Abstract
microRNAs (miRNA) encode small RNA molecules of ~22nts in length that regulate the deadenylation, translation, and decay of their target mRNAs. The identification of miRNAs in plants and animals has uncovered a new layer of gene regulation with important implications for development, cellular homeostasis and disease. Because each miRNA is predicted to regulate several hundred genes, a major challenge in the field remains to elucidate the precise roles for each miRNA and to understand the physiological relevance of individual miRNA-target interactions in vivo. Despite the wide variety of biological contexts where miRNAs function, a common theme emerges, whereby miRNAs shape gene expression within both spatial and temporal dimensions by removing messages from previous cellular states as well as modulating the levels of actively transcribed genes. This review will focus on the role that the teleost Danio rerio (zebrafish) has played in shaping our understanding of miRNA function in vertebrates.
Introduction
Cellular functions underlying embryonic development depend on the precise spatial and temporal regulation of gene expression. Over the last two decades microRNAs (miRNAs) have emerged as novel and widespread regulators of gene expression. Computational and experimental analyses indicate that individual miRNAs can basepair and selectively mediate the repression of hundreds of different target genes [1–4]. Further, current estimates suggest that between 25% and 70% of human genes are directly regulated by miRNAs [4–6]. Functional analyses have shown that miRNAs shape gene expression within a myriad of developmental and physiological contexts. Despite the large lists of predicted targets, we have yet to uncover the functions of the vast majority of miRNAs. A major challenge lies in identifying physiologically relevant targets of each miRNA and determining how the regulation of those targets influences cellular behavior during development and homeostasis. While previous reviews provide an overview of miRNA biogenesis, target recognition, and function[7–11], this review will examine the developmental roles of miRNAs, and the role that the teleost Danio rerio (zebrafish) has played in shaping our understanding of miRNA function in vertebrates.
Global overview of miRNA function in animals
Mature miRNAs are generated from longer transcripts through their sequential cleavage by RNAse III enzymes Drosha and Dicer [8]. Since disruption of Dicer activity abrogates miRNA biogenesis, this approach has been utilized in several animal models to assess the global role of miRNAs during embryonic development. In zebrafish, multiple mutant alleles of dicer exist that are predicted to eradicate function [12]. Due to a strong maternal Dicer contribution, zygotic mutants develop normally and are indistinguishable from wild-type siblings during embryogenesis[12]. However, dicer zygotic mutant fish die after 7–10 days, suggesting that miRNA activity is required during postembryonic stages. To completely remove dicer function, maternal and zygotic mutant (MZdicer) embryos have been generated via germline transplantation, whereby mutant germ cells are transplanted within wild-type hosts [13]. Complete loss of Dicer function leads to multiple developmental defects. However, although they develop abnormally, MZdicer embryos undergo fertilization and proceed through embryogenesis. This finding indicates that miRNA activity is not autonomously required for germ cell development and maintenance in the fish. Wild-type hosts possessing dicer mutant germlines can be crossed over multiple years and consistently give rise to viable oocytes and sperm. This result is in stark contrast to that observed in other animals. In mice, absence of maternally provided Dicer results in chromosome segregation defects and a disruption in oocyte maturation[14]. It is possible that these defects are not specifically due to the loss of miRNA activity, as dicer mediates the biogenesis of other small regulatory RNAs, including endogenous small interfering RNAs (endo-siRNAs) [14]. This argument is strengthened by an analysis of dgcr8 function in mice. Interestingly, dgcr8-deficient oocytes, which are predicted to retain siRNA processing yet lack miRNA activity, undergo normal oocyte maturation and early embryonic development (prior to embryonic day E6.5) [15, 16], suggesting that the defects observed in dicer mouse oocytes are likely due to the loss of siRNA activity rather than canonical miRNAs. Some of the defects observed in dicer oocytes appear to be associated, in part, with a failure to silence transposon-derived sequence elements [14].
In zebrafish, the MZdicer embryonic phenotype allows for the analysis of miRNA function through several developmental stages. MZdicer embryos generate anterior-posterior and dorsal-ventral body axes and are able to differentiate multiple cell types including haematopoietic, muscle and neuronal lineages [13]. Despite this, MZdicer embryos display severe morphogenesis defects during gastrulation, neural tube formation, heart and somite development, and die by day 5 post fertilization. A similar theme emerges from studies in mice. Although zygotic loss of dicer leads to early embryonic lethality (prior to embryonic day E8.5) in the mouse [17, 18], conditional deletion studies of Dicer function have allowed an assessment of miRNA function during later developmental stages. Interestingly, loss of dicer does not impede cell differentiation, but impacts subsequent morphogenesis and homeostasis in multiple contexts, such as limb or skin development [19–24]. Together, these results suggest that cells lacking miRNA function retain the ability to adopt different cellular fates. However, cellular properties such as cell growth and cell movement are often compromised.
This trend appears to be mostly consistent with that observed in invertebrates. Although several invertebrate miRNAs display striking phenotypes upon their loss, in most cases, these phenotypes do not involve a failure to establish major cell lineages. For example, loss of the let-7 miRNA, a critical regulator of developmental timing in C. elegans, results in reiterated seam cell divisions and a delay in their terminal differentiation [25]. Although let-7 influences the number and timing of seam cells present in the adult, it is not necessarily required for their differentiation per se. Perhaps the strongest example of a miRNA playing an active role in cell fate specification is the C.elegans lsy-6 miRNA, which is expressed in a restricted fashion within a distinct class of chemosensory neurons asymmetrically positioned along the left side of the body midline (ASEL neuron class) [26]. Loss of lsy-6 function disrupts left-right asymmetry such that ASEL neurons adopt similar expression profiles and chemosensory properties of their bilateral neighbors on the right side (ASER neuron class). In this striking example, a miRNA plays an instructive role in the diversification of the neuronal cell lineage into more specialized cell types. Importantly, lsy-6 activity is both necessary and sufficient for ASEL-specific terminal differentiation.
Let-7 and lsy-6 were isolated from genetic screens designed to uncover readily identifiable phenotypes. A more systematic study of miRNA function in C. elegans suggests that most miRNAs, when removed individually, are not essential for development [27]. It is likely that, in some of these cases, the failure to observe a phenotype is due to functional redundancy among miRNA paralogs.
Taken together, knockout studies in fish, mice, and invertebrates provide important insights into miRNA function in animals. Most notably, miRNAs do not appear to be obligatory for the establishment of major cell lineages in vivo. However, miRNAs play important roles during the terminal differentiation and maturation of various cell types within a particular cell lineage, as well as regulate the cellular properties (e.g. growth, movement, apoptosis) of differentiated cells during morphogenesis and homeostasis.
miR-430 shapes temporal expression patterns during MZT
The MZdicer mutant phenotypes set the stage for studying miRNA function in the fish. However, the challenge lies in identifying the underlying miRNAs responsible for these phenotypes. Surprisingly, reintroduction of one miRNA, miR-430, into the MZdicer background is able to rescue a significant portion of the morphogenetic defects that occur during early embryogenesis, including defects in gastrulation and brain morphogenesis [13]. These findings indicate that miR-430, in particular, plays a critical role during early zebrafish development.
miR-430 is abundantly expressed at the onset of zygotic transcription, during a period when developmental control is being transferred from maternally provided gene products to those transcribed zygotically. The maternal-to-zygotic transition (MZT) occurs in all animals, and consists of both the initiation of gene transcription and the active destabilization of maternal mRNAs [28]. It has been known for over two decades that the clearance of maternal instructions depends on zygotic transcription [29]. Indeed, inhibiting transcription leads to the stabilization of maternal transcripts, yet the factors that normally facilitate their degradation have remained mostly elusive. Microarray analyses of MZdicer mutants coupled with target validation assays reveal that zygotically expressed miR-430 has hundreds of direct targets. Intriguingly, the miR-430 target pool shows a four-fold enrichment for maternal genes. Conversely, maternal transcripts are strongly enriched for miR-430 target sites. These results indicate that miR-430 induces the clearance of a large fraction of maternal mRNAs in zebrafish [2]. miR-430-mediated mRNA degradation is achieved through the accelerated deadenylation of target transcripts, and has provided an entry point for understanding the molecular mechanisms behind miRNA-mediated target mRNA turnover. In Xenopus, the orthologue of miR-430 (miR-427) also facilitates the deadenylation of maternal transcripts [30]. It is important to note that multiple mammalian orthologues of miR-430 (miR-302, miR-372, miR-516–520) are expressed during early embryogenesis and could potentially regulate the clearance of maternal transcripts in mammals. How do other animals which lack miR-430 orthologs mediate MZT? Interestingly, in Drosophila, a different miRNA family, miR-309, in an example of convergent evolution, is employed in the clearance of maternal transcripts during its MZT [31].
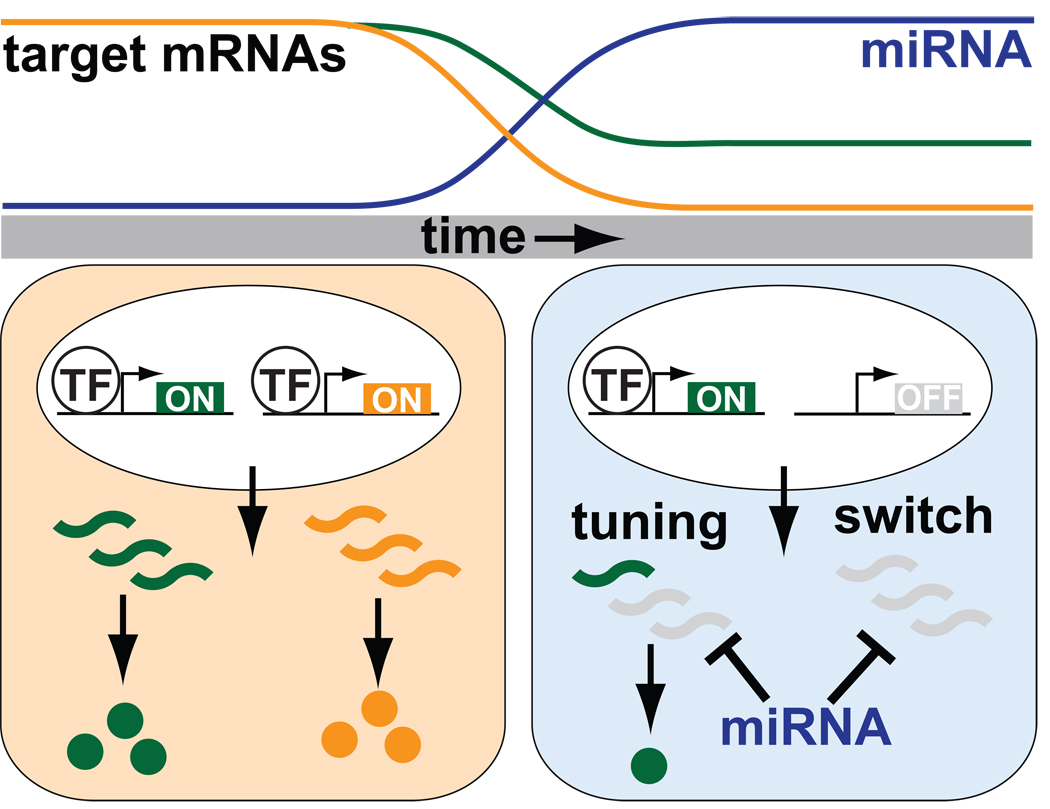
It remains unknown to what degree excess maternal mRNAs contribute to the early phenotypic defects in MZdicer embryos. However, reintroduction of the miR-430 duplex into MZdicer embryos rescues gastrulation and neural tube defects, indicating that these particular phenotypes are specifically due to miR-430 depletion [13]. Gastrulation defects could potentially be due to the misregulation of a few key targets, or, alternatively, could be a more general consequence of overall maternal mRNA accumulation. Further, it remains to be determined whether miR-430 coordinates early morphogenetic movements by precisely regulating the steady-state levels of particular transcripts (tuning function), or whether miR-430 simply reduces the maternal mRNA pool to inconsequential levels, thus clearing the slate for zygotic activation (switch function). Given the sheer number of putative targets, it is likely that both modes of regulation are utilized, depending on the particular transcript and its responsiveness to miR-430 directed repression.
A role for miRNAs in orchestrating major developmental transitions was first attributed to the founding C.elegans miRNAs, lin-4 and let-7 [25, 32, 33]. Dramatic upregulation of let-7 expression at the end of larval development represses larval-specific gene expression and facilitates the progression into adulthood. In hindsight, the employment of miRNAs to shape the temporal dynamics underlying developmental transitions makes sense. First, post-transcriptional repression by miRNAs is a rapid mechanism to repress protein output. Second, a miRNA can target large numbers of genes and thereby shape expression profiles on a global scale. Finally, the degree of repression conferred by a miRNA can be individually tailored for each transcript based on the number and accessibility of target sites within their 3’UTRs. This inherent versatility allows the same miRNA to both remove previously transcribed mRNAs that are no longer needed, as well as precisely regulate steady-state levels of transcripts that are still being transcribed (Figure 1).
Figure 1.
miRNAs regulate developmental transitions. Onset of miRNA activity (in blue cell) can serve to remove mRNAs that originate from a previous transcriptional stage (red), as well as tune the expression levels of actively transcribed genes (green).
miRNAs modulate expression levels of actively transcribed genes
An understanding of miRNA function requires the identification of biologically relevant targets. This goal is hampered by two major limitations. First, miRNAs can regulate a large number of targets. For example, as alluded to above, over 300 transcripts are responsive to miR-430 activity in vivo [2]. Second, the degree of repression observed is often quite modest, with most targets displaying a 2- to 3- fold reduction in transcript levels. To better discern how miRNAs participate within genetic regulatory networks, it is important to consider target repression, however subtle, within the context of other regulatory processes in the cell. Specifically, what is the relationship between the transcription of a specific target and its repression by a miRNA within any given tissue? This is particularly important when discerning whether a miRNA “tunes” the steady-state levels of an actively transcribed target mRNA or, alternatively, removes unwanted transcripts that originate from a previous stage and/or transcriptional noise. Initial studies based on target prediction algorithms observed that miRNAs and their targets tend to be expressed in a mutually-exclusive manner within complementary domains [1, 34, 35]. These findings have lent support to a “fail-safe” model of miRNA regulation, in which miRNAs ensure that transcript levels of transcriptionally repressed target genes do not accumulate (due to spurious or “noisy” transcription).
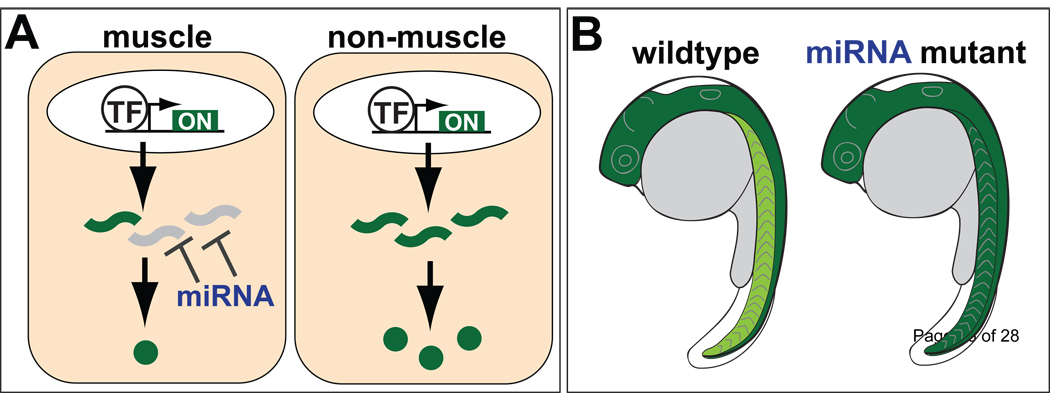
Expression profiling studies in zebrafish have addressed this question by assessing tissue-specific target gene expression in the presence or absence of miR-1/206 and miR-133 in muscle, and miR-124 in neurons [36, 37]. These studies have led to important insights into the primary modes of regulation conferred by miRNAs. First, many target genes are expressed in the same tissue as the miRNA, albeit at lower levels than that observed in other tissues. Second, miRNA-mediated repression has a significant effect on the expression levels of most targets within miRNA-expressing cells (Figure 2). For example, miR-1 is a muscle-specfic miRNA, and miR-1 targets tend to be present at higher levels outside of muscle compared to non-targets. However, this tendency is lost upon knockdown of miR-1 [36]. Taken together, these findings indicate that miRNAs play instructive roles in shaping gene expression of actively transcribed genes. Interestingly, instructive interactions (co-expression of miRNA and target) are preferentially conserved between species compared to interactions that are more in line with a “fail-safe” mode of regulation [37].
Figure 2.
miRNAs play instructive roles in shaping gene expression. (A) In this example, a miRNA and its targets are co-expressed in muscle. By repressing an actively transcribed gene, the miRNA modulates final protein output within muscle. (B) In the presence of the miRNA, target expression levels are lower in muscle (light green) compared to non-muscle (dark green). In the absence of miRNA activity, target expression levels are similar in both tissues.
Restricted expression of miRNAs shapes expression domain boundaries during patterning and organogenesis
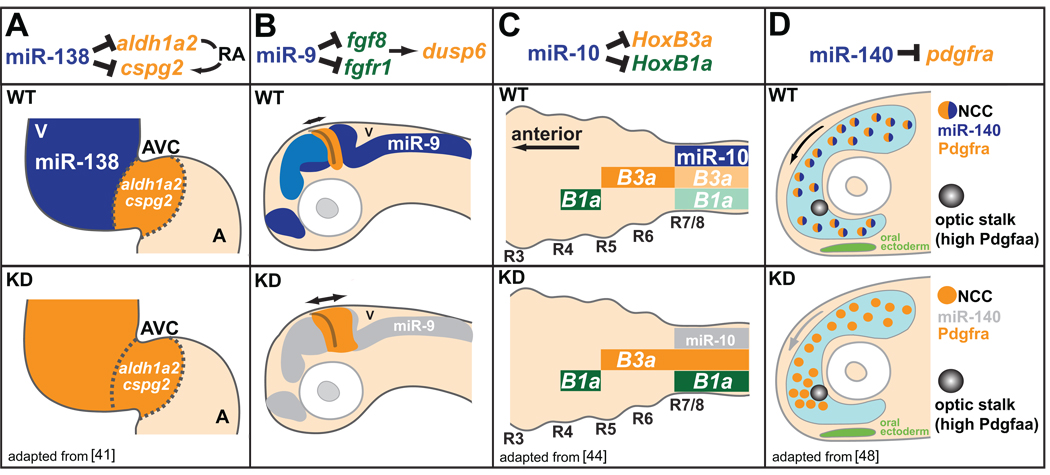
The majority of miRNAs are expressed in restricted spatial fashions within particular organs [38, 39]. It is tempting to speculate that, in these cases, miRNAs define functionally distinct domains by repressing subsets of more broadly expressed genes (established by early transcriptional activators)[40]. An example of miRNA-mediated refinement of expression domains is illustrated by miR-138 function during heart development [41] (Figure 3A). The two-chambered heart of the zebrafish is composed of distinct atrial and ventricular chambers connected by an atrio-ventricular canal (AVC). Ventricle-specific expression of miR-138 within cardiac muscle marks the spatial extent of AVC-specific gene expression. Specifically, knockdown of miR-138 results in the expansion of AVC-specific markers including notch1B and cspg2 into the ventricle, and results in maturation defects within the ventricle and pericardial edema. In addition, aldh1a2, an enzyme involved in retinoic acid (RA) signaling, is directly repressed by miR-138 in the ventricle, suggesting that increased RA signaling, normally confined to the AVC, is responsible, at least in part, for the miR-138 knockdown phenotype. A similar example is observed during gut development in the fish. High miR-145 expression in intestinal smooth muscle promotes muscle cell maturation through the downregulation of the transcription factor gata6, which is expressed in both muscle and epithelial cells of the gut [42]. In both cases, the expression of a miRNA helps to “lock in” terminal fates that are distinct from the fates of neighboring cells in which the miRNA is absent or present at low levels.
Figure 3.
miRNAs shape gene expression boundaries and migratory behaviors. (A) miR-138 (expression in blue) represses RA signaling in the ventricle (V) by targeting atrioventricular canal (AVC)-specific genes aldh1a2 and cspg2. miR-138 knockdown causes ventricular expression of AVC-specific genes (orange), and results in defects in ventricle morphology and function. A, atrium. (B) miR-9 expression (blue) adjacent to the mid-hindbrain boundary (MHB) delimits the extent of FGF signaling originating from the MHB. Knockdown of miR-9 leads to increased expression of FGF target genes, and the ectopic expansion of the FGF target dusp6 (orange). (C) miR-10 (expression in blue) dampens HoxB3a (orange) and HoxB1a (green) posterior expression in the nervous system. Knockdown of miR-10 results in increased expression of HoxB3a and HoxB1a within posterior domains. (D) miR-140 represses pdgfra expression within a subset of neural crest cells (NCCs) involved in palatogenesis to regulate migratory response to the Pdgfaa attractant (light blue). Knockdown of miR-140 results in increased expression of pdgfra, and defects in NCC migration around the optic stalk, a key source of Pdgfaa, leading to palate clefting.
The restricted expression patterns of miRNAs can also refine expression boundaries in more complex ways. miR-9 expression within regions surrounding the mid-hindbrain boundary (MHB) serves to both delimit the spatial extent of the MHB by antagonizing FGF signaling (Figure 3B), as well as promote neurogenesis near the MHB by repressing antineurogenic genes[43]. In another example, miR-10 contributes to anterior-posterior patterning by refining Hox gene expression in the fish embryo [44](Figure 3C). Knock-down of miR-10 activity results in the ectopic expression of HoxB1a and HoxB3a within the Hox-4 expression domain. Interestingly, miR-10 and HoxB4 derive from a common polycistronic transcript, and act synergistically to repress HoxB1a and HoxB3a expression within the Hox-4 domain. This latter example highlights how a miRNA can integrate into a transcriptional genetic hierarchy to reinforce expression domain boundaries. In all of these cases, a common theme emerges: miRNA regulation shapes gene expression post-transcriptionally to delimit spatial and temporal boundaries during embryogenesis.
miRNAs shape response to signaling gradients
miRNAs can also shape domain boundaries, not through their restricted expression pattern, but by modulating a cell’s response to extracellular ligands. For example, miRNA-mediated repression of cell surface receptors or downstream effectors can modify the level of signaling output produced after exposure to a specific morphogen concentration. This “tuning” activity, however subtle, can have a dramatic impact where target expression lies near a threshold point. Several examples in zebrafish implicate miRNAs in the regulation of signal pathways. For example, miR-214 influences the propensity of somite cells to adopt slow muscle fate through the modulation of Hedgehog response [45]. During germ layer specification, miR-430 represses the Nodal signaling ligands lefty and squint to modulate mesendoderm induction [46]. Interestingly, lefty and squint function as antagonist and agonist of the Nodal pathway, respectively, indicating that miRNA regulation is not always unidirectional, but can serve to dampen and balance positive and negative regulatory inputs together to achieve optimal output.
miRNA regulation can also modulate a cell’s response to migratory cues. A striking example in fish is observed for miR-140 during craniofacial morphogenesis [47, 48](Figure 3D). miR-140 is expressed in skeletogenic precursors, including a subset of neural crest cells whose migration is directed by platelet-derived growth factor (Pdgf) signaling. By repressing the expression of the Pdgf receptor (pdgfra) , miR-140 dampens the sensitivity of these cells to the attracting properties of the Pdgf ligand (Pdgfaa). The functional significance of this interaction becomes apparent for cells located close to the optic stalk, which is a key source of the Pdgf ligand. Rather than bypassing the optic stalk to reach their appropriate destination, migrating cells with disrupted miR-140 activity become trapped, ultimately leading to palate clefting.
miRNAs can function at multiple levels in a signal transduction cascade. An intriguing possibility is that cells receiving a common signal can generate very different transcriptional responses based on the differential expression of a miRNA. In this scenario, the miRNA can either target a core component of the signaling pathway, and thus dampen or accentuate the overall response, or selectively modulate the expression of particular downstream target genes. The above examples illustrate how miRNAs can tune a cell’s response to external cues, whether to set threshold requirements for cell fate commitment, or to mediate the push and pull forces of migratory signals.
miRNAs mediate cellular homeostasis
Much of miRNA research has focused on understanding how miRNAs impact developmental processes. However, miRNAs have also been shown to maintain cellular homeostasis within differentiated tissues in a variey of contexts, including insulin secretion [49] and response [50], fat metabolism [51], muscle growth [52], and hair follicle organization[23]. Recent studies in zebrafish have begun to implicate specific miRNAs in tissue maintenance and homeostasis. For example, disruption of miR-1 leads to defects in the sarcomeric organization of actin in fast skeletal muscle[36]. Several putative targets of miR-1 are regulators of actin polymerization and filament formation, suggesting that miR-1 activity is important for modulating actin dynamics in muscle cells. Another miRNA, miR-126, is required to maintain vascular integrity[53, 54]. Disruption of miR-126 results in compromised endothelial tube organization and vessel lumen collapse.
miRNAs can also regulate the properties of very specialized cells. miR-8 expression in zebrafish ionocytes, cells responsible for pH and ion homeostasis, modulates the apical trafficking of ion transport proteins [55]. Reduced expression of miR-8 by morpholino knockdown results in a decreased ability to tolerate changes in pH and salt concentration. It is unclear whether miR-8 expression is dynamically regulated in response to osmotic stress. However, dynamic expression of a miRNA in response to DNA damage-induced stress has been observed for another zebrafish miRNA, miR-125b, resulting in the derepression of p53, a miR-125b target [56]. These studies add to a growing list of functions for miRNAs in countering environmental stresses [51, 57–59].
Many miRNAs are expressed in differentiated tissues. In zebrafish, the majority of miRNAs remain expressed in the adult [38, 60]. It is tempting to speculate that these miRNAs provide robustness to the gene expression profiles that underlie cellular homeostasis and tissue maintenance. If this is the case, then the phenotypic outcomes of miRNA loss are likely to become more apparent under stressed conditions. It is important to note that, in many cases, it is difficult to dissect cell differentiation defects from compromised cell maintenance. For example, miR-126 is proposed to modulate signaling by VEGF, which is critical for both vascular development and homeostasis. Given the large number of targets under miRNA control, it is likely that the same miRNA can function to both establish cell identity, as well as maintain that identity during later stages. Conditional deletion techniques, as well as other emerging technologies [41, 61], will allow the dissection of the post-differentiation functions of miRNAs.
miRNAs and Regeneration
In addition to maintaining tissue-specific properties, miRNAs likely govern cell proliferation and growth within mature tissues. Zygotic dicer mutants which are indistinguishable from their wild-type siblings due to maternally provided dicer activity undergo growth arrest and die after two weeks, suggesting that miRNA function during postembryonic stages is critical for tissue homeostasis [12]. The potential for miRNAs to regulate proliferation and growth in adult tissues is supported by studies of zebrafish regeneration. Zebrafish possess an impressive ability to repair several organs (e.g. heart, liver), as well as re-grow amputated fins [62]. Interestingly, the expression of several miRNAs is altered (both up- and down-regulated) upon caudal fin amputation, suggesting that miRNAs could dynamically regulate the proliferative potential of regenerating tissue [63, 64]. In support of this hypothesis, disruption of Dicer activity prevents regeneration [64]. Although the functions of miRNAs during regeneration are likely extensive, two miRNAs, miR-133 and miR-203, appear to impact this process by modulating FGF and Wnt signaling cascades, respectively. In both cases, downregulation of the miRNA is associated with increased proliferation at the site of amputation [63, 64]. Future work should clarify to what degree these, and other miRNAs regulate growth potential and cell homeostasis in both uninjured and regenerating tissues.
miRNA target repression and 3’UTR accessibility
Target repression is dependent on the ability of a miRNA to recognize and bind sequences within the target transcript. A fundamental challenge is to understand the parameters that dictate target site efficacy. In addition to basepairing constraints (reviewed in [7, 8]), target-miRNA interactions can be influenced in vivo by a variety of other regulatory processes. In some cell types, the presence of the miRNA and its target in the same tissue does not necessarily result in repression. For example, a subset of miR-430 targets in zebrafish, including nanos and tdrd7, are repressed by miR-430 in somatic cells, but are resistant to miR-430 activity in primordial germ cells (PGCs), resulting in their germline-specific expression [65]. This differential regulation is due to the presence of the germline-specific RNA binding protein Dnd1, which binds to cis-elements within the 3’UTRs of protected targets [66]. Although the mechanistic details are still unclear, Dnd1 appears to prevent access of the miRNA to its target. More recently, another RNA-binding protein, Dazl, has been shown to also protect tdrd7 mRNA from the repressive effects of miR-430 [67]. In this case, rather than preventing physical interaction, Dazl appears to antagonize miRNA activity by inducing the polyadenylation of target mRNAs, suggesting that multiple mechanisms can be employed to counter miRNA activity (Figure 4).
Figure 4.
Dnd1 and Dazl prevent miRNA-mediated repression in primary germ cells (PGCs). In somatic cells (top panel), the RISC complex directs the deadenylation and translational inhibition of target mRNAs. In PGCs, Dnd1 prevents the interaction between the RISC complex and its target mRNA (middle panel). In PGCs, Dazl antagonizes miRNA-mediated repression by inducing the polyadenylation of the target mRNA (bottom panel).
The modulation of miRNA-mediated repression by UTR-binding proteins is not unique to the germline. In stressed human cells, miR-122-mediated repression of cat-1 mRNA is alleviated by HuR, an AU-rich-element-binding protein [68]. These examples illustrate how miRNA activity can be differentially regulated depending on the cell type and the specific mRNA target. It is also important to consider other regulatory mechanisms that can modify miRNA activity in vivo [69]. From the level of miRNA processing [70, 71]to RISC activity [72], it is likely that a variety of cofactors will be identified that modify the repressive effects of miRNAs in both tissue- and target-specific manners, further underscoring the importance of understanding miRNA regulation and target site efficacy within the endogenous context of the cell.
Target conservation and evolutionary implications of miRNA function
miRNAs have a profound influence on organismal development and physiology. How have miRNAs impacted animal evolution? Phylogenetic analysis indicates that miRNAs were present in the earliest metazoan ancestors [73, 74]. In addition, the continual accrual of miRNAs within animal genomes over evolutionary time suggests that miRNAs have played important roles in shaping animal evolution. The vertebrate lineage in particular has seen a large expansion in its miRNA repertoire that is correlated with increases in morphological complexity [75].
Although miRNAs show robust conservation, target site sequences are often not conserved, indicating that the target pool recognized by a particular miRNA can vary dramatically across lineages [1]. Many of these non-conserved targets are likely functional, as demonstrated by their derepression in the absence of the miRNA [2, 36, 37, 76]. Further evidence for the functional relevance of non-conserved targets comes from SNP density mapping in humans. Up to 50% of non-conserved targets sites are under significant negative selection in cases where the miRNA and target mRNA are expressed in the same tissue [77]. Taken together, these findings indicate that extensive lineage-specific rewiring of miRNA networks has occurred during evolution.
The function of a miRNA can be modified through both changes in its target pool as well as changes in its temporal and spatial expression. A prominent example is the deeply conserved miR-1, which has retained an ancestral connection to muscle development in both flies and vertebrates, yet has undergone extensive rewiring of its targets leading to the regulation of very different aspects of mesoderm development within each group [36, 52, 78]. Changes in the target pool can reshape how a miRNA influences specific developmental pathways. For example, the regulation of Nodal signaling by the miR-430 family (miR-430/427/302) is conserved in fish, Xenopus, and humans [79]. However, while direct repression of Lefty family members (antagonist ligands) is conserved in all three species, only fish and Xenopus target Nodal family members (activator ligands). It is possible this difference has important consequences for mesendoderm induction in humans, as the alleviation of miR-430 repression of the Nodal activator squint (through protection of a single target site in the squint mRNA 3’UTR) results in increased mesendoderm specification in the fish [46].
Given the ease with which mRNAs can come under control of a given miRNA (through the acquisition of a single or few nucleotide mutations within their 3’UTR sequences), it is reasonable to assume that novel miRNA-target interactions are constantly being sampled within the fitness landscape. Recent analyses suggest that, even among individuals within a species, polymorphic target sites can have a significant impact on gene expression [77, 80]. Most importantly, target site polymorphisms have been shown to underlie detectable phenotypic variation. A single nucleotide change that results in the creation of a miR-1/206 target site in the 3’UTR of the myostatin gene has been attributed to muscular hypertrophy in Texel sheep [81]. Target site polymorphisms have also been associated with human disease. A rare SNP linked to Tourette’s syndrome alters a miR-189 binding site in the Tourette’s candidate gene SLITRK1, presumably resulting in a hypomorphic allele [82]. Both of these examples illustrate the potential power of single nucleotide changes to modify development and physiology within a species. Given the degree of target site nonconservation across multiple species, it is tempting to speculate that changes in miRNA-target interactions have profoundly influenced morphological diversity within different animal lineages.
Summary and Future Prospects
As outlined above, miRNAs have been shown to impact a wide range of developmental processes in zebrafish, whether orchestrating major developmental transitions (e.g. miR-430 and MZT), shaping expression domains during embryonic patterning (e.g. miR-10), or dictating protein output within particular tissues to ensure proper cell homeostasis (e.g. miR-1). It is clear that miRNAs can play instructive roles in a variety of ways. As “tuners” of gene expression, miRNAs can modulate signaling pathways to direct cell migratory behavior (e.g. miR-140) as well as impact cell fate decisions (e.g. miR-214).
A major challenge in the field is to identify the most biologically relevant miRNA:target interactions. The restricted expression patterns of many fish miRNAs allow for the detailed analysis of target gene expression within and outside of miRNA-expressing domains. Further, the functional importance of specific miRNA:target interactions in vivo can be assessed through the use of target protectors [46]. Most 3’UTRs contain target sites for multiple miRNAs. Target protectors and miRNA-sensitive reporters will allow the dissection of 3’UTR sequences to assess the influence of combinatorial interactions of miRNAs on transcript homeostasis, as well as uncover novel regulatory motifs and trans-acting factors that regulate miRNA activity in vivo.
Despite the wide range of developmental and physiological contexts within which miRNAs function, common themes have started to emerge. miRNAs shape gene expression within a variety of spatial and temporal contexts to both remove messages that remain from previous cellular states as well as modulate the levels of actively transcribed genes. Future work in the coming years will be geared toward understanding how the post-transcriptional repression of physiological targets by miRNAs impacts development, tissue homeostasis and disease.
Table 1.
Functions and target genes of zebrafish miRNAs
| miRNA | Targets | Function | Reference |
|---|---|---|---|
| miR-1 | Pfn21 Atp6v1ba Cnn3a |
Muscle development and Maintenance |
[36] |
| miR-8 | Nherf-1 | Osmotic stress response | [55] |
| miR-9 | Fgf8 Fgfr1 Cnpy1 Her5 Her9 |
Mid-hindbrain development |
[43] |
| miR-10 | HoxB3a HoxB1a |
Neural patterning | [44] |
| miR-15a-1 | - | Inner ear development | [83] |
| miR-18a | - | Inner ear development | [83] |
| miR-30a | - | Liver development | [84] |
| miR-125b | p53 | DNA-damage response | [56] |
| miR-126 | Spred1 PIK3R2 |
Vasculature development And maintenance |
[53] |
| miR-133 | Mps1 | Muscle development and maintenance Caudal Fin Regeneration |
[36] [63] |
| miR-138 | Aldh1a2 cspg2 |
Heart development | [41] |
| miR-140 | pdgfra | Neural crest migration | [47] |
| miR-144 | KLFD | Erythropoiesis | [85] [86] |
| miR-145 | Gata-6 | Gut development | [42] |
| miR-200 | - | Olfactory neurogenesis | [24] |
| miR-203 | Lef1 | Caudal fin regeneration | [64] |
| miR-214 | Su(fu) | Muscle development, Hedgehog signaling |
[45] |
| miR-375 | - | Pancreatic islet development |
[87] |
| miR-430 | Squint Lefty Nanos Tdr7 |
Mesendoderm induction, Nodal signaling Germ cell development |
[46] [65] |
| miR-451 | Gata-2 | Erythropoiesis | [88] |
Acknowledgements
Thanks to A. Staton, C. Stahlhut and D. Cifuentes for comments on the manuscript. AJG is a Pew Fellow Lois and Franklin Top Yale Scholar and his laboratory is funded by grants from NIH (GM 081602-01, MH089956), Muscular Dystrophy Association and the Pew Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Farh KK, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310(5755):1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 2.Giraldez AJ, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312(5770):75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 3.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 4.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Friedman RC, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 7.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38 Suppl:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Staton A, AJ G. MicroRNAs in Development and Disease. Encyclopedia of Life Sciences (ELS) 2008 [Google Scholar]
- 10.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 11.Plasterk RH. Micro RNAs in animal development. Cell. 2006;124(5):877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 12.Wienholds E, et al. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet. 2003;35(3):217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 13.Giraldez AJ, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308(5723):833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 14.Murchison EP, et al. Critical roles for Dicer in the female germline. Genes Dev. 2007;21(6):682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, et al. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39(3):380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suh N, et al. MicroRNA function is globally suppressed in mouse oocytes and early embryos. 2010 doi: 10.1016/j.cub.2009.12.044. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernstein E, et al. Dicer is essential for mouse development. Nat Genet. 2003;35(3):215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 18.Kanellopoulou C, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19(4):489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis TH, et al. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci. 2008;28(17):4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris KS, et al. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci U S A. 2006;103(7):2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harfe BD, et al. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102(31):10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andl T, et al. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16(10):1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yi R, et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38(3):356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 24.Choi PS, et al. Members of the miRNA-200 family regulate olfactory neurogenesis. Neuron. 2008;57(1):41–55. doi: 10.1016/j.neuron.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 26.Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426(6968):845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- 27.Miska EA, et al. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3(12):e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136(18):3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- 29.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982;30(3):687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- 30.Lund E, et al. Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. Rna. 2009;15(12):2351–2363. doi: 10.1261/rna.1882009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bushati N, et al. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol. 2008;18(7):501–506. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- 32.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 33.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75(5):855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 34.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 35.Stark A, et al. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution. Cell. 2005;123(6):1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 36.Mishima Y, et al. Zebrafish miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization. Genes Dev. 2009;23(5):619–632. doi: 10.1101/gad.1760209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shkumatava A, et al. Coherent but overlapping expression of microRNAs and their targets during vertebrate development. Genes Dev. 2009;23(4):466–481. doi: 10.1101/gad.1745709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wienholds E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309(5732):310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 39.Kapsimali M, et al. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007;8(8):R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319(5871):1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- 41.Morton SU, et al. microRNA-138 modulates cardiac patterning during embryonic development. Proc Natl Acad Sci U S A. 2008;105(46):17830–17835. doi: 10.1073/pnas.0804673105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeng L, Carter AD, Childs SJ. miR-145 directs intestinal maturation in zebrafish. Proc Natl Acad Sci U S A. 2009;106(42):17793–17798. doi: 10.1073/pnas.0903693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leucht C, et al. MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat Neurosci. 2008;11(6):641–648. doi: 10.1038/nn.2115. [DOI] [PubMed] [Google Scholar]
- 44.Woltering JM, Durston AJ. MiR-10 represses HoxB1a and HoxB3a in zebrafish. PLoS One. 2008;3(1):e1396. doi: 10.1371/journal.pone.0001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flynt AS, et al. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet. 2007;39(2):259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318(5848):271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 47.Eberhart JK, et al. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet. 2008;40(3):290–298. doi: 10.1038/ng.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clouthier DE. MicroRNAs in facial development. Nat Genet. 2008;40(3):268–269. doi: 10.1038/ng0308-268. [DOI] [PubMed] [Google Scholar]
- 49.Poy MN, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432(7014):226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 50.Teleman AA, Maitra S, Cohen SM. Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes Dev. 2006;20(4):417–422. doi: 10.1101/gad.374406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu P, et al. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13(9):790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 52.Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19(19):2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fish JE, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15(2):272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang S, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15(2):261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flynt AS, et al. miR-8 microRNAs regulate the response to osmotic stress in zebrafish embryos. J Cell Biol. 2009;185(1):115–127. doi: 10.1083/jcb.200807026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le MT, et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23(7):862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X, et al. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137(2):273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Rooij E, et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316(5824):575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 59.Leung AK, Sharp PA. microRNAs: a safeguard against turmoil? Cell. 2007;130(4):581–585. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 60.Chen PY, et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev. 2005;19(11):1288–1293. doi: 10.1101/gad.1310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Loya CM, et al. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat Methods. 2009;6(12):897–903. doi: 10.1038/nmeth.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Poss KD, Keating MT, Nechiporuk A. Tales of regeneration in zebrafish. Dev Dyn. 2003;226(2):202–210. doi: 10.1002/dvdy.10220. [DOI] [PubMed] [Google Scholar]
- 63.Yin VP, et al. Fgf-dependent depletion of microRNA-133 promotes appendage regeneration in zebrafish. Genes Dev. 2008;22(6):728–733. doi: 10.1101/gad.1641808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thatcher EJ, et al. Regulation of zebrafish fin regeneration by microRNAs. Proc Natl Acad Sci U S A. 2008;105(47):18384–18389. doi: 10.1073/pnas.0803713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mishima Y, et al. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol. 2006;16(21):2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kedde M, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131(7):1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 67.Takeda Y, et al. DAZL relieves miRNA-mediated repression of germline mRNAs by controlling poly(A) tail length in zebrafish. PLoS One. 2009;4(10):e7513. doi: 10.1371/journal.pone.0007513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bhattacharyya SN, et al. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125(6):1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 69.Ding XC, Weiler J, Grosshans H. Regulating the regulators: mechanisms controlling the maturation of microRNAs. Trends Biotechnol. 2009;27(1):27–36. doi: 10.1016/j.tibtech.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 70.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320(5872):97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davis BN, et al. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454(7200):56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hammell CM, et al. nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell. 2009;136(5):926–938. doi: 10.1016/j.cell.2009.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grimson A, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455(7217):1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wheeler BM, et al. The deep evolution of metazoan microRNAs. Evol Dev. 2009;11(1):50–68. doi: 10.1111/j.1525-142X.2008.00302.x. [DOI] [PubMed] [Google Scholar]
- 75.Heimberg AM, et al. MicroRNAs and the advent of vertebrate morphological complexity. Proc Natl Acad Sci U S A. 2008;105(8):2946–2950. doi: 10.1073/pnas.0712259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet. 2006;38(12):1452–1456. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- 78.Zhao Y, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129(2):303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 79.Rosa A, Spagnoli FM, Brivanlou AH. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell. 2009;16(4):517–527. doi: 10.1016/j.devcel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 80.Kim J, Bartel DP. Allelic imbalance sequencing reveals that single-nucleotide polymorphisms frequently alter microRNA-directed repression. Nat Biotechnol. 2009;27(5):472–477. doi: 10.1038/nbt.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clop A, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38(7):813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- 82.Abelson JF, et al. Sequence variants in SLITRK1 are associated with Tourette's syndrome. Science. 2005;310(5746):317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- 83.Friedman LM, et al. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc Natl Acad Sci U S A. 2009;106(19):7915–7920. doi: 10.1073/pnas.0812446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hand NJ, et al. The microRNA-30 family is required for vertebrate hepatobiliary development. Gastroenterology. 2009;136(3):1081–1090. doi: 10.1053/j.gastro.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fu YF, et al. Mir-144 selectively regulates embryonic alpha-hemoglobin synthesis during primitive erythropoiesis. Blood. 2009;113(6):1340–1349. doi: 10.1182/blood-2008-08-174854. [DOI] [PubMed] [Google Scholar]
- 86.Du TT, et al. Experimental validation and complexity of miRNA-mRNA target interaction during zebrafish primitive erythropoiesis. Biochem Biophys Res Commun. 2009;381(4):688–693. doi: 10.1016/j.bbrc.2009.02.122. [DOI] [PubMed] [Google Scholar]
- 87.Kloosterman WP, et al. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5(8):e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pase L, et al. miR-451 regulates zebrafish erythroid maturation in vivo via its target gata2. Blood. 2009;113(8):1794–1804. doi: 10.1182/blood-2008-05-155812. [DOI] [PMC free article] [PubMed] [Google Scholar]