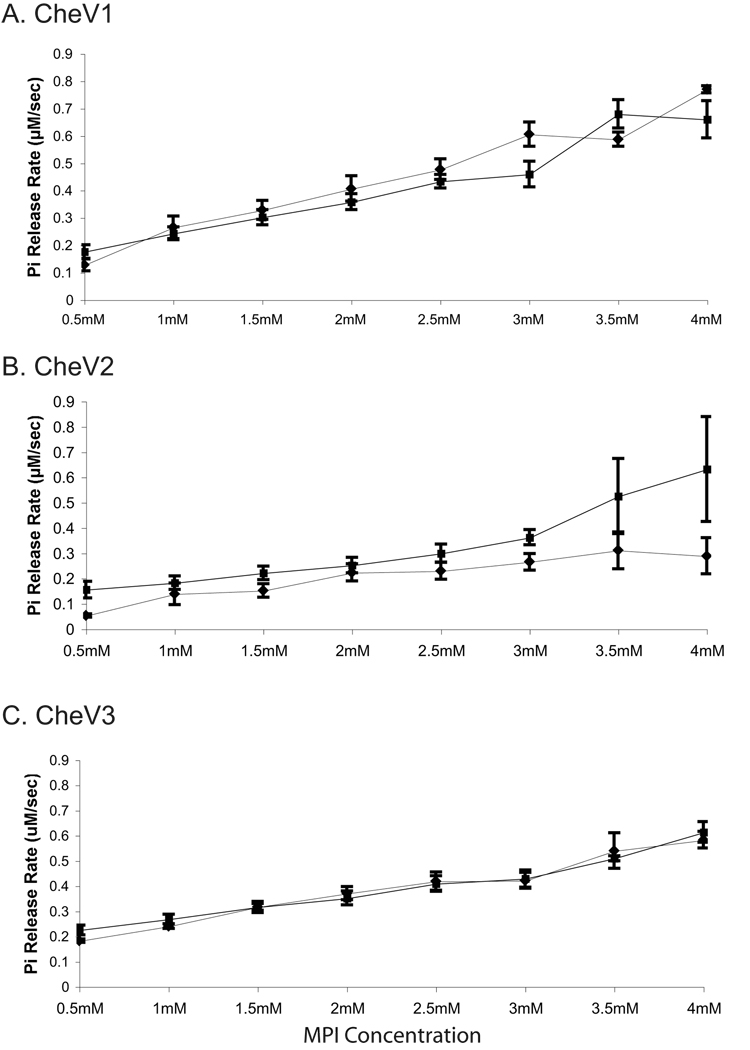
CheZHP was also tested for its phosphatase activity on other H. pylori chemotaxis proteins that have REC domains: CheV1, CheV2, and CheV3. Under our experimental conditions, phosphate release rates by the CheV proteins are comparatively lower than that of CheYHP (Fig. 5 as compared to Fig. 1; 0.2–0.7 µM /sec versus 1.4 µM /sec), suggesting that while MPI is a phosphodonor for each, it may be a poor substrate. We also determined the optimal Mg2+ concentration for phosphorylation and autodephosphorylation of each CheV, and found it to be similar to CheYHP (data not shown). The low phosphate release by CheV proteins could be due either to poor phosphorylation or to slow release, both of which have precedent. The dephosphorylation of Bacillus subtilis CheV was shown to be slowed by the presence of the N-terminal CheW domain (Karatan et al., 2001), and others have had difficulty detecting phosphorylated H. pylori CheV proteins (Jiménez-Pearson et al., 2005). The phosphate release rates for CheV1 and CheV3 do not exhibit saturation, as would be expected with Michaelis-Menton kinetics, and suggesting that the affinity of the CheV REC domain for MPI is very weak (Bourret, 2010). This lack of saturation would predict that phosphorylation is the rate limiting step, and thus that phosphatase assays might be quite insensitive. Indeed, we did not detect any enhanced phosphate release when CheZHP was added to either the CheV1 or CheV3 reactions. CheV2, on the other hand, reaches saturation at 3.5 mM MPI, and has relatively lower phosphate release rate of 0.25 µM /sec compared to 0.5 µM /sec of CheV1 and CheV3 (Fig. 5B). Previous work showed that CheV2 was phosphorylated by acetyl phosphate (Pittman et al., 2001, Jiménez-Pearson et al., 2005). The addition of CheZHP to the reaction containing CheV2 increased the phosphate release rate by 1.6 fold, suggesting that CheZHP has some phosphatase activity on CheV2-P (Fig. 5B). Taken together, these results suggest that CheZHP is primarily a phosphatase for CheYHP-P and CheAY-P, with weak activity on CheV2-P.
Figure 5. Dephosphorylation of the CheV proteins.
Shown is the amount of phosphate released using MPI as a phosphodonor. ♦ CheV alone, ■ CheV with CheZHP. Each reaction contains 5 µM of the indicated CheV proteins s with and without 1 µM CheZHP. (A) CheV1, (B) CheV2, and (C) CheV3. The data represents an average of three experiments, and error bars show standard error.
Domain analysis of HP0170/CheZHP
Our biochemical analysis strongly supported that CheZHP is a phosphatase despite its limited homology to E. coli CheZ. To search for distant homologs of CheZHP, the amino acid sequence of HP0170 from H. pylori strain SS1 was used in a PSI-BLAST query. This analysis identified 268 proteins including E. coli CheZ after the 3rd iteration (Table 1). These proteins had been annotated as either hypothetical proteins, CheZ, probable phosphatase, or MCP and were found in the phylum Proteobacteria (classes α, β, γ, δ, and ε), phylum Aquificae (class Aquificae), phylum Deferribacteres (class Deferribacteres), phylum Nitrospirae (class Nitrospira), and phylum Bacteroidetes (class Sphingobacteria). Representative of each genus were used to generate an amino acid sequence alignment (Fig. 6,Table S1). Of note, all CheZ-bearing classes have annotated chemotaxis genes in their genomes, supporting that these CheZ orthologs act in this pathway. The large spaces between groups of conserved residues are result of incorporating sequences that are longer than the majority of the proteins, therefore no conservation was observed (Fig. 6). We found several CheZ regions with high levels of conservation with the most significant corresponding to the E. coli CheZ active site and the C-terminal end. The active site residues corresponding to Gln 147 and Asp143 of E. coli CheZ are 100% conserved among these proteins. The co-crystal structure of CheZ with CheY-BeF3−Mg2+showed residue Asp143 forms a salt bridge with CheY indicating the importance of this residue for CheZ function, and the CheZ active site residue Gln147 is inserted into the CheY active site and to accelerate the dephosphorylation of CheY-P (Zhao et al., 2002). The C-terminal sequences of CheZ proteins are also conserved supporting the importance of this region in binding to CheY-P, as shown for E. coli CheZ (Blat & Eisenbach, 1996). There are a few other moderately conserved regions that correspond to amino acids 43–86 of E. coli CheZ. Within this region, residues 67–71 create part of the CheY binding surface located in the middle of the CheZ four-helix bundle (Zhao et al., 2002). Other important regions of E. coli CheZ do not seem to be as conserved. For example, the helix-turn-helix hairpin of CheZ that has been shown to be involved in its polar localization via interactions with CheA-short does not show significant conservation, as noted by others (Cantwell et al., 2003). CheA-short is a variant of CheA arising from initiation at an internal methionine, Met97; while sequence alignment of H. pylori CheAY with E. coli CheA shows that Met97 appears to be conserved (data not shown), we do not yet know if CheA-short exists in H. pylori or any bacteria outside of E. coli and its close relative Salmonella enterica (McNamara & Wolfe, 1997).
Table 1.
Selected proteins for PSI-BLAST 3rd iteration.
| Annotation | Bacterial strain | Score (Bits) | E-value |
|---|---|---|---|
| Chemotaxis phosphatase, CheZ |
Desulfatibaciillum alkenivorans | 44.7 | 0.009 |
| Putative chemotaxis phosphatase, CheZ |
Hydrogenobaculum Sp. YO4AAS1 | 40.8 | 0.13 |
| Chemotaxis phosphatase, CheZ |
Shewanella sediminis HAW-EB3 | 38.5 | 0.6 |
Discussion
The primary finding of this study is that the HP0170 protein has phosphatase activity toward CheYHP-P, but also acts on two other chemotaxis REC domain proteins, CheAY and CheV2. Furthermore, CheZHP appears to employ the same mechanism as E. coli CheZ, based on the results of a phosphate release assay using various CheZHP mutants. The co-crystal structure of E. coli CheZ with CheY-BeF3−Mg2+ showed two amino acids to be paramount for phosphatase function. The first residue, Gln147, inserts itself into the active site of CheY-P and orients a water molecule for nucleophilic attack, while the second residue, Asp143, forms a salt bridge with Lys109 of CheY-P to stabilize the interaction between proteins (Zhao et al., 2002). These two residues, as shown in Figure 6, are completely conserved amongst CheZ orthologs. In our original identification of CheZHP via a genetic screen that found cheZ loss-of-function mutations, one of the suppressor mutants had a D189N mutation, corresponding to Asp143 of E. coli CheZ, indicating its importance for the function of CheZHP (Terry et al., 2006). Phosphate release assays with CheZHP D189N and CheZHP Q193R resulted in inactivation of the CheZHP phosphatase activity. The C-terminus of CheZ was proposed to interact directly with CheY-P and bring it to CheZ active site (Silversmith, 2005). Here we found that a CheZHP mutant lacking these C-terminal amino acids, CheZHPΔC12, showed no increase in dephosphorylation of CheYHP-P. The results from these three mutants indicate that CheZHP uses the same mechanism to dephosphorylate CheYHP-P as E. coli CheZ. Overall, the secondary structure prediction of CheZHP is alpha helical, similar to E. coli CheZ, and thus it seems likely that the common portion folds similarly to E. coli CheZ. The N-terminal variable portion may form a separate domain that folds independently, but we have not analyzed this idea further. In CheZHP, this region is highly charged and contains an amino acid repeat of lysines and glutamic acids, as noted previously (Terry et al., 2006).
We found that CheZHP has additional phosphatase activity on CheAY. The H. pylori chemotaxis system has an advantage over that of E. coli in that we could test whether CheZHP can dephosphorylate a range of other REC-containing chemotaxis proteins. Prior to this, CheZ had only been tested on CheY and CheB, and was found to not dephosphorylate CheB (Hess et al., 1988). To our knowledge, there have been no tests of CheZ, or any of the chemotaxis phosphatase, on other REC-containing chemotaxis proteins such as CheV or CheAY. In the case of CheAY, CheZHP appears to act on the REC domain, as the truncated variant that lacks the REC domain, CheA’, was not affected by the presence of CheZHP (Fig. 4). Many microbes have a REC domain attached to the C terminus of CheA (Wuichet et al., 2007), akin to hybrid histidine kinases (Gao & Stock, 2009). In hybrid histidine kinases, the additional REC domain typically functions to transfer phosphoryl groups to a second unlinked histidine kinase, but this reaction has not been studied in CheAY proteins (Gao & Stock, 2009). An alternate function suggested by our findings along with those of Dagmar Beier's group, is that the CheAY REC domain functions to funnel phosphates away from the phosphorylated histidine with facilitated removal by a phosphatase such as CheZ, akin to a phosphate sink. Thus a model for phosphate flow in the H. pylori chemotaxis system is that CheAY autophosphorylates, and then passes phosphoryl groups to CheYHP or, as a second choice, to its own REC domain (also referred to as CheY2) (Jiménez-Pearson et al., 2005). Previous work showed that CheYHP can retrophosphorylate CheAY, and thus phosphoryl groups can flow back from CheYHP to CheAY and finally to the REC domain of CheAY. If CheZHP targets the CheAY REC domain, phosphoryl groups would be removed from the system and not move to CheYHP, thus promoting a phosphate sink-type function.
CheZHP furthermore has weak activity on CheV2, but we did not detect any activity toward CheV1 and CheV3. The phenotypes associated with loss of cheV2 are fundamentally different than those associated with cheV1 and cheV3 mutants, supporting that there are key differences in these proteins (Lowenthal et al., 2009). Specifically, cheV2 null mutants display a hyper-switching phenotype, while cheV1 and cheV3 null mutants are biased toward smooth swimming (Lowenthal et al., 2009). It is not yet known if there are different phenotypes associated with phosphorylation mutants of these CheV proteins, and furthermore, whether our observed CheV2-CheZHP interaction occurs in vivo. The CheV slower autodephosphorylation rate and resistance to CheZHP suggests that a quick turnover rate for most CheVs may not be required for chemotaxis, as suggested by others (Karatan et al., 2001, Szurmant & Ordal, 2004). The stability of their phosphorylation state has suggested that their role is more likely to be involved in adaptation (Szurmant & Ordal, 2004). We were unable to identify any significant conservation of known CheZ-interacting residues (Zhu et al., 1997, McEvoy et al., 1999) on CheYHP, CheAY, or CheV2, pointing out that we do not yet have a complete grasp on the molecular basis for protein-protein interactions of the CheZ family.
An additional known E. coli CheZ protein-protein interaction is with a CheA variant, CheA-short, to drive CheZ's localization to the chemoreceptor patch (Cantwell et al., 2003). The CheZ regions responsible for this localization are located near the apical hairpin turn of the four-helix bundle, residues 80–120 in E. coli CheZ (Cantwell & Manson, 2009). This region is not well conserved amongst various CheZHP orthologs including CheZHP, suggesting that the mechanism for cellular localization of CheZHP might be different. Our preliminary observations show that CheZHP localizes to the pole and can be observed in both soluble and membrane fractions independent of chemoreceptors, CheAY or CheW. (P. Lertsethtakarn, Jenny Draper, and K.M. Ottemann, unpublished). Additionally, CheA-short has not been shown to exist in H. pylori, therefore, it is possible that CheZHP might localize via different mechanism or with different chemotaxis proteins. The CheZHP N-terminus is the most variable region of the protein and is about 40 amino acids longer than E. coli CheZ; indeed, it seems like the C-terminal portion forms a CheZ domain that is consistent between orthologs, with an additional domain or extension on the N-terminal end (Fig. 6). In E. coli CheZ, the N-terminus forms the majority of the extended coiled-coil structure (Zhao et al., 2002), but limited activity has been ascribed to it. There are, however, several point mutations in this region that enhance CheZ phosphatase activity, suggesting this region contributes to phosphatase function (Huang & Stewart, 1993, Sanna & Simon, 1996). The low conservation level in this region, however, suggests that its function might differ between bacteria, or that it simply is not under positive selection. The N-terminal region might be responsible for CheZ's ability to have several substrates in addition to the proposed function of the C-terminus as well as functioning as an interface for its cellular localization.
The identification of CheZHP expands the number of classes of bacteria that harbor the CheZ phosphatase. A database search for CheZHP orthologs using CheZHP from H. pylori SS1 recovered a large number of Gram-negative bacteria with CheZHP orthologs. This analysis strongly suggests that CheZ-type phosphatases are not restricted to the proteobacteria, as previously hypothesized (Wuichet et al., 2007). We found CheZ-orthologs in several non-proteobacter classes, but not in the Gram-positive organisms or archaea, indicating that CheZ is not as widely distributed as CheC-type phosphatases (Muff & Ordal, 2008). Interestingly, some of the identified proteins were annotated as methyl-accepting chemoreceptors (MCP) suggesting the CheZ domain might be fused to other chemotaxis proteins, although this awaits experimental analysis. The high level of conservation of the active site and the CheY-P binding domain (Fig.6) suggests that CheZ function is well conserved among the orthologs. There is little conservation outside of these regions, however, making identification of CheZ orthologs difficult. In general, CheZ and CheC-family phosphatase proteins have been noted to display very low amounts of conservation from organism to organism (Muff & Ordal, 2008). The basis for this low sequence conservation in this family is not yet known. Our work adds the knowledge, however, that remote CheZ proteins can retain function, and thus that CheZ family members play significant roles in chemotaxis systems of a wide variety of bacteria.
Experimental procedures
Molecular Biology Manipulations
H. pylori strain SS1 (Lee et al., 1997) genomic DNA (Promega Wizard DNA Preparation Kit) was used as a template for amplification of cheV1 (hp0019), cheV2 (hp0616), cheV3 (hp0393), cheYHP (hp1067), and cheZHP (hp0170). Gene numbers are from H. pylori strain 26695 (Tomb et al., 1997). All primers are listed in Supplemental Table 2. These genes were cloned into pGEX-6p-2 for overexpression of GST-tagged proteins to facilitate protein purification. The plasmids are pGEX-6p-2-cheV1, pGEX-6p-2-cheV2, pGEX-6p-2-cheV3, pGEX-6p-2-cheYHP, and pGEX-6p-2-cheZHP. pTrc-CheAY and pTrc-CheA’ (Jiménez-Pearson et al., 2005) were generous gifts from Dagmar Beier (University of Würzburg, Germany).
pGEX-6p-2-cheYHPD53A, pGEX-6p-2-cheZHPD189N, and pGEX-6p-2-cheZHPQ193R were generated by site directed mutagenesis using PfuTurbo DNA polymerase following the manufacturer’s protocol (Stratagene). pGEX-6p-2-cheY and pGEX-6p-2-cheZHP were used as templates and primers are listed in Supplemental Table 2. To generate the variant of CheZHP with 12 amino acids removed from the C-terminus, CheZHPΔC12, pGEX-6p-2-cheZHP was used as a template for inverse PCR (iPCR) using primers hp0170iPCRfor2 and hp0170iPCRrev2.
Protein purification
All proteins were over-expressed in E. coli BL21 (Promega). Cultures were grown in LB or 2XYT plus ampicillin to OD600 0.5–0.8 and induced with 0.1–1 mM of IPTG for 4–16 hours at 37°C or 30°C. Cells were harvested by centrifugation, lysed by sonication and then centrifuged at 206,020g for 45 minutes to remove membranes and unlysed cells. The supernatant was applied to a GST Prep 16/10 column (GE Healthcare) for purification of GST-tagged proteins or a His Prep FF16/10 (GE Healthcare) for purification of His-tagged CheAY or CheA’. GST-tagged proteins were treated with Prescission protease® to cleave GST, followed by repurification on a GST Prep 16/10 column to remove GST. Proteins were dialyzed in dialysis buffer (50 mM Hepes, pH 7.6, 50 mM KCl, and 20–30% glycerol) and stored in −80°C.
Phosphate release assay
Phosphate release was monitored using the EnzChek ® Phosphate Assay Kit (Invitrogen) as described by Silversmith et al. (Silversmith et al., 2001). To determine the autodephosphorylation rate of CheYHP, CheV1, CheV2, CheV3, and CheZHP we utilized 450µl reaction containing buffer (100 mM Hepes, pH 7 and 20 mM MgCl2), 1 mM MESG (2-amino-6-mercapto-7-methyl-purine riboside), 0.5 U PNP (purine nucleoside phosphorylase), and various concentration of phosphodonor monophosphoimidazole (MPI), a kind gift of Ruth Silversmith (University of North Carolina, Chapel Hill). The reaction was allowed to equilibrate at room temperature until the OD360 remained stable, after which 5 µM of each protein (except CheZHP in which 1 µM was used) was mixed into the reaction. The change in absorbance at OD360 was monitored for 10 minutes at room temperature. To monitor dephosphorylation activity of CheZHP on CheYHP-P and CheV-P, the reaction was set up as above but equilibrated in the presence of 1 µM CheZHP. CheYHP or one of the CheV proteins was added as above and the reaction was monitored for 10 minutes. To determine the optimal metal concentrations for CheYHP and the CheV proteins the reaction was performed as above with the replacement of 20 mM MgCl2 with several concentrations either MgCl2, KCl, MnCl2, or no added metal (data not shown). To determine phosphatase release rates, we calculated the amount of phosphate released at each time point using the extinction coefficient at 360nm of 0.0091 µM −1 cm−1 at pH 7 (Silversmith et al., 2001). The release rate was determined by performing a linear fit (Excel, Microsoft), to calculate a slope of plotted phosphate release as a function of time. The first two time points were excluded from this analysis due to slight fluctuation of OD360 after protein addition.
Phosphorylation assays with [γ-32P]ATP
A labeled ATP solution was created by mixing 2 mM unlabelled ATP with 11 µM [γ-32P] ATP (10Ci/mMole) (PerkinElmer LAS Inc), to yield a final ATP solution of 2.01 mM. Reactions containing either 2 µM CheAY or 6 µM CheA’, 43 or 114 µM labeled ATP solution, 20 mM MgCl2, 50 mM KCl, 50 mM Tris/HCl pH 7.5, were incubated for 10 minutes at room temperature. CheYHP or CheYHP+CheZHP was then added to the reaction at 2:4 or 2:4:1 ratio of CheA:CheY:CheZ. Samples were collected and stopped in 2X SDS sample buffer (Scopes & Smith, 2006).
To determine if CheZHP affected CheAY, a mixture of 200 µM unlabeled ATP plus 3 µM [γ-32P] ATP (10Ci/mMol) was used to phosphorylate CheAY for 10 minutes at room temperature, followed by a “chase” with 10 mM unlabelled ATP in the presence or absence of CheZHP. For CheA', 12 µM CheA' was phosphorylated with a mixture of 400 µM unlabeled ATP plus 6 µM [γ-32P]ATP (10Ci/mMol) at room temperature for 45 minutes, followed by ATP removal using a PD SpinTrap G-25 column (GE Healthcare). Purified CheA’-P was allowed to sit at room temperature for five minutes and the reactions were chased with unlabeled ATP as described above. Samples were loaded onto 12% or 15% SDS-PAGE to separate the protein mixture. Gels were dried and exposed to X-ray film (KODAK BioMax XAR Film) to visualize the radioactive bands or exposed to a phosphoroimager cassette and scanned to obtain the image with Typhoon TRIO (Amersham Biosicences). ImageQuant 5.2 (GE Healthcare) was used to quantify radioactive bands to calculate phosphorylated half life of each protein.
Sequence alignment and analysis
Amino acid sequence from HP0170 H. pylori strain SS1 (GenBank accession number GU223223) was used for PSI-BLAST analysis. The 2nd iteration yielded sequences annotated as CheZ under ‘Sequences with E-values worse than threshold.’ Three proteins, annotated as CheZ or putative CheZ, were selected for the 3rd iteration as shown in Table 1. This resulted in 268 proteins identified in the next iteration. A representative from each genus with the highest E-value was selected for multiple sequence alignment with ClustalW and the alignment file was input into Jalview to generate graphical representation of the level of amino acids conservation (Chenna et al., 2003, Waterhouse et al., 2009). A 20 amino acid region surrounding the active site and the area corresponding to the C-terminus 14 amino acids were the most highly conserved, and were selected from the alignment and input into WebLogo (Crooks et al., 2004).
Supplementary Material
Acknowledgements
The authors are grateful to Ruth Silversmith for numerous conversations, advice, CheZ enthusiasm and especially for providing MPI; Anonymous reviewer two for pointing out the need to for sufficient MgCl2 during cold ATP chase experiments; Dagmar Beier for the generous gift of CheA expressing plasmids; Susan Williams, Jenny Draper, Lisa Collison, Chad Saltikov, and Fitnat Yildiz for comments on the manuscript. The described project was supported by Grant Numbers AI050000 (to K.M.O.) from the National Institutes of Allergy and Infectious Disease (NIAID) at the National Institutes of Health. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
References
- Blat Y, Eisenbach M. Conserved C-Terminus of the phosphatase CheZ is a binding domain for the chemotactic response regulator CheY. Biochemistry. 1996;35:5679–5683. doi: 10.1021/bi9530447. [DOI] [PubMed] [Google Scholar]
- Boesch KC, Silversmith RE, Bourret RB. Isolation and characterization of nonchemotactic CheZ mutants of Escherichia coli. Journal of Bacteriology. 2000;182:3544–3552. doi: 10.1128/jb.182.12.3544-3552.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret RB. Receiver domain structure and function in response regulator proteins. Current Opinion in Microbiology. 2010;13:142–149. doi: 10.1016/j.mib.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantwell B, Draheim R, Weart R, Nguyen C, Stewart R, Manson M. CheZ phosphatase localizes to chemoreceptor patches via CheA-short. Journal of Bacteriology. 2003;185:2354–2361. doi: 10.1128/JB.185.7.2354-2361.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantwell BJ, Manson MD. Protein domains and residues involved in the CheZ/CheAs interaction. Journal of Bacteriology. 2009;191:5838–5841. doi: 10.1128/JB.00280-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Research. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covacci A, Telford JL, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: A Sequence Logo Generator. Genomic Research. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foynes S, Dorrell N, Ward SJ, Stabler RA, McColm AA, Rycroft AN, Wren BW. Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infection and Immunity. 2000;68:2016–2023. doi: 10.1128/iai.68.4.2016-2023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Stock AM. Biological insights from structures of two-component proteins. Annual Review of Microbiology. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess JF, Oosawa K, Kaplan N, Simon MI. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell. 1988;53:79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- Huang C, Stewart RC. CheZ mutants with enhanced ability to dephosphorylate CheY, the response regulator in bacterial chemotaxis. Biochimica et Biophysica Acta. 1993;1202:297–304. doi: 10.1016/0167-4838(93)90019-n. [DOI] [PubMed] [Google Scholar]
- Jiménez-Pearson M, Delany I, Scarlato V, Beier D. Phosphate flow in the chemotactic response system of Helicobacter pylori. Microbiology. 2005;151:3299–3311. doi: 10.1099/mic.0.28217-0. [DOI] [PubMed] [Google Scholar]
- Kandulski A, Selgrad M, Malfertheiner P. Helicobacter pylori infection: a clinical overview. Digestive and Liver Diseases. 2008;40:619–626. doi: 10.1016/j.dld.2008.02.026. [DOI] [PubMed] [Google Scholar]
- Karatan E, Saulmon MM, Bunn MW, Ordal GW. Phosphorylation of the response regulator CheV is required for adaptation of attractants during Bacillus subtilis chemotaxis. The Journal of Biological Chemistry. 2001;276:43618–43626. doi: 10.1074/jbc.M104955200. [DOI] [PubMed] [Google Scholar]
- Lee A, O'Rourke J, De Ungria MC, Robertson B, Daskalopoulos G, Dixon MF. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology. 1997;112:1386–1397. doi: 10.1016/s0016-5085(97)70155-0. [DOI] [PubMed] [Google Scholar]
- Lowenthal AC, Simon C, Fair AS, Mehmood K, Terry K, Anastasia Stephanie, Ottemann KM. A fixed-time diffusion analysis method determines that the three cheV genes of Helicobacter pylori differentially affect motility. Microbiology. 2009;155:1181–1191. doi: 10.1099/mic.0.021857-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy MM, Bren A, Eisenbach M, Dahlquist FW. Identification of the binding interfaces on CheY for two of its targets, the phosphatase CheZ and the flagellar switch protein FliM. Journal of Molecular Biology. 1999;289:1423–1433. doi: 10.1006/jmbi.1999.2830. [DOI] [PubMed] [Google Scholar]
- McNamara BP, Wolfe AJ. Coexpression of the long and short forms of CheA, the chemotaxis histidine kinase, by members of the family Enterobacteriaceae. Journal Bacteriology. 1997;179:1813–1818. doi: 10.1128/jb.179.5.1813-1818.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muff TJ, Foster RM, Liu PJY, Ordal GW. CheX in the three-phosphatase system of bacterial chemotaxis. Journal of Bacteriology. 2007;189:7007–7013. doi: 10.1128/JB.00896-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muff TJ, Ordal GW. The diverse CheC-type phosphatase: chemotaxis and beyond. Molecular Microbiology. 2008;70:1054–1061. doi: 10.1111/j.1365-2958.2008.06482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazy Y, Motaleb MA, Guarnieri MT, Charon NW, Zhao R, Silversmith RE. Identical phosphatase mechanisms achieved through distinct modes of binding phosphorprotein substrate. Proceedings of the National Academy of Sciences. 2010 doi: 10.1073/pnas.0911185107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman M, Goodwin M, Kelly D. Chemotaxis in the human gastric pathogen Helicobacter pylori: different roles for CheW and the three CheV paralogues, and evidence for CheV2 phosphorylation. Microbiology. 2001;147:2493–2504. doi: 10.1099/00221287-147-9-2493. [DOI] [PubMed] [Google Scholar]
- Sanna MG, Simon MI. In vivo and in vitro characterization of Escherichia coli protein CheZ gain- and loss-of-function mutants. Journal of Bacteriology. 1996;178:6275–6280. doi: 10.1128/jb.178.21.6275-6280.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopes RK, Smith JA. In: Analysis of proteins. In: Current protocols in molecular biology. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. USA: John Wiley & Sons, Inc; 2006. pp. 10.12A.31. [Google Scholar]
- Silversmith R, Smith J, Guanga G, Les J, Bourret R. Alteration of a nonconserved active site residue in the chemotaxis response regulator CheY affects phosphorylation and interaction with CheZ. The Journal of Biological Chemistry. 2001;276:18478–18484. doi: 10.1074/jbc.M011418200. [DOI] [PubMed] [Google Scholar]
- Silversmith RE. High mobility of carboxyl-terminal region of bacterial chemotaxis phosphatase CheZ is diminished upon binding divalent cation or CheY-P substrate. Biochemistry. 2005;44:7768–7776. doi: 10.1021/bi0501636. [DOI] [PubMed] [Google Scholar]
- Silversmith RE. Auxiliary phosphatases in two-component signal transduction. Current Opinion in Microbiology. 2010;12:1–7. doi: 10.1016/j.mib.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Schmitt R. Phosphotransfer between CheA, CheY1, and CheY2 in chemotaxis signal transduction chain of Rhizobium meliloti. Biochemistry. 1998;37 doi: 10.1021/bi972330a. [DOI] [PubMed] [Google Scholar]
- Szurmant H, Ordal GW. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiology and Molecular Biology Reviews. 2004;68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry K, Go A, Ottemann K. Proteomic mapping of a suppressor of non-chemotactic cheW mutants reveals that Helicobacter pylori contains a new chemotaxis protein. Molecular Microbiology. 2006;61:871–882. doi: 10.1111/j.1365-2958.2006.05283.x. [DOI] [PubMed] [Google Scholar]
- Terry K, Williams SM, Lynn C, Ottemann KM. Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infection and Immunity. 2005;73:803–811. doi: 10.1128/IAI.73.2.803-811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomb JF, White O, Kerlavage ARC, A R, Suttion GG, Flesichmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quachenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, J.C V. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nature Review: Molecular Cell Biology. 2004;5 doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2 - a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuichet K, Alexander RP, Zhulin IB. Comparative Genomic and Protein Sequence Analyses of a Complex System Controlling Bacterial Chemotaxis. Methods in Enzymology. 2007;422:1–31. doi: 10.1016/S0076-6879(06)22001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Collins E, Bourret R, Silversmith R. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nature Structural Biology. 2002;9:570–575. doi: 10.1038/nsb816. [DOI] [PubMed] [Google Scholar]
- Zhu X, Volz K, Matsumura P. The CheZ-binding Surface of CheY Overlaps the CheA- and FliM-binding Surfaces. The Journal of Biological Chemistry. 1997;272:23758–23764. doi: 10.1074/jbc.272.38.23758. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.