Abstract
Context
Polymorphisms in DNA repair genes have been reported contributing factors in head and neck cancer risk but studies have shown conflicting results.
Objective
To clarify the impact of DNA repair gene polymorphisms in head and neck cancer risk.
Method
A meta-analysis including 30 case-control studies was performed.
Results
Marginally statistically significant association was found for XRCC1 codon 399 (for Caucasians only), XPD Asp312Asn and XRCC1 codon 194 variants and head and neck cancer.
Conclusion
Assessments of the effects of smoking, alcohol, HPV and race/ethnicity on the association between DNA repair gene polymorphisms and head and neck cancer are needed.
Keywords: DNA repair genes, XP, ERCC1, XRCC1, XRCC3
INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) is the eighth most common cancer worldwide(Parkin et al., 2005), with approximately 48,010 new cases expected in the United States during 2009(Jemal et al., 2009) that varies in terms of incidence, mortality and survival by race. Among the documented risk factors associated with head and neck cancer, smoking and alcohol consumption are by far the main factors, followed by diet, poor oral health, exposure to human papillomavirus (HPV), exposure to environmental carcinogens, and genetic polymorphisms in carcinogen metabolizing enzymes, like alcohol dehydrogenases (ADH) and glutathione-S-transferase (GST) and DNA repair genes(Scully and Bagan, 2009).
Appropriate recognition and repair of DNA damage requires the integrity of the DNA damage response and repair machinery to maintain a normal cellular functionality. A defective DNA damage response can result in apoptosis or may lead to genomic instability, unregulated cell growth and an increased cancer risk(Hoeijmakers, 2001). There is considerable variation in the way an individual responds to DNA damage. While some individuals have proper DNA repair capacity (DRC), patients with a defective DNA damage response, such as those with Xeroderma Pigmentosum, are more susceptible to cancer. Phenotypically normal individuals with reduced DRC may also have increased cancer risk; these subjects if properly identified, could be targeted for intervention programs(Li et al., 2009).
There are three major pathways involved in DNA damage repair, depending on the type and magnitude of the damage, base excision repair (BER), nucleotide excision repair (NER) and double strand break (DSB) repair by the homologous recombination or nonhomologous endjoining pathways. Several molecular epidemiologic studies have evaluated the association of head and neck cancer with polymorphisms in DNA repair genes, such as XPA(An et al., 2007, Bau et al., 2007, Hall et al., 2007, Sugimura et al., 2006, Abbasi et al., 2009), XPC(Kietthubthew et al., 2006, Shen et al., 2001, Sugimura et al., 2006, Wang et al., 2007, Yang et al., 2005), XPD(An et al., 2007, Bau et al., 2007, Harth et al., 2008, Huang et al., 2005, Kietthubthew et al., 2006, Majumder et al., 2007, Ramachandran et al., 2006, Sturgis et al., 2002a, Sturgis et al., 2002b, Sturgis et al., 2000, Rydzanicz et al., 2005, Gajecka et al., 2005b), XPF(Sugimura et al., 2006), ERCC1(An et al., 2007, Sturgis et al., 2000, Sugimura et al., 2006), XRCC1(Demokan et al., 2005, Harth et al., 2008, Kietthubthew et al., 2006, Kowalski et al., 2009, Majumder et al., 2005, Olshan et al., 2002, Ramachandran et al., 2006, Sturgis et al., 1999, Tae et al., 2004, Varzim et al., 2003, Yen et al., 2008, Rydzanicz et al., 2005, Gajecka et al., 2005b, Csejtei et al., 2009) and XRCC3(Benhamou et al., 2004, Huang et al., 2005, Kietthubthew et al., 2006, Majumder et al., 2007, Werbrouck et al., 2008, Yen et al., 2008, Gajecka et al., 2005b, Rydzanicz et al., 2005). However, the results are conflicting rather than conclusive.
Given the large amount of data already published, it is important to perform a systematic review and meta-analysis of the current literature to assess the association of polymorphisms in DNA repair genes and head and neck cancer. A recent study by Vineis et al.(Vineis et al., 2009) provided a field synopsis of the association of variants in DNA repair genes and cancer risk. Although data for head and neck cancer were reported in this study, the focus of the study was to evaluate the associations between DNA repair gene variants in different types of cancers. Here, we present a meta-analysis with an updated literature review giving rise to a larger number of studies and additional data for a genetic polymorphism each in XPC and ERCC1 that were not reported in the previous review. The association between the DNA damage repair genes, XPA, XPC, XPD, XPF, ERCC1, XRCC1 and XRCC3 with oral, pharyngeal and laryngeal cancer was evaluated. Associations according to race and head and neck sub-site were also evaluated when possible.
METHODS
Literature search and selection criteria
A bibliographical search was performed in both MEDLINE and EMBASE to identify studies that evaluated DNA repair gene polymorphisms and oral, pharyngeal and laryngeal cancer up to January 15, 2010. The search terms used were: (oral or buccal or mouth or “head and neck” or pharyngeal or pharynx or oropharyngeal or laryngeal or larynx) and (cancer or neoplasms or tumor or carcinoma or carcinogenesis) and (“xeroderma pigmentosum complementation group A” XPA or “xeroderma pigmentosum complementation group C” or XPC or “xeroderma pigmentosum complementary group D” or XPD or “xeroderma pigmentosum complementation group F” or XPF or ERCC1 or “X-ray repair cross complementing protein 1” or XRCC1 or “X-ray repair cross complementing protein 3” or XRCC3). The literature cited from the selected articles was manually reviewed in order to detect articles that might have been missed in the search. The inclusion criteria for the selection of papers were the following: (1) the papers should be written in English or Spanish, (2) the papers should be case-control studies assessing the association between polymorphisms in DNA damage response genes and oral, pharyngeal and laryngeal cancer, including at least one of the following genes: XPA, XPC, XPD, XPF, ERCC1, XRCC1 and XRCC3, (3) studies must provide data to calculate crude odds ratios for oral, pharyngeal and laryngeal cancer. The exclusion criteria were (1) studies on nasopharyngeal cancer, (2) studies that included only cases and no controls, (3) studies with overlapping patient populations, (4) studies that evaluated response to treatment, secondary tumors or recurrence. From each study, the following information was extracted and tabulated: author’s last name, country where the study was conducted, year of publication, race/ethnicity of the study population, and genotyping information from cases and controls. The literature search yielded 58 publications. The following studies were excluded: two were published in Chinese(Yang et al., 2008b, Wen et al., 2007) and one in Polish(Rusin et al., 2008); three did not report gene polymorphisms(Cheng et al., 2002, Wei et al., 2005, Yang et al., 2006); two assessed the association between gene polymorphisms and survival(Grau et al., 2009, Handra-Luca et al., 2007); seven evaluated treatment efficacy(Bozec et al., 2007, Carles et al., 2006, Fountzilas et al., 2009, Jun et al., 2008, Kornguth et al., 2005, Quintela-Fandino et al., 2006, Werbrouck et al., 2009); one assessed the potential of gene polymorphisms as predictive and prognostic markers(Koh et al., 2009); two studies were conducted in patients only(Geisler et al., 2005, Hsieh et al., 2003); one study evaluated the potential of gene polymorphisms as risk modifiers of the association of oral contraceptives and oral cancer risk(Applebaum et al., 2009); one did not evaluate the genes of interest(Gajecka et al., 2005a); three studies evaluated the risk of oral leukoplakia(Majumder et al., 2009, Wang et al., 2007, Yang et al., 2008a); two studies evaluated the risk of secondary primary neoplasms(Gal et al., 2005); two reported on haplotypes only(Michiels et al., 2007, Majumder et al., 2009) and one included lung cancer under upper-aerodigestive tract (UADT) cancer(Buch et al., 2005). Therefore, 30 studies that included 19,343 individuals (7,291 cases and 12,052 controls) were considered for the meta-analysis.
Statistical analysis
The association between DNA damage response gene polymorphisms and oral, pharyngeal and laryngeal cancer risk was assessed by calculating odds ratios (OR) with 95% confidence intervals (CI). The combined ORs were calculated under the dominant, recessive, and additive genetic model for each polymorphism using the meta-analysis technique. Stratified combined ORs were calculated for each gene association according to race when data were provided by three or more studies. The majority of studies included more than one head and neck sub-site, and there were some studies conducted in Asian populations that included only oral cavity sub-site cases. Therefore, it was only possible to evaluate subsite-specific meta-ORs for the Asian studies. The between-study heterogeneity was determined by performing the Χ2-basedQ statistics test, and it was considered significant for a P<0.10(Whitehead and Whitehead, 1991). When significant heterogeneity was observed (Q-test p-value<0.10), the meta-OR was not reported in the results. The fixed-effect model was used under the assumption of homogeneity between studies. The I2 statistic was used as a confirmatory test for heterogeneity(Higgins et al., 2003, Ioannidis et al., 2007), with I2 <25%, 25–50%, and >50% representing low, moderate and high degree of heterogeneity, respectively. To explore between-study heterogeneity, stratification by race and control source was conducted. When heterogeneity could not be resolved, meta-ORs were not reported. To detect potential publication bias the Harbord test was employed. The Harbord test is a modified linear regression test for funnel plot asymmetry and is a measurement of small-study effect, considering significance at p< 0.10(Harbord et al., 2006). Genotype frequencies in the control populations according to race were calculated and tests on the equality of proportions was performed for Asian and Caucasian control populations in order to compare differences in genotype frequencies between the two groups. All of the statistical tests were performed using STATA SE (version 10) software (StataCorp LP, College Station, TX).
RESULTS
Thirty publications were included in this meta-analysis with a total of 19,343 subjects (7,291 cases and 12,052 controls). Some of the publications reported on multiple gene polymorphisms. There were five studies with results on the XPA 5′-UTR (A23G), one on XPD exon 8 C23047T, one on XPD exon 8 C23051T, six on XPD exon 6 codon 156 C22541A, twelve on XPD exon 23 codon 751 C35931A, six on XPD Asp312Asn, four on XPC-PAT, one on XPC Ala499Val, three on XPC exon 15 Lys939Gln, one on XPF 5′-UTR T2063A, four on ERCC1 3′UTR C8092A, fifteen on XRCC1 codon 194, sixteen on XRCC1 codon 399, four on XRCC1 exon 9 codon 280 Arg280His, and eight on XRCC3 Thr241Met. The study-specific OR, meta OR and heterogeneity statistics for each polymorphism are shown in Table 1.
Table 1.
Publications reporting DNA damage response gene polymorphisms and the risk of oral, pharyngeal and laryngeal cancer.
| Author (year) Country |
Ethnic Groups (n) | Recruitment Period | Control Source | Tumor Sites | Cases (n) | Controls (n) | Crude OR (95% CI) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| XPA 5′-UTR (A23G) | A/A | A/G | G/G | A/A | A/G | G/G | A/G vs. A/A | G/G vs. A/A | A/G + G/G vs. A/A | ||||
|
An J (2007)(An et al., 2007) USA |
C (829) | 1995–2005 | Hospital | Oral cavity, pharynx, larynx | 110 | 360 | 359 | 128 | 346 | 380 | 1.21 (0.90–1.62) | 1.09 (0.82–1.47) | 1.15 (0.88–1.52) |
|
Bau DT (2007)(Bau et al., 2007) Taiwan |
A (259) | 1997–2005 | Healthy | Oral Cavity | 38 | 84 | 32 | 29 | 53 | 23 | 1.20 (0.66–2.18) | 1.06 (0.51–2.18) | 1.16 (0.66–2.05) |
|
Hall J (2007)(Hall et al., 2007) Romania, Poland, Russia, Slovakia and Czeck Republic |
C (1690) | 2000–2002 | Hospital | Oral cavity, pharynx, larynx | 75 | 247 | 275 | 294 | 1125 | 891 | 0.86 (0.33–1.69) | 1.20 (0.90–1.61) | 1.01 (0.77–1.33) |
|
Sugimura T (2006)(Sugimura et al., 2006) Japan |
A (363) | 1988–2004 | Hospital | Oral cavity | 23 | 65 | 34 | 74 | 105 | 62 | 1.99 (1.13–3.49) | 1.76 (0.94–3.30) | 1.91 (1.12–3.24) |
|
Abbasi R (2009)(Abbasi et al., 2009) Germany |
C (1026) | 1998–2000 | Healthy | Larynx | 30 | 109 | 107 | 72 | 281 | 291 | 0.93 (0.59–1.50) | 0.88 (0.55–1.43) | 0.91 (0.58–1.43) |
| META | 1.15 (0.96–1.36) | 1.12 (0.95–1.32) | |||||||||||
| P, Q test | 0.090 | 0.523 | 0.260 | ||||||||||
| P, Eggers test | 0.441 | 0.867 | 0.546 | ||||||||||
|
XPD Exon 6 C22541A Codon 156 |
C/C | C/A | A/A | C/C | C/A | A/A | C/A vs. C/C | A/A vs. C/C | C/A + AA vs. C/C | ||||
|
Majumder M (2007)(Majumder et al., 2007) India |
A (699) | 1999–2005 | Healthy | Oral cavity | 88 | 156 | 64 | 124 | 191 | 73 | 1.52 (0.91–2.56) | 0.82 (0.34–1.95) | 1.36 (0.82–2.22) |
|
Kietthubthew S (2006)(Kietthubthew et al., 2006) Thai |
A (304) | 1998–1999 | Hospital | Oral cavity | 45 | 52 | 9 | 82 | 62 | 20 | 1.15 (0.81–1.62) | 1.23 (0.80–1.90) | 1.17 (0.85–1.63) |
| Sturgis EM (2000)(Sturgis et al., 2000) | NHW(685) | 1995–1999 | Healthy | Oral cavity, pharynx, hypopharynx | 62 | 97 | 30 | 154 | 241 | 101 | 0.99 (0.68–1.45) | 0.73 (0.44–1.22) | 0.92 (0.64–1.32) |
|
Abbasi R (2009)(Abbasi et al., 2009) Germany |
C (1026) | 1998–2000 | Healthy | Larynx | 79 | 28 | 41 | 199 | 309 | 139 | 1.04 (0.74–1.45) | 0.74 (0.48–1.14) | 0.95 (0.69–1.30) |
|
Rydzanicz M (2005)(Rydzanicz et al., 2005) Poland |
C (325) | --- | Healthy | Oral cavity and larynx | 73 | 82 | 26 | 54 | 69 | 20 | 0.88 (0.55–1.41) | 0.96 (0.49–1.90) | 0.90 (0.57–1.41) |
|
Gajecka M (2005)(Gajecka et al., 2005b) Poland |
C (615) | --- | Healthy | Larynx | 127 | 127 | 32 | 105 | 171 | 43 | 0.61 (0.43–0.87) | 0.62 (0.36–1.04) | 0.61 (0.44–0.85) |
| META | 0.84 (0.68–1.04) | ||||||||||||
| P, Q test | 0.052 | 0.402 | 0.067 | ||||||||||
| P, Eggers test | 0.467 | 0.841 | 0.495 | ||||||||||
|
XPD Exon 23 A35931C Codon 751 |
A/A | A/C | C/C | A/A | A/C | C/C | A/C vs. A/A | C/C vs. A/A | A/C + C/C vs. A/A | ||||
|
An J (2007)(An et al., 2007) USA |
C (829) | 1995–2005 | Hospital | Oral cavity, pharynx, larynx | 330 | 394 | 105 | 358 | 386 | 110 | 1.10 (0.90–1.35) | 1.03 (0.76–1.40) | 1.09 (0.90–1.33) |
|
Bau DT (2007)(Bau et al., 2007) Taiwan |
A (259) | 1997–2005 | Healthy | Oral cavity | 134 | 18 | 2 | 89 | 15 | 1 | 0.79 (0.38–1.66) | 1.32 (0.11–14.86) | 0.83 (0.41–1.69) |
|
Hart V (2008)(Harth et al., 2008) Germany |
C (612) | 1996–1998 | Healthy | Oral cavity, pharynx, larynx | 111 | 154 | 47 | 108 | 149 | 43 | 1.00 (0.71–1.42) | 1.06 (0.65–1.73) | 1.02 (0.73–1.42) |
| Huang WY (2005)(Huang et al., 2005) | W (2,250) B (258) O (186) |
1997–2006 | Healthy | Oral cavity, pharynx, larynx | 240 | 235 | 69 | 345 | 325 | 105 | 1.03 (0.82–1.31) | 0.94 (0.66–1.33) | 1.02 (0.82–1.27) |
|
Kietthubthew S (2006)(Kietthubthew et al., 2006) Thai |
A (304) | 1998–1999 | Hospital | Oral cavity | 83 | 21 | 1 | 126 | 36 | 2 | 0.88 (0.48–1.62) | 0.75 (0.06–8.5) | 0.88 (0.49–1.59) |
|
Majumder M (2007)(Majumder et al., 2007) India |
A (699) | 1999–2005 | Healthy | Oral cavity | 158 | 125 | 26 | 190 | 158 | 40 | 0.95 (0.69–1.30) | 0.78 (0.45–1.33) | 0.92 (0.68–1.24) |
|
Ramachandran S((2006),Ramachandran et al., 2006) India |
A (220) | --- | Healthy | Oral cavity | 49 | 46 | 15 | 71 | 31 | 8 | 2.15 (1.20–3.85) | 2.71 (1.06–6.90) | 2.27 (1.32–3.90) |
|
Sturgis EM (2000)(Sturgis et al., 2000) USA |
NHW (685) | 1995–1999 | Healthy | Oral cavity, pharynx, hypopharynx | 75 | 83 | 31 | 218 | 221 | 57 | 1.09 (0.75–1.57) | 1.58 (0.94–2.63) | 1.19 (0.85–1.68) |
|
Abbasi R (2009)(Abbasi et al., 2009) Germany |
C (1026) | 1998–2000 | Healthy | Larynx | 95 | 117 | 34 | 250 | 280 | 114 | 1.09 (0.79–1.51) | 0.78 (0.50–1.23) | 1.01 (0.75–1.36) |
| Matullo G (2006)(Matullo et al., 2006) France, Italy, Spain, United Kingdom, The Nederlands, Greece, Germany, Denmark |
C (1176) | 1993–1998 | Healthy | Oral cavity, pharynx, larynx | 34 | 29 | 9 | 397 | 504 | 193 | 0.90 (0.56–1.45) | 0.54 (0.25–1.15) | 0.80 (0.51–1.27) |
|
Rydzanicz M (2005)(Rydzanicz et al., 2005) Poland |
C (325) | --- | Healthy | Oral cavity, larynx | 0.89 (0.55–1.46) | 0.94 (0.50–1.78) | 0.91 (0.57–1.43) | ||||||
|
Gajecka M (2005)(Gajecka et al., 2005b) Poland |
C (615) | --- | Healthy | Larynx | 0.61 (0.43–0.87) | 0.65 (0.41–1.05) | 0.62 (0.44–0.87) | ||||||
| META | 1.01 (0.91–1.12) | 0.96 (0.82–1.11) | |||||||||||
| P, Q test | 0.126 | 0.184 | 0.000 | ||||||||||
| P, Eggers test | 0.719 | 0.720 | 0.446 | ||||||||||
| XPD Asp312Asn | G/G | G/A | A/A | G/G | G/A | A/A | G/A vs. G/G | A/A vs. G/G | G/A + A/A vs. G/G | ||||
|
An J (2007)(An et al., 2007) USA |
C (771) | 1995–2005 | Hospital | Oral cavity, pharynx, larynx | 330 | 395 | 104 | 370 | 386 | 98 | 1.14 (0.93–1.40) | 1.18 (0.86–1.62) | 1.16 (0.95–1.40) |
|
Hart V (2008)(Harth et al., 2008) Germany |
C (612) | 1996–1998 | Healthy | Oral cavity, pharynx, larynx | 113 | 158 | 40 | 101 | 145 | 52 | 0.97 (0.68–1.38) | 0.68 (0.42–1.12) | 0.90 (0.64–1.25) |
|
Majumder M (2007)(Majumder et al., 2007) India |
A (699) | 1999–2005 | Healthy | Oral cavity | 152 | 119 | 34 | 205 | 146 | 36 | 1.09 (0.79–1.51) | 1.27 (0.76–2.12) | 1.13 (0.84–1.53) |
|
Sturgis EM (2002a)(Sturgis et al., 2002b) USA |
NHW (626) | 1995–2001 | Healthy | Oral cavity, pharynx, larynx | 123 | 165 | 25 | 142 | 135 | 36 | 1.41 (1.01–1.96) | 0.80 (0.45–1.40) | 1.28 (0.93–1.76) |
|
Abbasi R (2009)(Abbasi et al., 2009) Germany |
C (1026) | 1998–2000 | Healthy | Larynx | 93 | 119 | 34 | 258 | 304 | 82 | 1.08 (0.79–1.49) | 1.15 (0.72–1.83) | 1.10 (0.81–1.49) |
| Matullo G (2006)(Matullo et al., 2006) France, Italy, Spain, United Kingdom, The Nederlands, Greece, Germany, Denmark |
C (1176) | 1993–1998 | Healthy | Oral, pharyngeal, laryngeal | 32 | 46 | 4 | 418 | 506 | 170 | 1.18 (0.74–1.89) | 0.30 (0.10–0.88) | 0.97 (0.61–1.53) |
| META | 1.14 (1.01–1.29) | 1.11 (0.99–1.25) | |||||||||||
| P, Q test | 0.772 | 0.075 | 0.717 | ||||||||||
| P, Eggers test | 0.987 | 0.100 | 0.350 | ||||||||||
| XPD Exon 8 C23047G | C/C | C/G | G/G | C/C | C/G | G/G | C/G vs. C/C | G/G vs. C/C | C/G + G/G vs. C/C | ||||
|
Sturgis EM (2002b)(Sturgis et al., 2002a) USA |
NHW (580) | 1995–1998 | Hospital | Oral cavity, oro/hypopha rynx, larynx | 179 | 0 | 1 | 393 | 7 | 0 | --- | --- | 0.31 (0.03–2.57) |
| XPD Exon 8 C23051G | C/C | C/G | G/G | C/C | C/G | G/G | C/G vs. C/C | C/G vs. C/C | C/G + G/G vs. C/C | ||||
|
Sturgis EM (2002b)(Sturgis et al., 2002a) USA |
NHW (580) | 1995–1998 | Hospital | Oral cavity, oro/hypopha rynx, larynx | 24 | 2 | 0 | 110 | 5 | 0 | --- | --- | 6.70 (0.27–165.11) |
| XPC PAT | +/+ | +/− | −/− | +/+ | +/− | −/− | +/− vs. +/+ | −/− vs. +/+ | (+/−) + (−/−) vs. +/+ | ||||
|
Kietthubthew S (2006)(Kietthubthew et al., 2006) Thai |
A (304) | 1998–1999 | Hospital | Oral cavity | 60 | 36 | 10 | 89 | 66 | 9 | 0.80 (0.48–1.36) | 1.64 (0.63–4.29) | 0.91 (0.56–1.49) |
|
Shen H (2001)(Shen et al., 2001) USA |
NHW (294) AA (178) HA (103) NCh (109) |
1995–1999 | Healthy | Oral cavity, pharynx, larynx | 102 | 135 | 50 | 141 | 133 | 37 | 1.40 (0.98–1.99) | 1.86 (1.13–3.06) | 1.50 (1.08–2.09) |
|
Sugimura T (2006)(Sugimura et al., 2006) Japan |
A (363) | 1988–2004 | Hospital | Oral cavity | 42 | 63 | 17 | 78 | 128 | 35 | 0.91 (0.56–1.47) | 0.90 (0.45–1.79) | 0.91 (0.57–1.44) |
|
Yang M (2005)(Yang et al., 2005) South Korea |
A (155) | --- | Hospital | Oral cavity, oro/hypopharynx, larynx | 35 | 29 | 9 | 38 | 33 | 11 | 0.95 (0.48–1.87) | 0.88 (0.32–2.39) | 0.94 (0.50–1.76) |
| META | 1.09 (0.86–1.37) | 1.39 (0.99–1.97) | 1.14 (0.92–1.43) | ||||||||||
| P, Q test | 0.270 | 0.287 | 0.186 | ||||||||||
| P, Eggers test | 0.205 | 0.441 | 0.134 | ||||||||||
|
XPC Exon 15 Lys939Gln |
Lys/Lys | Lys/Gln | Gln/Gln | Lys/Lys | Lys/Gln | Gln/Gln | Lys/Gln vs. Lys/Lys | Gln/Gln vs. Lys/Lys | Lys/Gln + Gln/Gln vs. Lys/Lys | ||||
|
An J (2007)(An et al., 2007) USA |
C (771) | 1995–2005 | Hospital | Oral cavity, pharynx, larynx | 312 | 339 | 118 | 315 | 425 | 114 | 0.94 (0.77–1.16) | 1.10 (0.90–1.34) | 0.97 (0.79–1.18) |
|
Kietthubthew S (2006)(Kietthubthew et al., 2006) Thai |
A (304) | 1998–1999 | Hospital | Oral cavity | 59 | 37 | 10 | 87 | 67 | 10 | 0.81 (0.48–1.36) | 0.85 (0.61–1.19) | 0.90 (0.55–1.47) |
|
Abbasi R (2009)(Abbasi et al., 2009) Germany |
C (1026) | 1998–2000 | Healthy | Larynx | 83 | 120 | 45 | 230 | 329 | 88 | 1.01 (0.73–1.40) | 1.41 (0.91–2.19) | 1.10 (0.81–1.49) |
| META | 0.94 (0.80–1.12) | 1.17 (0.92–1.49) | 0.99 (0.85–1.16) | ||||||||||
| P, Q test | 0.787 | 0.470 | 0.737 | ||||||||||
| P, Eggers test | 0.661 | 0.467 | 0.987 | ||||||||||
| XPC Exon 15 Ala499Val | Ala/Ala | Ala/Val | Val/Val | Ala/Ala | Ala/Val | Val/Val | Ala/Val vs. Ala/Ala | Val/Val vs. Ala/Ala | Ala/Val + Val/Val vs. Ala/Ala | ||||
|
An J (2007)(An et al., 2007) USA |
C (771) | 1995–2005 | Hospital | Oral cavity, pharynx, larynx | 455 | 293 | 91 | 454 | 342 | 58 | 0.85 (0.69–1.04) | 1.56 (1.09–2.23) | 0.96 (0.79–1.16) |
| XPF 5′-UTR (T2063A) | T/T | T/A | A/A | T/T | T/A | A/A | T/A vs. T/T | A/A vs. T/T | T/A + A/A vs. T/T | ||||
|
Sugimura T (2006)(Sugimura et al., 2006) Japan |
A (363) | 1988–2004 | Hospital | Oral cavity | 66 | 47 | 9 | 119 | 101 | 21 | 0.83 (0.53–1.33) | 0.77 (0.33–1.78) | 0.83 (0.53–1.28) |
|
ERCC1 3′UTR C 8092A |
C/C | C/A | A/A | C/C | C/A | A/A | C/A vs. C/C | A/A vs. C/C | C/A + A/A vs. C/C | ||||
|
An J (2007)(An et al., 2007) USA |
C (771) | 1995–2005 | Hospital | Oral cavity, pharynx, larynx | 455 | 326 | 48 | 485 | 315 | 54 | 1.10 (0.90–1.34) | 0.94 (0.62–1.42) | 1.08 (0.89–1.31) |
|
Sturgis EM (2002a)(Sturgis et al., 2002b) USA |
NHW (685) | 1995–1999 | Healthy | Oral cavity, pharynx, larynx | 183 | 116 | 14 | 172 | 127 | 14 | 0.85 (0.61–1.19) | 0.93 (0.43–2.02) | 0.87 (0.63–1.19) |
|
Sugimura T (2006)(Sugimura et al., 2006) Japan |
A (363) | 1988–2004 | Hospital | Oral cavity | 75 | 30 | 17 | 130 | 94 | 17 | 0.55 (0.33–0.91) | 1.73 (0.83–3.59) | 0.73 (0.47–1.14) |
|
Abbasi R (2009)(Abbasi et al., 2009) Germany |
C (1026) | 1998–2000 | Healthy | Larynx | 146 | 87 | 15 | 392 | 218 | 37 | 1.07 (0.78–1.46) | 1.08 (0.58–2.04) | 1.07 (0.80–1.45) |
| META | 1.07 (0.80–1.43) | 1.00 (0.87–1.14) | |||||||||||
| P, Q test | 0.063 | 0.546 | 0.322 | ||||||||||
| P, Eggers test | 0.112 | 0.420 | |||||||||||
| XRCC1 Exon 6 Codon 194 | C/C | C/T | T/T | C/C | C/T | T/T | C/T vs. C/C | T/T vs. C/C | C/T + T/T vs. C/C | ||||
|
Demokan S (2005)(Demokan et al., 2005) Turkey |
A (1936) | --- | Healthy | Oral cavity | 78 | 14 | 3 | 88 | 8 | 2 | 1.97 (0.78–4.95) | 1.69 (0.27–10.39) | 1.92 (0.83–4.44) |
|
Hart V (2008)(Harth et al., 2008) Germany |
C (612) | 1996–1998 | Healthy | Oral cavity, pharynx, larynx | 217 | 40 | 1 | 259 | 39 | 2 | 1.22 (0.76–1.97) | 0.59 (0.05–6.62) | 1.19 (0.75–1.91) |
|
Kowalski M (2009)(Kowalski et al., 2009) Poland |
C (216) | --- | Hospital | Head and neck cancr/NOS | 71 | 21 | 0 | 102 | 22 | 0 | 1.37 (0.70–2.98) | --- | 1.37 (0.70–2.68) |
|
Kietthubthew S (2006)(Kietthubthew et al., 2006) Thai |
A (304) | 1998–1999 | Hospital | Oral cavity | 40 | 50 | 16 | 77 | 67 | 20 | 1.43 (0.84–2.43) | 1.54 (0.71–3.29) | 1.46 (0.89–2.40) |
|
Majumder M (2005)(Majumder et al., 2005) India |
A (658) | 1999–2004 | Healthy | Oral cavity | 249 | 58 | 3 | 285 | 57 | 6 | 1.16 (0.77–1.74) | 0.57 (0.14–2.31) | 1.11 (0.75–1.64) |
|
Olshan AF (2002)(Olshan et al., 2002) USA |
--- | 1994–1997 | Hospital | Oral cavity, pharynx, larynx | 82 | 16 | 0 | 135 | 26 | 0 | 1.01 (0.51–2.00) | --- | 1.01 (0.51–2.00) |
|
Ramachandran S((2006),Ramachandran et al., 2006) India |
A (220) | --- | Healthy | Oral cavity | 66 | 37 | 7 | 90 | 19 | 1 | 2.65 (1.40–5.02) | 9.54 (1.14–79.46) | 3.00 (1.62–5.56) |
|
Sturgis EM (1999)(Sturgis et al., 1999) USA |
NHW (565) MA (39) AA (23) |
1995–1998 | Hospital | Oral cavity, oro/hypopha rynx, larynx | 180 | 22 | 1 | 363 | 61 | 0 | 0.72 (0.43–1.22) | 6.04 (0.24–149.04) | 0.76 (0.46–1.27) |
|
Tae K (2004)(Tae et al., 2004) Korea |
A (315) | 1997–2001 | Hospital | Oral cavity, oro/hypopha rynx, larynx | 59 | 52 | 9 | 101 | 39 | 5 | 2.28 (1.35–3.85) | 3.08 (0.98–9.62) | 2.37 (1.43–3.93) |
|
Yen CY (2008)(Yen et al., 2008) Taiwan |
A (2010) | --- | Hospital | Oral squamous cell carcinoma/NOS | 48 | 40 | 15 | 54 | 35 | 9 | 1.28 (0.70–2.33) | 1.87 (0.75–4.67) | 1.41 (0.81–2.45) |
|
Varzim G (2003)(Varzim et al., 2003) Portugal |
C (266) | 1998–1999 | Healthy | Larynx | 80 | 8 | 0 | 160 | 18 | 0 | 0.88 (0.37–2.13) | --- | 0.89 (0.37–2.13) |
| Matullo G (2006)(Matullo et al., 2006) France, Italy, Spain, United Kingdom, The Nederlands, Greece, Germany, Denmark |
C (1176) | 1993–1998 | Healthy | Oral cavity, pharynx, larynx | 78 | 4 | 0 | 951 | 141 | 2 | 0.34 (1.24–0.96) | 2.42 (0.11–50.93) | 0.34 (0.12–0.95) |
|
Rydzanicz M (2005)(Rydzanicz et al., 2005) Poland |
C (325) | --- | Healthy | Oral cavity and larynx | 165 | 16 | 1 | 129 | 14 | 0 | 0.89 (0.42–1.90) | 2.35 (0.09–58.10) | 0.95 (0.45–2.00) |
|
Csejtei A (2009)(Csejtei et al., 2009) Hungary |
C (211) | 2000–2003 | Healthy | Head and neck cancer/NOS | 96 | 11 | 1 | 85 | 15 | 2 | 0.65 (0.28–1.49) | 0.44 (0.04–4.97) | 0.63 (0.28–1.38) |
|
Gajecka M (2005)(Gajecka et al., 2005b) Poland |
C (615) | --- | Healthy | Larynx | 262 | 27 | 1 | 291 | 33 | 1 | 0.91 (0.53–1.55) | 1.11 (0.07–17.85) | 0.91 (0.54–1.55) |
| META | 1.69 (1.10–2.58) | ||||||||||||
| P, Q test | 0.018 | 0.646 | 0.005 | ||||||||||
| P, Eggers test | 0.621 | 0.902 | 0.535 | ||||||||||
|
XRCC1 Exon 10 Codon 399 |
G/G | G/A | A/A | G/G | G/A | A/A | G/A vs. G/G | A/A vs. G/G | G/A + A/A vs. G/G | ||||
|
Demokan S (2005)(Demokan et al., 2005) Turkey |
A (1936) | --- | Healthy | Oral cavity | 42 | 41 | 12 | 39 | 46 | 13 | 0.82 (0.45–1.51) | 0.85 (0.34–2.10) | 0.83 (0.47–1.48) |
|
Hart V (2008)(Harth et al., 2008) Germany |
C (612) | 1996–1998 | Healthy | Oral cavity, pharynx, larynx | 114 | 166 | 30 | 143 | 121 | 36 | 1.72 (1.22–2.41) | 1.04 (0.60–1.79) | 1.57 (1.13–2.16) |
| Huang WY (2005)(Huang et al., 2005) | W (2,250) B (258) O (186) |
1997–2006 | Healthy | Oral cavity, pharynx, larynx | 266 | 219 | 40 | 338 | 338 | 81 | 0.82 (0.65–1.04) | 0.62 (0.41–0.94) | 0.79 (0.63–0.98) |
|
Kowalski M (2009)(Kowalski et al., 2009) Poland |
C (216) | --- | Hospital | Head and neck cancer/NOS | 37 | 44 | 11 | 49 | 53 | 22 | 1.09 (0.61–1.97) | 0.66 (0.28–1.53) | 0.97 (0.56–1.68) |
|
Kietthubthew S (2006)(Kietthubthew et al., 2006) Thai |
A (304) | 1998–1999 | Hospital | Oral cavity | 55 | 45 | 6 | 67 | 74 | 23 | 0.74 (0.44–1.23) | 0.31 (0.12–0.83) | 0.64 (0.39–1.05) |
|
Majumder M (2005)(Majumder et al., 2005) India |
A (658) | 1999–2004 | Healthy | Oral cavity | 135 | 143 | 32 | 158 | 163 | 27 | 1.02 (0.84–1.28) | 1.38 (0.79–2.43) | 1.08 (0.79–1.47) |
| Li C (2007)(Li et al., 2007) | NHW (1684) | 1995–2003 | Hospital | Oral cavity, oro/hypopha rynx, larynx | 335 | 374 | 121 | 360 | 385 | 109 | 1.04 (0.84–1.28) | 1.19 (0.88–1.60) | 1.08 (0.89–1.31) |
|
Ramachandran S((2006),Ramachandran et al., 2006) India |
A (220) | --- | Healthy | Oral cavity | 46 | 48 | 16 | 73 | 33 | 4 | 2.30 (1.29–4.10) | 6.34 (1.99–20.17) | 2.75 (1.59–4.75) |
|
Sturgis EM (1999)(Sturgis et al., 1999) USA |
NHW (565) MA (39) AA (23) |
1995–1998 | Hospital | Oral cavity, oro/hypopha rynx, larynx | 94 | 77 | 32 | 181 | 197 | 46 | 0.75 (0.52–1.08) | 1.33 (0.80–2.24) | 0.86 (0.62–1.21) |
|
Tae K (2004)(Tae et al., 2004) Korea |
A (315) | 1997–2001 | Hospital | Oral cavity, oro/hypopha rynx, larynx | 69 | 51 | 9 | 86 | 64 | 7 | 0.99 (0.611–1.61) | 1.60 (0.56–4.52) | 1.05 (0.66–1.68) |
|
Varzim G (2003)(Varzim et al., 2003) Portugal |
C (266 | 1998–1999 | Healthy | Larynx | 37 | 40 | 11 | 80 | 80 | 18 | 1.08 (0.62–1.86) | 1.32 (0.56–4.07) | 1.13 (0.67–1.89) |
| Matullo G (2006)(Matullo et al., 2006) France, Italy, Spain, United Kingdom, The Nederlands, Greece, Germany, Denmark |
C (1176) | 1993–1998 | Healthy | Oral cavity, pharynx, larynx | 34 | 38 | 10 | 484 | 482 | 128 | 1.02 (0.92–1.12) | 1.11 (0.53–2.31) | 1.12 (0.71–1.77) |
|
Rydzanicz M (2005)(Rydzanicz et al., 2005) Poland |
C (325) | --- | Healthy | Oral cavity and larynx | 63 | 98 | 21 | 59 | 63 | 21 | 1.46 (0.91–2.34) | 0.13 (0.05–0.33) | 1.33 (0.84–2.08) |
|
Csejtei A (2009)(Csejtei et al., 2009) Hungary |
C (211) | 2000–2003 | Healthy | Head and neck cancer/NOS | 50 | 47 | 11 | 53 | 41 | 8 | 1.22 (0.69–2.15) | 1.46 (0.54–3.92) | 1.25 (0.73–2.16) |
|
Gajecka M (2005)(Gajecka et al., 2005b) Poland |
C (615) | --- | Healthy | Larynx | 106 | 153 | 34 | 124 | 145 | 50 | 1.23 (0.87–1.74) | 0.80 (0.48–1.32) | 1.12 (0.81–1.56) |
| META | |||||||||||||
| P, Q test | 0.014 | 0.000 | 0.004 | ||||||||||
| P, Eggers test | 0.360 | 0.868 | 0.355 | ||||||||||
| XRCC1 Exon 9 Codon 280 | G/G | G/A | A/A | G/G | G/A | A/A | G/A vs. G/G | A/A vs. G/G | G/A + A/A vs. G/G | ||||
|
Hart V (2008)(Harth et al., 2008) Germany |
C (612) | 1996–1998 | Healthy | Oral cavity, pharynx, larynx | 283 | 28 | 1 | 270 | 30 | 0 | 0.89 (0.51–1.53) | 2.86 (0.11–70.57) | 0.92 (0.54–1.58) |
|
Majumder M (2005)(Majumder et al., 2005) India |
A (658) | 1999–2004 | Healthy | Oral cavity | 228 | 79 | 3 | 264 | 81 | 3 | 1.12 (0.79–1.61) | 1.15 (0.23–5.79) | 1.13 (0.79–1.61) |
|
Ramachandran S((2006),Ramachandran et al., 2006) India |
A (220) | --- | Healthy | Oral cavity | 77 | 31 | 2 | 83 | 26 | 1 | 1.28 (0.70–2.35) | 2.15 (0.19–24.25) | 1.32 (0.73–2.39) |
|
Tae K (2004)(Tae et al., 2004) Korea |
A (315) | 1997–2001 | Hospital | Oral cavity, oro/hypopha rynx, larynx |
113 | 21 | 1 | 139 | 29 | 0 | 0.89 (0.48–1.64) | 3.68 (0.14–91.38) | 0.93 (0.51–1.71) |
| META | 1.06 (0.83–1.35) | 1.74 (0.55–5.52) | 1.08 (0.85–1.37) | ||||||||||
| P, Q test | 0.749 | 0.901 | 0.790 | ||||||||||
| P, Eggers test | 0.648 | 0.003 | 0.737 | ||||||||||
| XRCC3 Thr241Met | Thr/Thr | Thr/Met | Met/Me t | Thr/Thr | Thr/Met | Met/Met | Thr/Met vs. Thr/Thr | Met/Met vs. Thr/Thr | Thr/Met + Met/Met vs. Thr/Thr | ||||
|
Kietthubthew S (2006)(Kietthubthew et al., 2006) Thai |
A (304) | 1998–1999 | Hospital | Oral cavity | 83 | 22 | 1 | 140 | 23 | 1 | 1.61 (0.84–3.07) | 1.68 (0.10–27.32) | 1.62 (0.86–3.04) |
|
Majumder M (2005)(Manuguerra et al., 2006) India |
A (658) | 1999–2004 | Healthy | Oral cavity | 201 | 97 | 12 | 220 | 120 | 8 | 0.88 (0.63–1.22) | 1.09 (0.48–2.49) | 0.90 (0.66–1.24) |
| Huang WY (2005)(Huang et al., 2005) | W (2,250) B (258) O (186) |
1997–2006 | Healthy | Oral cavity, pharynx, larynx | 232 | 223 | 61 | 329 | 334 | 97 | 0.94 (0.74–1.20) | 1.41 (0.95–2.10) | 1.02 (0.81–1.28) |
|
Yen CY (2008)(Yen et al., 2008) Taiwan |
A (2010 | --- | Hospital | Oral squamous cell carcinoma/NOS | 96 | 7 | 0 | 89 | 9 | 0 | 0.72 (0.25–2.01) | --- | 0.72 (0.26–2.02) |
|
Werbrouck J (2008)(Werbrouck et al., 2008) Belgium |
C (309) | 2004–2006 | Healthy | Oral cavity, pharynx, larynx | 69 | 59 | 29 | 44 | 75 | 33 | 0.50 (0.30–0.83) | 0.63 (0.33–1.20) | 0.54 (0.34–0.87) |
|
Benhamou S (2004)(Benhamou et al., 2004) France |
C (422) | 1988–1992 | Healthy | Oral cavity, pharynx, larynx | 42 | 54 | 23 | 47 | 89 | 30 | 0.67 (0.39–1.16) | 1.11 (0.54–2.28) | 0.77 (0.46–1.28) |
| Matullo G (2006)(Matullo et al., 2006) France, Italy, Spain, United Kingdom, The Nederlands, Greece, Germany, Denmark |
C (1176) | 1993–1998 | Healthy | Oral cavity, pharynx, larynx | 29 | 39 | 14 | 383 | 544 | 167 | 0.95 (0.58–1.56) | 1.11 (0.57–2.15) | 0.98 (0.62–1.57) |
|
Rydzanicz M (2005)(Rydzanicz et al., 2005) Poland |
C (325) | --- | Healthy | Oral cavity, larynx | 22 | 71 | 89 | 14 | 71 | 58 | 0.64 (0.30–1.34) | 0.98 (0.46–2.06) | 0.79 (0.39–1.60) |
|
Gajecka M (2005)(Gajecka et al., 2005b) Poland |
C (615) | --- | Healthy | Larynx | 135 | 125 | 33 | 144 | 131 | 47 | 1.02 (1.72–1.43 | 0.75 (0.45–1.24) | 0.95 (0.69–1.30) |
| Shen H (2002)(Shen et al., 2002) USA |
NHW (721) | 1995–2001 | Healthy | Oral cavity, pharynx, larynx | 150 | 159 | 58 | 141 | 170 | 43 | 0.88 (0.64–1.21) | 1.27 (0.80–2.00) | 0.96 (0.71–1.29) |
| META | 0.89 (0.78–1.01) | 1.07 (0.88–1.31) | 0.93 (0.82–1.05) | ||||||||||
| P, Q test | 0.277 | 0.521 | 0.370 | ||||||||||
| P, Eggers test | 0.457 | 0.641 | 0.486 | ||||||||||
Abbreviations: C= Caucasians, A= Asians, NHW= Non-Hispanic whites, MA= Mexican-Americans, AA= African-Americans, HA= Hispanic-Americans, NCh= Native Chinese, B= Black, O= Others, NOS= not otherwise specified,
XPA polymorphisms
XPA 5′-UTR A23G
Five publications reported data on XPA 5′-UTR A23G, for a total of 1,959 cancer cases and 4,279 controls. The source of controls was mostly from hospital populations (three out of five studies). Three of the studies were conducted in Caucasian populations while the other two studies were conducted in Asian populations. There was no significant difference in the frequency of the XPA 5′-UTR A23G heterozygous polymorphism between Caucasians and Asians (45.7% vs. 44.5%, p=0.667).
Overall, there was evidence of moderate between-study heterogeneity for the heterozygous variant (A/G) (Q statistics: 8.03 p= 0.090; I2= 50%, 95% CI: 0–82). Stratification by control source did not resolve heterogeneity. For Caucasian populations moderate heterogeneity was observed (Q statistics: 2.74, p= 0.256; I2= 26%, 95% CI: 0–92). There was no association between the heterozygous variant and oral, pharyngeal and laryngeal cancer risk and no evidence of small-study effect (p=0.858). For the homozygous and combined variants of XPA 5′-UTR A23G, overall there was no association with oral, pharyngeal and laryngeal cancer risk. No evidence of between study heterogeneity was observed for the homozygous variant (G/G) (Q statistics: 3.21; p= 0.523; I2= 0%, 95% CI: 0–79), and low heterogeneity was observed for the combined variants (A/G and G/G alleles) (Q statistics: 5.28; p= 0.260; I2= 24%, 95% CI: 0–69). There was no evidence of a small-study effect (homozygous: p=0.867; combined: p=0.546).
Race-specific analysis could only be performed for Caucasians, there was a marginal association between the homozygous variant and oral, pharyngeal and laryngeal cancer risk (meta-OR: 1.11, 95% CI: 0.91–1.34) and no heterogeneity was observed between the studies (Q statistics: 1.23; p= 0.541; I2= 0%, 95% CI: 0–90). For the combined variant, no association was found for this population (meta-OR: 1.05, 95% CI: 0.88–1.26) and there was no heterogeneity between the studies (Q statistics: 0.90; p= 0.638; I2= 0%, 95% CI: 0–90).
XPD polymorphisms
XPD Exon 6 C22541A Codon 156
For the XPD exon 6 C22541A codon 156 polymorphism, six studies involved 1,337 cases and 2,283 controls. The source of controls was mostly healthy population (five out of six studies). Two of the studies were conducted in Asian populations and four studies were conducted in Caucasian populations. There was no significant difference in the frequency of the XPD exon 6 C22541A codon 156 heterozygous polymorphism between Caucasians and Asians (45.8% vs. 45.7%, p=0.967).
Large between-study heterogeneity was observed for the heterozygous and the combined variants and this was not resolved after stratification by race and control source. For the XPD exon 6 C22541A codon 156 homozygous variant, no association was observed and there was no evidence of between-study heterogeneity. No evidence of a small-study effect on any of the variants (p=0.467; p=0.841; p=0.495).
Race-specific analyses could only be performed for Caucasians. A significant inverse association was seen between the homozygous variant and oral, pharyngeal and laryngeal cancer risk (AA, meta-OR: 0.74, 95% CI: 0.57–0.95). There was no heterogeneity between these studies (Q statistics: 1.04; p= 0.791; I2= 0%, 95% CI: 0–85).
XPD Exon 23 A35931C Codon 751
Twelve studies reporting the exon 23 A35931C polymorphism of XPD were conducted to evaluate its association with oral, pharyngeal and laryngeal cancer risk, for a total of 3,289 cases and 5,135 controls. The source of controls was mostly from the healthy population (ten out of twelve studies). Seven studies were conducted in Caucasian populations, four studies were performed in Asian populations which included oral cavity cases only, and one study was conducted in a mixed population comprised of Caucasians, African-Americans and Hispanics. There was significant difference in the frequency of the XPD exon 23 A35931C codon 751 heterozygous polymorphism between Caucasians and Asians (31.3% vs. 44.7%, p<0.0001).
There was no association between XPD Exon 23 A35931C heterozygous or homozygous variants with oral, pharyngeal and laryngeal cancer risk. Moderate between-studies heterogeneity was observed and there was no evidence of a small-study effect (p=0.976; p=0.684, respectively). For the combined variants, large heterogeneity was observed (A/C + C/C, Q statistics: 61.32; p= 0.000; I2= 82%, 95% CI: 70–89) and stratification by race and control source did not resolve heterogeneity.
For the race-specific analysis, no independent associations were observed in Caucasians for the heterozygous (meta-OR: 1.00, 95% CI: 0.90–1.12) and homozygous (meta-OR: 0.94, 35% CI: 0.81–1.10) variants. There was low and moderate heterogeneity observed between studies, respectively. For Asians, large heterogeneity was seen for the heterozygous variant (Q statistics: 7.02; p= 0.071; I2= 57%, 95% CI: 0–86). The meta-OR for the homozygous variant did not show an association with oral cavity cancer risk (meta-OR: 1.06, 95% CI: 0.68–1.66). There was moderate heterogeneity between these studies (Q statistics: 5.26; p= 0.154; I2= 43%, 95% CI: 0–81).
XPD Asp312Asn
Six studies reported data on the XPD Asp312Asn polymorphism for a total of 2,103 cases and 3,719 controls. Five studies were conducted in Caucasian populations and one in an Asian population. There was significant difference in the frequency of the XPD Asp312Asn heterozygous polymorphism between Caucasians and Asians (37.5% vs. 44.3%, p=0.010).
A marginally significant association was observed between the XPD Asp312Asn heterozygous and combined variants and oral, pharyngeal and laryngeal cancer risk (G/A, meta-OR: 1.14, 95% CI: 1.01–1.29 and G/A + A/A, meta-OR: 1.11, 95% CI: 0.99–1.25, respectively). There was no indication of between-studies heterogeneity for these studies (heterozygous, G/A, Q statistics: 2.53; p= 0.772; I2= 0%, 95% CI: 0–75, combined variant, G/A + A/A, Q statistics: 2.89; p= 0.717; I2= 0%, 95% CI: 0–75) and no evidence of a small-study effect (p=0.987 and p=0.350, respectively). For the homozygous variant (A/A), moderate between-studies heterogeneity was observed (Q statistics: 10.01; p= 0.075; I2= 50%, 95% CI: 0–80). Stratification by race and control source did not resolve the observed heterogeneity, and there was no evidence of a small-study effect (p=0.100).
Race-specific analyses could only be performed for Caucasians. There was no change in the marginally significant association between the heterozygous and combined variants and oral, pharyngeal and laryngeal cancer risk, and no heterogeneity was observed between the studies (data not shown).
XPD Exon 8 C23047G and C23051G
Only one study evaluated the association between both polymorphisms XPD Exon 8 C23047G and C23051G and oral, pharyngeal and laryngeal cancer risk. This study was conducted in a non-Hispanic White population including 180 cases and 400 controls. No significant association was evident when estimating the crude ORs for the combined variants.
XPC polymorphisms
XPC-PAT
Four publications reported data for XPC-PAT, for a total of 588 cases and 798 controls. The source of controls was mostly a hospital population (three out of four studies). Three studies were conducted in Asian populations and one study was performed in a mixed population comprised of non-Hispanic Whites, African-American, Hispanic-Americans and Asians. There was no significant difference in the frequency of the XPC-PAT heterozygous polymorphism between Caucasians and Asians (46.6% vs. 42.8%, p=0.293).
Overall, no significant association was reported for heterozygous or combined variants but a marginally significant association for the homozygous variant (−/−, meta-OR: 1.39, 95% CI: 0.99–1.97) was observed. There was no evidence of heterogeneity between the studies for the heterozygous and homozygous variants and for the combined variants, moderate heterogeneity was observed between the studies. No small-study effect was seen for any of the variants (homozygous: p=0.205; heterozygous: p=0.441; combined: p=0.134).
Similar to the overall population, no association was observed for the heterozygous and combined variants in Asians (−/+, meta-OR: 0.88, 95% CI: 0.65–1.21; −/+ and −/−, meta-OR: 0.92, 95% CI: 0.68–1.23). However, the marginally significant association no longer remained for the homozygous variant in the Asian population (−/−, meta-OR: 1.05, 95% CI: 0.65–1.71) and no heterogeneity observed between any of the Asian studies (data not shown).
XPC Exon 15 Lys939Gln
For the XPC Exon 15 Lys939Gln polymorphism, three studies reported data for 1,192 cases and 1,787 controls. Most of the controls were drawn from a hospital population (two out of three studies). Two studies were performed in Caucasians, while one study was conducted in Asians. There was no significant difference in the frequency of the XPD exon 6 C22541A codon 156 heterozygous polymorphism between Caucasians and Asians (40.9% vs. 46.5%, p=0.170).
Overall, no association of this polymorphism with oral, pharyngeal and laryngeal cancer was reported for heterozygous, homozygous or the combined variants. There was no evidence of heterogeneity between the studies for the any of the variants; nor was there a small-study effect (p=0.661; p=0.467; p=0.987, respectively)
XPC Exon 15 Ala499Val
One study reported data for XPC Exon 15 Ala499Val for a total of 829 cases and 854 controls. This study was conducted in Caucasians, while using hospital population as a source for controls. It reported a significant association between the homozygous variant and oral, pharyngeal and laryngeal cancer risk (OR: 1.56, 95% CI: 1.09–2.23).
XPF polymorphisms
XPF 5′-UTR T2063A
Only one study reported data on XPF 5′-UTR T2063A (122 cases and 241 controls). It was conducted in Asians and used hospital population as a source of controls. No association with the risk of oral, pharyngeal and laryngeal cancer was reported. No other XPF gene polymorphisms were reported in the studies reviewed.
ERCC1 polymorphisms
ERCC1 3′UTR C8092A
Four studies were reported for a total of 1,521 cases and 2,177 controls. Three studies were conducted in Caucasians, and one in an Asian population. Regarding the source of controls, two studies used hospital-based controls, while the remaining two used a healthy population. There was no significant difference in the frequency of the ERCC1 3′UTR C8092A heterozygous polymorphism between Caucasians and Asians (39.0% vs. 34.1%, p=0.132).
Overall, large heterogeneity between studies was detected for the heterozygous variant, and there was no evidence of a small-study effect (p=0.112). After stratification by race, homogeneity was obtained for the Caucasian population but no association was observed (C/A, meta-OR: 1.04, 95% CI: 0.89–1.21; Q statistics: 1.69; p= 0.429, I2= 0%, 95% CI: 0–90). For the homozygous and combined variants, no association was reported. There was no between-studies heterogeneity detected for these variants, and no evidence of a small-study effect (p=0.420; p=0.144, respectively). For Caucasians, no association was observed for the homozygous or combined variants (A/A, meta-OR: 0.98, 95% CI: 0.72–1.34; and C/A + A/A, meta-OR: 1.03, 95% CI: 0.98–1.19), and no heterogeneity was observed between studies (data not shown).
XRCC1 polymorphisms
XRCC1 Exon 6 Codon 194
Fifteen studies were reviewed on the association of XRCC1 exon 6 codon 194, for a total of 2,330 cases and 3,834 controls. Six studies were performed in Asians (four of these included oral cavity cases only), seven studies were performed in Caucasians, one study was performed in a mixed population of non-Hispanic whites, African-Americans and Mexican-Americans, and one study was conducted in a mixed population of White and non-Whites. Nine studies used healthy population controls and the remaining six studies used hospital population controls. There was significant difference in the frequency of the XRCC1 exon 6 codon 194 heterozygous polymorphism between Caucasians and Asians (22.8% vs. 13.0%, p<0.0001).
Moderate heterogeneity was seen between the studies that reported data for the heterozygous variant, (C/T, Q statistics: 27.21; p= 0.018; I2= 49%, 95% CI: 7–72), while large between-study heterogeneity was observed for the combined variant and there was no small-study effect (heterozygous: p=0.621; combined: p=0.535). For the heterozygous variant, stratification by control source did not resolve heterogeneity. There were differences in association of the C/T variant according to race. An increased association for the C/T variant and oral, pharyngeal and laryngeal cancer risk was observed only for the Asian population (Asians, meta-OR: 1.59, 95% CI: 1.27–1.99; Caucasians, meta-OR: 0.92, 95% CI: 0.74–1.14). Moderate heterogeneity was still observed for the Asian population (C/T, Q statistics: 7.44; p= 0.190; I2= 33%, 95% CI: 0–73), while homogeneity was obtained for the Caucasian population (C/T, Q statistics: 7.83; p= 0.451; I2= 0%, 95% CI: 0–65). Race stratification was also performed to evaluate the source of heterogeneity for the combined variants. No association was found for Caucasians (C/T + TT variants: meta-OR: 0.92, 95% CI: 0.74–1.14) and the studies were homogeneous (C/T + TT, Q statistics: 7.71; p= 0.462; I2= 0%, 95% CI: 0–65). For Asians, large heterogeneity remained and this was not resolved when the Asian studies were limited to oral cavity cases only. For the homozygous variants, overall, a significant increased risk of oral, pharyngeal and laryngeal cancer was observed (meta-OR: 1.69, 95% CI: 1.10–2.58). There was no between-study heterogeneity (T/T, Q statistics: 7.38; p= 0.496; I2= 0%, 95% CI: 0–64) and no small-study effect (p=0.902).
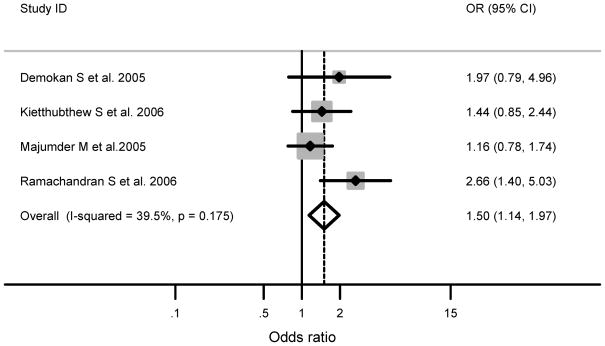
Tumor site-specific analysis was possible for oral cavity studies, and all of these studies were conducted in Asian populations. Similar to the overall results for the heterozygous variant in Asian populations irrespective of tumor site, a significantly increased association was still observed (meta-OR: 1.50, 95% CI: 1.14–1.97) for oral cavity studies and moderate between-study heterogeneity remained (Q statistics: 4.96; p= 0.175; I2= 40%, 95% CI: 0–79) (Figure 1).
Figure 1.
Published case-control studies that included only oral cavity cases in Asian populations show a significant association of the XRCC1 exon 6 codon 194 (C/T) heterozygous variant and the risk of oral cavity cancer. The shaded boxes represent the study-specific odds ratio, and the horizontal lines represent the confidence intervals; the size of each box depict how each study is weighted in the analysis, the diamond represents the meta-OR and its width represents the CI for the meta-OR.
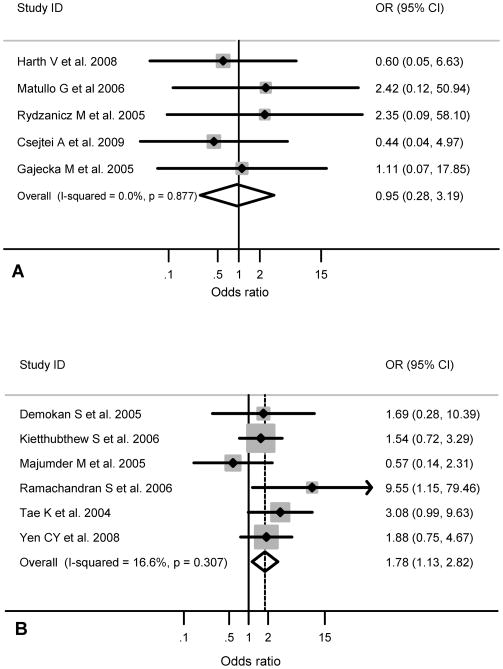
Race-specific analyses revealed no association of the homozygous variant and cancer risk for Caucasians and there was no heterogeneity between the studies (T/T, Q statistics: 1.21; p= 0.876; I2= 0%, 95% CI: 0–79) (Figure 2a). In contrast, the meta-OR was significantly associated between the homozygous variant and oral, pharyngeal and laryngeal cancer in Asians (meta-OR: 1.78, 95% CI: 1.13–2.82), and there was no between-study heterogeneity (TT, Q statistics: 6.00, p= 0.306; I2= 17%, 95% CI: 0–79) (Figure 2b). When the Asian studies were limited to oral cavity cases, there was in increased, but non-significant association between the homozygous variant and oral cavity cancer risk (Asian, oral cavity, meta-OR: 1.50, 95% CI: 0.82–2.74) with moderate heterogeneity between these studies (Q statistics: 4.78; p= 0.189; I2= 37%, 95% CI: 0–78).
Figure 2.
(A) Published case-control studies show non-significant association of the XRCC1 exon 6 codon 194 (T/T) homozygous variant and the risk of head and neck cancer Caucasian populations. The shaded boxes represent the study-specific odds ratio, and the horizontal lines represent the confidence intervals; the size of the boxes depict how each study is weighted in the analysis, the diamond represents the meta-OR and its width represents the CI for the meta-OR. (B) Published case-control studies show a significant association of the XRCC1 exon 6 codon 194 (T/T) homozygous variant and the risk of head and neck cancer Asian populations. The shaded boxes represent the study-specific odds ratio, and the horizontal lines represent the confidence intervals; the size of the boxes depict how each study is weighted in the analysis, the diamond represents the meta-OR and its width represents the CI for the meta-OR.
XRCC1 Exon 10 Codon 399
Sixteen studies reported the association of XRCC1 exon 10 codon 399 and oral, pharyngeal and laryngeal cancer for a total of 3,582 cases and 5,347 controls. Five studies were conducted in Asians (four of these included oral cavity cases only); seven studies were conducted in Caucasians, one study was conducted in non-Hispanic whites, one study was performed in a mix population of non-Hispanic whites, African-Americans and Mexican-Americans, and one study were conducted in a mix population of whites and nonwhites. The majority of the studies used healthy control populations (ten out of fifteen studies). There was no significant difference in the frequency of the XRCC1 exon 10 codon 399 heterozygous polymorphism between Caucasians and Asians (42.8% vs. 44.5%, p=0.340).
For all of the studies, moderate between-study heterogeneity was observed in the heterozygous variant, while large heterogeneity was observed in the homozygous and combined variants. There was no small-study effect observed for any of these variants (heterozygous: p=0.360; homozygous: p=0.868; combined: p=0.355). Race and tumor-site stratification did not resolve the observed heterogeneity for the heterozygous variant but homogeneity was obtained after stratification by controls source. For the studies that used hospital controls no association between G/A variant and oral, pharyngeal and laryngeal cancer risk was observed [Hospital (G/A, meta-OR: 0.95, 95% CI: 0.82–1.11; Q statistics: 3.56; p= 0.469; I2= 0%, 95% CI: 0–79)] but there was large heterogeneity for the studies that used healthy controls. For the homozygous variant, stratification by race, control source, and limiting to oral cavity cases, did not resolve heterogeneity. For the combined variants G/A + AA, race-specific analyses revealed large between-study heterogeneity for Asians (G/A + AA, Q statistics: 16.27; p= 0.003; I2= 75%, 95% CI: 40–90) which was not resolved when the analysis was limited to oral cavity Asian cases only (data not shown). In contrast, a marginal association between G/A + AA variant and oral, pharyngeal and laryngeal cancer risk was observed for Caucasians (meta-OR: 1.14, 95% CI: 1.01–1.27) and homogeneity was observed (Q statistics: 7.63; p= 0.470; I2= 0%, 95% CI: 0–65).
XRCC1 Exon 9 Codon 280
Four publications reported data on XRCC1 exon 9 codon 280, for a total of 879 cases and 926 controls. The source of controls was mostly a healthy population (three out of four studies). Three studies were conducted in Asian populations, while one study was conducted in a Caucasian population. There was significant difference in the frequency of the XRCC1 exon 6 codon 194 heterozygous polymorphism between Caucasians and Asians (21.7% vs. 10.0%, p<0.0001).
Overall, there was no association between XRCC1 exon 9 codon 280 and the risk of oral, pharyngeal and laryngeal cancer. There was no evidence of between-study heterogeneity for any of the variants. No evidence of a small-study effect was observed for heterozygous and combined variants (p=0.634 and p=0.749, respectively) but a small study-effect was observed for the homozygous variant (p=0.003).
Similarly, no association was observed for XRCC1 exon 9 codon 280 and the risk of oral, pharyngeal and laryngeal cancer, after limiting the analysis to the Asian population (G/A, meta-OR: 1.11, 95% CI: 0.84–1.46; A/A, meta-OR: 1.62, 95% CI: 0.47–5.57; and G/A + A/A, meta-OR: 1.12, 95% CI: 0.86–1.47) and there was no heterogeneity between studies for any of the variants (data not shown).
XRCC3 polymorphisms
XRCC3 Thr241Met
Ten studies reported on XRCC3 Thr241Met, for a total of 2,235 cases and 3,601 controls. Six studies were conducted in Caucasian populations, three studies were conducted in Asian populations, and one study was conducted in a mixed population of Whites and non-Whites. The source of controls was primarily healthy populations (seven out of nine studies). There was significant difference in the frequency of the XRCC3 Thr241Met heterozygous polymorphism between Caucasians and Asians (24.9% vs. 48.4%, p<0.0001).
Overall, there was no association between XRCC3 Thr241Met heterozygous, homozygous and combined variants and the risk of oral, pharyngeal and laryngeal cancer, no evidence of between-study heterogeneity for any of the variants and no evidence of a small-study effect (heterozygous: p=0.457; homozygous: p=0.641; combined: p=486). No independent associations were observed for Caucasians or Asians (data not shown). For the Caucasian studies, there was no to low heterogeneity between studies for all of the variants; and for the Asian studies, moderate between-study heterogeneity was observed (data not shown).
DISCUSSION
This meta-analysis of 30 case-control studies assessed the association of polymorphisms in DNA damage response genes with oral, pharyngeal and laryngeal cancer risk. A previous review by Vineis et al.(Vineis et al., 2009), evaluated the association of variants in DNA repair genes and cancer susceptibility in general, without in-depth analysis of head and neck cancer, given the broad scope of their paper. Here, we provide an updated systematic revision of the literature analyzing a larger number of studies and genetic polymorphisms. We have also reported results according to race and head and neck subsite, when possible.
There are three major pathways involved in DNA repair, depending on the type and magnitude of the damage. First, the base excision repair (BER) pathway repairs small base modifications, including oxidatively-induced lesions and single-strand breaks (SSBs), through exposure of the cells to reactive oxygen species (ROS), an endogenous toxic agent. For this pathway, we report results for three polymorphisms in the XRCC1 gene. The nucleotide excision repair (NER) pathway removes a broader spectrum of genomic damage, including bulky adducts induced by large polycyclic aromatic hydrocarbons, such as those present in benzo[α]pyrene in cigarette smoke, and crosslinks caused by UV-light photoproducts and chemotherapeutic agents. We have evaluated eleven polymorphisms in nucleotide excision repair genes XPA, XPC, XPD, XPF and ERCC1. Finally, single (SSBs) and double strand breaks (DSBs), endogenously produced by reactive oxygen species among other factors, can undergo either an error-prone (by non-homologous DNA end joining) or an error-free (by homologous recombination) repair process(Hakem, 2008). For this pathway, we have evaluated one polymorphism in the XRCC3 gene.
Although there is little evidence about the direct influence of genetic polymorphisms on the functionality of the BER pathway, recent publications with conflicting results have addressed the association between various polymorphisms in BER genes, such as XRCC1, and the risk of oral, pharyngeal and laryngeal cancer. Similar to Vineis et al.(Vineis et al., 2009), our meta-analysis revealed an almost two-fold statistically significant increased association between the XRCC1 codon 194 homozygous T/T variant and oral, pharyngeal and laryngeal cancer. We also report that this statistically significant two-fold increased risk was observed for Asian populations and for Asian oral cavity cancer cases. Comparison of the meta-ORs between Asians and Caucasians was not possible, since the Caucasian studies included more than one head and neck subsite, while the Asian studies were more homogeneous and included oral cavity cancer cases only. Therefore, studies that investigate this association between XRCC1 and cancer according to race and head and neck subsite are warranted.
XRCC1 is an important component in the BER, because it has the ability to interact with and serves as a scaffold for other key proteins that are responsible for strand incision at the DNA damage site, as well as DNA polymerase β and DNA ligase III, responsible for synthesis and re-joining of the DNA strand break, respectively(Altieri et al., 2008). Although the functional impact of the XRCC1 codon 194 polymorphisms remains unknown since it was first reported(Shen et al., 1998), it is plausible that changes in amino acid sequence at conserved sites may alter the functionality of the protein. This eventually could lead to a defective BER pathway, increased genomic instability and cancer risk.
XPC, XPA and XPD play important roles the nucleotide excision repair pathway. We observed marginal significant increased associations between XPD Asp312Asn heterozygotes and combined variants, as well as the XPC-PAT homozygous variant and the risk of oral, pharyngeal and laryngeal cancer. Our findings contrast with those reported by Vineis et al.(Vineis et al., 2009) and Manuguerra et al.(Manuguerra et al., 2006), who found no association. However, our meta-analysis included twice as many studies for each of these polymorphisms. Although no associations were seen between the XPA 5′-UTR homozygous and XPD codon 156 variants, for Caucasians, a marginally increased association and a significantly inverse association were observed, respectively. XPC is responsible for the detection of the DNA damage lesion, while XPA and XPD, along with other proteins are responsible for the local unwinding of the DNA helix and the demarcation of the lesion. The formation of the open complex enables incorporation of endonucleases to excise the damaged site and further gap filling and sealing by DNA polymerase δ and ligase I, respectively(Altieri et al., 2008). It has been reported that the XPD Asp312Asn variant in smokers is significantly correlated with increased aromatic DNA adduct levels(Hou et al., 2002), while another study found decreased DNA damage-induced apoptosis in lymphoblastoid cells(Seker et al., 2001). Although the effect of the XPD codon 156 variant on this pathway is unknown, based on our findings, it would be interesting to determine whether this polymorphism provides a gain of function on the XPD protein activity. The functional implication of the XPA 5′-UTR (A23G) and XPC-PAT variants are unknown. The NER pathway is responsible for removing bulky adducts generated from cigarette smoke, among other environmental carcinogens(Altieri et al., 2008). Cigarette smoke is one of the primary risk factors for head and neck cancer, leading to chromosomal instability(Reshmi and Gollin, 2005). Thus, further investigation of these polymorphisms in the context of tobacco dose is needed.
It is also important to consider our findings in the context of Human Papillomavirus (HPV), an additional independent risk factor for head and neck cancer. HPV distribution in head and neck cancer seems to be subsite-specific and associated with improved outcome. It has been reported that HPV is mainly distributed in the oropharynx, with the highest distribution in the tonsils(Ragin and Taioli, 2007). Patients with HPV-positive tumors are less likely to have subsequent tumors, recurrences, metastases and new primary tumors, which contrast with what is observed in patients with HPV-negative tumors(Ragin et al., 2004), and distinct molecular profiles are observed between HPV-positive and HPV-negative tumors(Ragin et al., 2006). Therefore, we were interested in exploring whether there was an association between DNA repair gene polymorphisms by anatomic sub-site and HPV status. This analysis could not be performed due to the lack of the studies reporting on gene polymorphisms by anatomic sub-site or HPV status. Further studies are needed to address this interesting question.
A recent report describes an unequal burden of head and neck cancer in the US, in which the disparities were greater in African-American males, who showed a higher incidence and mortality rate for head and neck cancer compared to Caucasians(Goodwin et al., 2008). Also, African-Americans have been reported to have a younger age of onset compared to Caucasians(Gourin and Podolsky, 2006), and a greater likelihood to be smokers(Arbes et al., 1999). Therefore, it is important to have a better understanding of health disparities in minority populations by knowing whether genetic polymorphisms can identify high-risk individuals in the population who could be targeted with chemoprevention strategies. Surprisingly, in our meta-analysis, just one study by Shen et al.(Shen et al., 2001) reported genetic polymorphisms by race (non-Hispanic Whites, African-Americans, Hispanic-Americans, and native Chinese) without finding significant ethnic differences among the four groups. We have also observed a lack of publications concerning African-Americans or individuals of African descent while evaluating other gene polymorphisms(Ragin et al., 2010). Future assessments of genetic polymorphisms in the DNA repair pathway in minority populations are needed.
There are some limitations to this meta-analysis. First, the majority of the studies did not report gene polymorphism by sub-site and smoking status. Therefore, we were unable to perform stratification by those variables, which may explain some negative results. Second, heterogeneity due to ethnic ancestry (mostly Caucasians and Asians) and the small number of studies per ethnic group for the majority of the gene polymorphisms may have limited the ability of this meta-analysis to find true associations. Nevertheless, while performing a summary estimate, an average of each OR is weighted for the precision of each study, thus reducing the possibility of a biased estimate. Furthermore, despite performing stratification by race, when possible, to further assess heterogeneity, at times heterogeneity could not be resolved possibly due to the variation in the PCR methodology (PCR-RFLP, sequencing, melting curve analysis, 5′-exonuclease assay, MALDITOF-MS) employed in some of the studies in the same subgroups. In addition, subsite-specific analyses could only be performed for oral cavity cases, but these studies were only found in Asian populations. Therefore, the level of heterogeneity according to head and neck subsite in each racial group was not comparable. Third, despite conducting a meta-analysis with an almost overall absence of publication bias, it was only observed for the XRCC1 exon 9 codon 280 homozygous variant. We have not included any unpublished data, which may lead to false-positive results and/or bias. The source of publication bias for that particular variant remains unknown. Despite these limitations, the current meta-analysis has also some advantages. First the overall number of studies and genetic polymorphisms included were consistently large, compared to previously conducted meta-analysis, which significantly increased the statistical power of the analysis. Second, each of the studies included in the meta-analysis met our inclusion criteria. Third, we did not detect publication bias in the overall estimate that yielded statistically significant associations; which indicates that the pooled results should be unbiased.
CONCLUSION
In conclusion, our meta-analysis supports the idea that polymorphisms in DNA repair genes, XRCC1 codon 194 and XPD codon 156 (in Caucasians), XPD Asp312Asn, may be associated with oral, pharyngeal and laryngeal cancer risk while borderline associations have been suggested for other DNA repair genes. The current meta-analysis also reflects the need for larger studies including minority populations like African-Americans and Hispanics, who happen to experience higher incidence, and worse survival rates for head and neck cancer compared to Caucasians. These larger studies should also include analysis of not only environmental risk factors, such as HPV infection and exposure to cigarette smoke, but also the possible role of gene-gene interactions. Based on our results, plausible candidates like XRCC1 and XPD gene polymorphisms should be included in future large-scale epidemiological studies that eventually will provide a better understanding of the contributions of environmental risk factors and genetic polymorphisms to the development of head and neck cancer and racial disparities in incidence and survival.
Acknowledgments
The authors wish to thank Dr. Emanuela Taioli for support and critical review of this manuscript. This work was supported in part by Grant Number KL2 RR024154-03 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and the NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the NCRR or NIH. This work was also supported in part by grant number R13CA130596A and the State University of New York Dean’s Research Initiative Award to CR. This work was also supported in part by a Fulbright Scholarship, sponsored by the United States Department of State to REFO.
Funding: This work was supported in part by Grant Number KL2 RR024154-03 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and the NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the NCRR or NIH. This work was also supported in part by grant number R13CA130596A and the State University of New York Dean’s Research Initiative Award to CR. This work was also supported in part by a Fulbright Scholarship, sponsored by the United States Department of State to REFO.
Footnotes
Declaration of interest: The authors have no commercial association that might pose or create a conflict of interest with the information presented this manuscript.
References
- Abbasi R, Ramroth H, Becher H, Dietz A, Schmezer P, Popanda O. Laryngeal cancer risk associated with smoking and alcohol consumption is modified by genetic polymorphisms in ERCC5, ERCC6 and RAD23B but not by polymorphisms in five other nucleotide excision repair genes. Int J Cancer. 2009;125:1431–9. doi: 10.1002/ijc.24442. [DOI] [PubMed] [Google Scholar]
- Altieri F, Grillo C, Maceroni M, Chichiarelli S. DNA damage and repair: from molecular mechanisms to health implications. Antioxid Redox Signal. 2008;10:891–937. doi: 10.1089/ars.2007.1830. [DOI] [PubMed] [Google Scholar]
- An J, Liu Z, Hu Z, Li G, Wang LE, Sturgis EM, El-Naggar AK, Spitz MR, Wei Q. Potentially functional single nucleotide polymorphisms in the core nucleotide excision repair genes and risk of squamous cell carcinoma of the head and neck. Cancer Epidemiol Biomarkers Prev. 2007;16:1633–8. doi: 10.1158/1055-9965.EPI-07-0252. [DOI] [PubMed] [Google Scholar]
- Applebaum KM, Nelson HH, Zens MS, Stukel TA, Spencer SK, Karagas MR. Oral contraceptives: a risk factor for squamous cell carcinoma? J Invest Dermatol. 2009;129:2760–5. doi: 10.1038/jid.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbes SJ, Jr, Olshan AF, Caplan DJ, Schoenbach VJ, Slade GD, Symons MJ. Factors contributing to the poorer survival of black Americans diagnosed with oral cancer (United States) Cancer Causes Control. 1999;10:513–23. doi: 10.1023/a:1008911300100. [DOI] [PubMed] [Google Scholar]
- Bau DT, Tsai MH, Huang CY, Lee CC, Tseng HC, Lo YL, Tsai Y, Tsai FJ. Relationship between polymorphisms of nucleotide excision repair genes and oral cancer risk in Taiwan: evidence for modification of smoking habit. Chin J Physiol. 2007;50:294–300. [PubMed] [Google Scholar]
- Benhamou S, Tuimala J, Bouchardy C, Dayer P, Sarasin A, Hirvonen A. DNA repair gene XRCC2 and XRCC3 polymorphisms and susceptibility to cancers of the upper aerodigestive tract. Int J Cancer. 2004;112:901–4. doi: 10.1002/ijc.20474. [DOI] [PubMed] [Google Scholar]
- Bozec A, Formento P, Lassalle S, Lippens C, Hofman P, Milano G. Dual inhibition of EGFR and VEGFR pathways in combination with irradiation: antitumour supra-additive effects on human head and neck cancer xenografts. Br J Cancer. 2007;97:65–72. doi: 10.1038/sj.bjc.6603791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch S, Zhu B, Davis AG, Odom D, Siegfried JM, Grandis JR, Romkes M. Association of polymorphisms in the cyclin D1 and XPD genes and susceptibility to cancers of the upper aero-digestive tract. Mol Carcinog. 2005;42:222–8. doi: 10.1002/mc.20086. [DOI] [PubMed] [Google Scholar]
- Carles J, Monzo M, Amat M, Jansa S, Artells R, Navarro A, Foro P, Alameda F, Gayete A, Gel B, Miguel M, Albanell J, Fabregat X. Single-nucleotide polymorphisms in base excision repair, nucleotide excision repair, and double strand break genes as markers for response to radiotherapy in patients with Stage I to II head-and-neck cancer. Int J Radiat Oncol Biol Phys. 2006;66:1022–30. doi: 10.1016/j.ijrobp.2006.06.029. [DOI] [PubMed] [Google Scholar]
- Cheng L, Sturgis EM, Eicher SA, Spitz MR, Wei Q. Expression of nucleotide excision repair genes and the risk for squamous cell carcinoma of the head and neck. Cancer. 2002;94:393–7. doi: 10.1002/cncr.10231. [DOI] [PubMed] [Google Scholar]
- Csejtei A, Tibold A, Koltai K, Varga Z, Szanyi I, Gobel G, Prantner I, Steffler D, Feher G, De Blasio A, Ember I, Kiss I. Association between XRCC1 polymorphisms and head and neck cancer in a Hungarian population. Anticancer Res. 2009;29:4169–73. [PubMed] [Google Scholar]
- Demokan S, Demir D, Suoglu Y, Kiyak E, Akar U, Dalay N. Polymorphisms of the XRCC1 DNA repair gene in head and neck cancer. Pathol Oncol Res. 2005;11:22–5. doi: 10.1007/BF03032401. [DOI] [PubMed] [Google Scholar]
- Fountzilas G, Bamias A, Kalogera-Fountzila A, Karayannopoulou G, Bobos M, Athanassiou E, Kalogeras KT, Tolis C, Tsekeris P, Papakostas P, Vamvouka C, Zaramboukas T, Kosmidis P, Zamboglou N, Misailidou D. Induction chemotherapy with docetaxel and cisplatin followed by concomitant chemoradiotherapy in patients with inoperable non-nasopharyngeal carcinoma of the head and neck. Anticancer Res. 2009;29:529–38. [PubMed] [Google Scholar]
- Gajecka M, Rydzanicz M, Jaskula-Sztul R, Kujawski M, Szyfter W, Szyfter K. CYP1A1, CYP2D6, CYP2E1, NAT2, GSTM1 and GSTT1 polymorphisms or their combinations are associated with the increased risk of the laryngeal squamous cell carcinoma. Mutat Res. 2005a;574:112–23. doi: 10.1016/j.mrfmmm.2005.01.027. [DOI] [PubMed] [Google Scholar]
- Gajecka M, Rydzanicz M, Jaskula-Sztul R, Wierzbicka M, Szyfter W, Szyfter K. Reduced DNA repair capacity in laryngeal cancer subjects. A comparison of phenotypic and genotypic results. Adv Otorhinolaryngol. 2005b;62:25–37. doi: 10.1159/000082460. [DOI] [PubMed] [Google Scholar]
- Gal TJ, Huang WY, Chen C, Hayes RB, Schwartz SM. DNA repair gene polymorphisms and risk of second primary neoplasms and mortality in oral cancer patients. Laryngoscope. 2005;115:2221–31. doi: 10.1097/01.mlg.0000183736.96004.f7. [DOI] [PubMed] [Google Scholar]
- Geisler SA, Olshan AF, Cai J, Weissler M, Smith J, Bell D. Glutathione S-transferase polymorphisms and survival from head and neck cancer. Head Neck. 2005;27:232–42. doi: 10.1002/hed.20141. [DOI] [PubMed] [Google Scholar]
- Goodwin WJ, Thomas GR, Parker DF, Joseph D, Levis S, Franzmann E, Anello C, Hu JJ. Unequal burden of head and neck cancer in the United States. Head Neck. 2008;30:358–71. doi: 10.1002/hed.20710. [DOI] [PubMed] [Google Scholar]
- Gourin CG, Podolsky RH. Racial disparities in patients with head and neck squamous cell carcinoma. Laryngoscope. 2006;116:1093–106. doi: 10.1097/01.mlg.0000224939.61503.83. [DOI] [PubMed] [Google Scholar]
- Grau JJ, Caballero M, Campayo M, Jansa S, Vargas M, Alos L, Monzo M. Gene single nucleotide polymorphism accumulation improves survival in advanced head and neck cancer patients treated with weekly paclitaxel. Laryngoscope. 2009;119:1484–90. doi: 10.1002/lary.20254. [DOI] [PubMed] [Google Scholar]
- Hakem R. DNA-damage repair; the good, the bad, and the ugly. EMBO J. 2008;27:589–605. doi: 10.1038/emboj.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J, Hashibe M, Boffetta P, Gaborieau V, Moullan N, Chabrier A, Zaridze D, Shangina O, Szeszenia-Dabrowska N, Mates D, Janout V, Fabianova E, Holcatova I, Hung RJ, McKay J, Canzian F, Brennan P. The association of sequence variants in DNA repair and cell cycle genes with cancers of the upper aerodigestive tract. Carcinogenesis. 2007;28:665–71. doi: 10.1093/carcin/bgl160. [DOI] [PubMed] [Google Scholar]
- Handra-Luca A, Hernandez J, Mountzios G, Taranchon E, Lacau-St-Guily J, Soria JC, Fouret P. Excision repair cross complementation group 1 immunohistochemical expression predicts objective response and cancer-specific survival in patients treated by Cisplatin-based induction chemotherapy for locally advanced head and neck squamous cell carcinoma. Clin Cancer Res. 2007;13:3855–9. doi: 10.1158/1078-0432.CCR-07-0252. [DOI] [PubMed] [Google Scholar]
- Harbord RM, Egger M, Sterne JA. A modified test for small-study effects in meta-analyses of controlled trials with binary endpoints. Stat Med. 2006;25:3443–57. doi: 10.1002/sim.2380. [DOI] [PubMed] [Google Scholar]
- Harth V, Schafer M, Abel J, Maintz L, Neuhaus T, Besuden M, Primke R, Wilkesmann A, Thier R, Vetter H, Ko YD, Bruning T, Bolt HM, Ickstadt K. Head and neck squamous-cell cancer and its association with polymorphic enzymes of xenobiotic metabolism and repair. J Toxicol Environ Health A. 2008;71:887–97. doi: 10.1080/15287390801988160. [DOI] [PubMed] [Google Scholar]
- Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–74. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- Hou SM, Falt S, Angelini S, Yang K, Nyberg F, Lambert B, Hemminki K. The XPD variant alleles are associated with increased aromatic DNA adduct level and lung cancer risk. Carcinogenesis. 2002;23:599–603. doi: 10.1093/carcin/23.4.599. [DOI] [PubMed] [Google Scholar]
- Hsieh LL, Chien HT, Chen IH, Liao CT, Wang HM, Jung SM, Wang PF, Chang JT, Chen MC, Cheng AJ. The XRCC1 399Gln polymorphism and the frequency of p53 mutations in Taiwanese oral squamous cell carcinomas. Cancer Epidemiol Biomarkers Prev. 2003;12:439–43. [PubMed] [Google Scholar]
- Huang WY, Olshan AF, Schwartz SM, Berndt SI, Chen C, Llaca V, Chanock SJ, Fraumeni JF, Jr, Hayes RB. Selected genetic polymorphisms in MGMT, XRCC1, XPD, and XRCC3 and risk of head and neck cancer: a pooled analysis. Cancer Epidemiol Biomarkers Prev. 2005;14:1747–53. doi: 10.1158/1055-9965.EPI-05-0162. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Patsopoulos NA, Evangelou E. Uncertainty in heterogeneity estimates in meta-analyses. BMJ. 2007;335:914–6. doi: 10.1136/bmj.39343.408449.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- Jun HJ, Ahn MJ, Kim HS, Yi SY, Han J, Lee SK, Ahn YC, Jeong HS, Son YI, Baek JH, Park K. ERCC1 expression as a predictive marker of squamous cell carcinoma of the head and neck treated with cisplatin-based concurrent chemoradiation. Br J Cancer. 2008;99:167–72. doi: 10.1038/sj.bjc.6604464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kietthubthew S, Sriplung H, Au WW, Ishida T. Polymorphism in DNA repair genes and oral squamous cell carcinoma in Thailand. Int J Hyg Environ Health. 2006;209:21–9. doi: 10.1016/j.ijheh.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Koh Y, Kim TM, Jeon YK, Kwon TK, Hah JH, Lee SH, Kim DW, Wu HG, Rhee CS, Sung MW, Kim CW, Kim KH, Heo DS. Class III beta-tubulin, but not ERCC1, is a strong predictive and prognostic marker in locally advanced head and neck squamous cell carcinoma. Ann Oncol. 2009;20:1414–9. doi: 10.1093/annonc/mdp002. [DOI] [PubMed] [Google Scholar]
- Kornguth DG, Garden AS, Zheng Y, Dahlstrom KR, Wei Q, Sturgis EM. Gastrostomy in oropharyngeal cancer patients with ERCC4 (XPF) germline variants. Int J Radiat Oncol Biol Phys. 2005;62:665–71. doi: 10.1016/j.ijrobp.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Kowalski M, Przybylowska K, Rusin P, Olszewski J, Morawiec-Sztandera A, Bielecka-Kowalska A, Pietruszewska W, Mlynarski W, Janusz S, Majsterek I. Genetic polymorphisms in DNA base excision repair gene XRCC1 and the risk of squamous cell carcinoma of the head and neck. J Exp Clin Cancer Res. 2009;28:37. doi: 10.1186/1756-9966-28-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wang LE, Wei Q. DNA repair phenotype and cancer susceptibility--a mini review. Int J Cancer. 2009;124:999–1007. doi: 10.1002/ijc.24126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder M, Indra D, Roy PD, Datta S, Ray JG, Panda CK, Roy B. Variant haplotypes at XRCC1 and risk of oral leukoplakia in HPV non-infected samples. J Oral Pathol Med. 2009;38:174–80. doi: 10.1111/j.1600-0714.2008.00690.x. [DOI] [PubMed] [Google Scholar]
- Majumder M, Sikdar N, Ghosh S, Roy B. Polymorphisms at XPD and XRCC1 DNA repair loci and increased risk of oral leukoplakia and cancer among NAT2 slow acetylators. Int J Cancer. 2007;120:2148–56. doi: 10.1002/ijc.22547. [DOI] [PubMed] [Google Scholar]
- Majumder M, Sikdar N, Paul RR, Roy B. Increased risk of oral leukoplakia and cancer among mixed tobacco users carrying XRCC1 variant haplotypes and cancer among smokers carrying two risk genotypes: one on each of two loci, GSTM3 and XRCC1 (Codon 280) Cancer Epidemiol Biomarkers Prev. 2005;14:2106–12. doi: 10.1158/1055-9965.EPI-05-0108. [DOI] [PubMed] [Google Scholar]
- Manuguerra M, Saletta F, Karagas MR, Berwick M, Veglia F, Vineis P, Matullo G. XRCC3 and XPD/ERCC2 single nucleotide polymorphisms and the risk of cancer: a HuGE review. Am J Epidemiol. 2006;164:297–302. doi: 10.1093/aje/kwj189. [DOI] [PubMed] [Google Scholar]
- Michiels S, Danoy P, Dessen P, Bera A, Boulet T, Bouchardy C, Lathrop M, Sarasin A, Benhamou S. Polymorphism discovery in 62 DNA repair genes and haplotype associations with risks for lung and head and neck cancers. Carcinogenesis. 2007;28:1731–9. doi: 10.1093/carcin/bgm111. [DOI] [PubMed] [Google Scholar]
- Olshan AF, Watson MA, Weissler MC, Bell DA. XRCC1 polymorphisms and head and neck cancer. Cancer Lett. 2002;178:181–6. doi: 10.1016/s0304-3835(01)00822-9. [DOI] [PubMed] [Google Scholar]
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- Quintela-Fandino M, Hitt R, Medina PP, Gamarra S, Manso L, Cortes-Funes H, Sanchez-Cespedes M. DNA-repair gene polymorphisms predict favorable clinical outcome among patients with advanced squamous cell carcinoma of the head and neck treated with cisplatin-based induction chemotherapy. J Clin Oncol. 2006;24:4333–9. doi: 10.1200/JCO.2006.05.8768. [DOI] [PubMed] [Google Scholar]
- Ragin CC, Langevin S, Rubin S, Taioli E. Review of studies on metabolic genes and cancer in populations of African descent. Genet Med. 2010;12:12–8. doi: 10.1097/GIM.0b013e3181c8e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragin CC, Reshmi SC, Gollin SM. Mapping and analysis of HPV16 integration sites in a head and neck cancer cell line. Int J Cancer. 2004;110:701–9. doi: 10.1002/ijc.20193. [DOI] [PubMed] [Google Scholar]
- Ragin CC, Taioli E. Survival of squamous cell carcinoma of the head and neck in relation to human papillomavirus infection: review and meta-analysis. Int J Cancer. 2007;121:1813–20. doi: 10.1002/ijc.22851. [DOI] [PubMed] [Google Scholar]
- Ragin CC, Taioli E, Weissfeld JL, White JS, Rossie KM, Modugno F, Gollin SM. 11q13 amplification status and human papillomavirus in relation to p16 expression defines two distinct etiologies of head and neck tumours. Br J Cancer. 2006;95:1432–8. doi: 10.1038/sj.bjc.6603394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S, Ramadas K, Hariharan R, Rejnish Kumar R, Radhakrishna Pillai M. Single nucleotide polymorphisms of DNA repair genes XRCC1 and XPD and its molecular mapping in Indian oral cancer. Oral Oncol. 2006;42:350–62. doi: 10.1016/j.oraloncology.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Reshmi SC, Gollin SM. Chromosomal instability in oral cancer cells. J Dent Res. 2005;84:107–17. doi: 10.1177/154405910508400203. [DOI] [PubMed] [Google Scholar]
- Rusin P, Markiewicz L, Majsterek I. Genetic predeterminations of head and neck cancer. Postepy Hig Med Dosw (Online) 2008;62:490–501. [PubMed] [Google Scholar]
- Rydzanicz M, Wierzbicka M, Gajecka M, Szyfter W, Szyfter K. The impact of genetic factors on the incidence of multiple primary tumors (MPT) of the head and neck. Cancer Lett. 2005;224:263–78. doi: 10.1016/j.canlet.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Scully C, Bagan J. Oral squamous cell carcinoma: overview of current understanding of aetiopathogenesis and clinical implications. Oral Dis. 2009;15:388–99. doi: 10.1111/j.1601-0825.2009.01563.x. [DOI] [PubMed] [Google Scholar]
- Seker H, Butkiewicz D, Bowman ED, Rusin M, Hedayati M, Grossman L, Harris CC. Functional significance of XPD polymorphic variants: attenuated apoptosis in human lymphoblastoid cells with the XPD 312 Asp/Asp genotype. Cancer Res. 2001;61:7430–4. [PubMed] [Google Scholar]
- Shen H, Sturgis EM, Khan SG, Qiao Y, Shahlavi T, Eicher SA, Xu Y, Wang X, Strom SS, Spitz MR, Kraemer KH, Wei Q. An intronic poly (AT) polymorphism of the DNA repair gene XPC and risk of squamous cell carcinoma of the head and neck: a case-control study. Cancer Res. 2001;61:3321–5. [PubMed] [Google Scholar]
- Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58:604–8. [PubMed] [Google Scholar]
- Sturgis EM, Castillo EJ, Li L, Eicher SA, Strom SS, Spitz MR, Wei Q. XPD/ERCC2 EXON 8 Polymorphisms: rarity and lack of significance in risk of squamous cell carcinoma of the head and neck. Oral Oncol. 2002a;38:475–7. doi: 10.1016/s1368-8375(01)00106-3. [DOI] [PubMed] [Google Scholar]
- Sturgis EM, Castillo EJ, Li L, Zheng R, Eicher SA, Clayman GL, Strom SS, Spitz MR, Wei Q. Polymorphisms of DNA repair gene XRCC1 in squamous cell carcinoma of the head and neck. Carcinogenesis. 1999;20:2125–9. doi: 10.1093/carcin/20.11.2125. [DOI] [PubMed] [Google Scholar]
- Sturgis EM, Dahlstrom KR, Spitz MR, Wei Q. DNA repair gene ERCC1 and ERCC2/XPD polymorphisms and risk of squamous cell carcinoma of the head and neck. Arch Otolaryngol Head Neck Surg. 2002b;128:1084–8. doi: 10.1001/archotol.128.9.1084. [DOI] [PubMed] [Google Scholar]
- Sturgis EM, Zheng R, Li L, Castillo EJ, Eicher SA, Chen M, Strom SS, Spitz MR, Wei Q. XPD/ERCC2 polymorphisms and risk of head and neck cancer: a case-control analysis. Carcinogenesis. 2000;21:2219–23. doi: 10.1093/carcin/21.12.2219. [DOI] [PubMed] [Google Scholar]
- Sugimura T, Kumimoto H, Tohnai I, Fukui T, Matsuo K, Tsurusako S, Mitsudo K, Ueda M, Tajima K, Ishizaki K. Gene-environment interaction involved in oral carcinogenesis: molecular epidemiological study for metabolic and DNA repair gene polymorphisms. J Oral Pathol Med. 2006;35:11–8. doi: 10.1111/j.1600-0714.2005.00364.x. [DOI] [PubMed] [Google Scholar]
- Tae K, Lee HS, Park BJ, Park CW, Kim KR, Cho HY, Kim LH, Park BL, Shin HD. Association of DNA repair gene XRCC1 polymorphisms with head and neck cancer in Korean population. Int J Cancer. 2004;111:805–8. doi: 10.1002/ijc.20338. [DOI] [PubMed] [Google Scholar]
- Varzim G, Monteiro E, Silva RA, Fernandes J, Lopes C. CYP1A1 and XRCC1 gene polymorphisms in SCC of the larynx. Eur J Cancer Prev. 2003;12:495–9. doi: 10.1097/00008469-200312000-00008. [DOI] [PubMed] [Google Scholar]
- Vineis P, Manuguerra M, Kavvoura FK, Guarrera S, Allione A, Rosa F, Di Gregorio A, Polidoro S, Saletta F, Ioannidis JP, Matullo G. A field synopsis on low-penetrance variants in DNA repair genes and cancer susceptibility. J Natl Cancer Inst. 2009;101:24–36. doi: 10.1093/jnci/djn437. [DOI] [PubMed] [Google Scholar]
- Wang Y, Spitz MR, Lee JJ, Huang M, Lippman SM, Wu X. Nucleotide excision repair pathway genes and oral premalignant lesions. Clin Cancer Res. 2007;13:3753–8. doi: 10.1158/1078-0432.CCR-06-1911. [DOI] [PubMed] [Google Scholar]
- Wei Q, Wang LE, Sturgis EM, Mao L. Expression of nucleotide excision repair proteins in lymphocytes as a marker of susceptibility to squamous cell carcinomas of the head and neck. Cancer Epidemiol Biomarkers Prev. 2005;14:1961–6. doi: 10.1158/1055-9965.EPI-05-0101. [DOI] [PubMed] [Google Scholar]
- Wen SX, Zhang XM, Tang PZ, Zhao D, Guo YL, Tan W, Lin DX. Association between genetic polymorphism in DNA repair genes XRCC3 and risks of laryngeal and hypopharyngeal carcinomas. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2007;42:856–9. [PubMed] [Google Scholar]
- Werbrouck J, De Ruyck K, Duprez F, Van Eijkeren M, Rietzschel E, Bekaert S, Vral A, De Neve W, Thierens H. Single-nucleotide polymorphisms in DNA double-strand break repair genes: association with head and neck cancer and interaction with tobacco use and alcohol consumption. Mutat Res. 2008;656:74–81. doi: 10.1016/j.mrgentox.2008.07.013. [DOI] [PubMed] [Google Scholar]
- Werbrouck J, De Ruyck K, Duprez F, Veldeman L, Claes K, Van Eijkeren M, Boterberg T, Willems P, Vral A, De Neve W, Thierens H. Acute normal tissue reactions in head-and-neck cancer patients treated with IMRT: influence of dose and association with genetic polymorphisms in DNA DSB repair genes. Int J Radiat Oncol Biol Phys. 2009;73:1187–95. doi: 10.1016/j.ijrobp.2008.08.073. [DOI] [PubMed] [Google Scholar]
- Whitehead A, Whitehead J. A general parametric approach to the meta-analysis of randomized clinical trials. Stat Med. 1991;10:1665–77. doi: 10.1002/sim.4780101105. [DOI] [PubMed] [Google Scholar]
- Yang H, Lippman SM, Huang M, Jack Lee J, Wang W, Spitz MR, Wu X. Genetic polymorphisms in double-strand break DNA repair genes associated with risk of oral premalignant lesions. Eur J Cancer. 2008a;44:1603–11. doi: 10.1016/j.ejca.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Kang MJ, Choi Y, Kim CS, Lee SM, Park CW, Lee HS, Tae K. Associations between XPC expression, genotype, and the risk of head and neck cancer. Environ Mol Mutagen. 2005;45:374–9. doi: 10.1002/em.20097. [DOI] [PubMed] [Google Scholar]
- Yang M, Kim WH, Choi Y, Lee SH, Kim KR, Lee HS, Tae K. Effects of ERCC1 expression in peripheral blood on the risk of head and neck cancer. Eur J Cancer Prev. 2006;15:269–73. doi: 10.1097/01.cej.0000195709.79696.0c. [DOI] [PubMed] [Google Scholar]
- Yang Y, Tian H, Zhang ZJ. Association of the XRCC1 and hOGG1 polymorphisms with the risk of laryngeal carcinoma. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2008b;25:211–3. [PubMed] [Google Scholar]
- Yen CY, Liu SY, Chen CH, Tseng HF, Chuang LY, Yang CH, Lin YC, Wen CH, Chiang WF, Ho CH, Chen HC, Wang ST, Lin CW, Chang HW. Combinational polymorphisms of four DNA repair genes XRCC1, XRCC2, XRCC3, and XRCC4 and their association with oral cancer in Taiwan. J Oral Pathol Med. 2008;37:271–7. doi: 10.1111/j.1600-0714.2007.00608.x. [DOI] [PubMed] [Google Scholar]