Abstract
The DNA of all cells is continually under assault from a wide range of DNA-damaging agents. To counter this threat to their genetic integrity, cells possess systems, collectively known as the DNA-damage response (DDR), to detect DNA damage, signal its presence and mediate its repair. Here, I provide an overview of the DDR and then describe how work in my laboratory and elsewhere has identified some of the key protein players that mediate cellular responses to the most cytotoxic form of DNA damage: the DNA double-strand break (DSB). Next, I discuss some of my laboratory’s recent work, which has revealed that the way cells respond to DSBs is modulated in a cell-cycle dependent manner to ensure that the cell uses the DSB repair system that is most suited to its cell-cycle stage. Finally, I explain how our increasing knowledge of the DDR is suggesting new avenues for treating cancer and provide an example of a DDR inhibitory drug that is showing promise in clinical trials.
Keywords: DNA repair, DNA damage, cell cycle, cancer, cancer therapy
Introduction
Although DNA within our cells is a relatively inert chemical, it is constantly being damaged by a large number of agents. Indeed, it has been estimated that each of the approximately 1013 cells in a human body experiences many thousands of DNA lesions every day [1]. Much of this damage is generated as a consequence of normal metabolism, with hydrolytic reactions and damage caused by reactive oxygen species accounting for a large proportion of DNA lesions [2]. In addition, environmental agents such as ionising radiation (IR), ultra-violet light (UV) and chemicals in tobacco smoke make major contributions to DNA-damage production. DNA lesions can prevent genome replication and transcription, and if they are not repaired or are repaired incorrectly, they can produce mutations or large-scale genome aberrations that may lead to cell malfunction or cell death [3-5]. To counter such threats, cells have evolved a variety of DNA-damage detection and repair systems that, fortunately, are able to accurately and efficiently repair the vast majority of the damage that our cells experience. Nevertheless, some lesions escape accurate repair, thus leading to corruption of the genetic information. It is generally agreed that the most cytotoxic of all DNA lesions are DNA double-strand breaks (DSBs), which are the principal cytotoxic lesions produced by IR and radio-mimetic chemicals, and which are also generated when the DNA replication apparatus encounters other DNA lesions, such as DNA single-strand breaks. It is paramount that cells accurately repair DSBs because failure to do so can lead to mutations or genome rearrangements; and indeed, a single unrepaired DSB can be sufficient to kill a cell [6, 7].
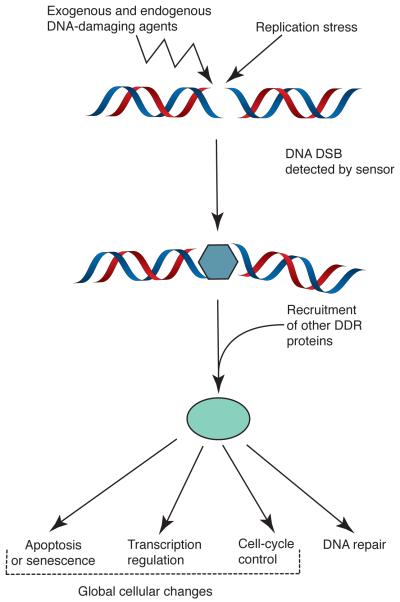
The way that cells react to DSBs is to trigger a complex and coordinated set of events that is often termed the DNA-damage response [8, 9]. As shown in Figure 1, DSBs are first detected by “sensor” proteins that then interact with further proteins to mediate DNA-repair processes. Furthermore, while repair is proceeding, the sensor/repair apparatus interacts with additional proteins to generate signalling mechanisms that, as I will explain below, rely heavily on the actions of DDR protein kinases. One of the key aspects of these DNA-damage-signalling events is the induction of so-called “checkpoint” mechanisms that slow down or stop cell-cycle progression while the damage persists, thereby helping to prevent the replication of damaged DNA or segregation of damaged chromosomes during mitosis. Additional aspects of the DDR-signalling mechanisms include alterations in chromatin structure and, particularly when the damage persists, the triggering of programmed cell death or long-term cell-cycle arrest known as senescence.
Figure 1.
Schematic representation of how cells respond to DSBs (see main text for details).
Clearly, the prime objective for cells experiencing DSBs is to mediate the repair of these highly toxic lesions. Studies in various experimental systems has led to the conclusion that, while there are many variations on the precise mechanisms of DSB repair, these events essentially boil-down into two largely distinct and complementary pathways: homologous recombination (HR) and non-homologous end-joining (NHEJ); [7, 10, 11]. In HR, after the DNA ends are first resected in the 5′ to 3′ direction by nucleases, the resulting 3′ single-stranded tails then invade the DNA double helix of a homologous undamaged partner molecule and are extended by the action of DNA polymerases that copy sequence information from the partner. Following branch-migration, the resulting DNA crossovers (Holliday junctions) can be resolved by various mechanisms to yield two intact, repaired DNA molecules. As a consequence, HR generally leads to accurate repair with no sequence loss or addition. By contrast, NHEJ does not need an undamaged partner DNA molecule and does not rely on extensive homologies between the two recombining ends. Instead, during NHEJ, sometimes after limited processing of the termini, the two DNA ends are ligated together. Consequently, NHEJ is often not error-free and small deletions of sequence are usually introduced. Initial work suggested that NHEJ is the predominant mechanism in higher eukaryotes but it is now established that HR also plays a crucial role in DSB repair in such organisms. Conversely, and as I will explain further below, although the NHEJ pathway was not identified through classical genetic approaches in unicellular eukaryotes such as the yeasts Schizosaccharomyces pombe and Saccharomyces cerevisiae, these organisms are now known to have an NHEJ apparatus that is evolutionarily conserved with that of higher eukaryotes.
The DNA-dependent protein kinase
Although I did not know it at the time, my entrance into the DDR field began in 1989, when I was working as a post-doctoral fellow on the control of human gene transcription in the laboratory of Robert Tjian in Berkeley, California. During this work, which I carried out with the assistance of an undergraduate, Judy MacDonald, we observed that transcription factor Sp1 present in human HeLa cell nuclear extracts was phosphorylated during in vitro transcription assays. Moreover, through defining the factor-requirements for phosphorylation, we learned that Sp1 phosphorylation only took place in the presence of DNA, and that phosphorylation ensued most efficiently when the DNA contained Sp1-binding sites. Furthermore, by partially purifying the kinase and carrying out biochemical experiments on it, I discovered that the kinase itself bound to DNA and was stimulated by DNA. Although the concept of a DNA-activated protein kinase was new to me, a search through the literature revealed that a kinase with such properties had been described previously in clam egg extracts by Carl Anderson and colleagues [12]. Upon contacting Carl and his collaborator Susan Lees-Miller at the Brookhaven National Laboratory, USA, I learned that they and the group of Tim Carter (St John’s University, New York, USA) had both independently partially purified this enzyme from human cells. Through exchanging kinase preparations, we were led to the conclusion that we were all working on the same enzyme; and later that year, three papers were published on this kinase [13-15], now known as the DNA-dependent protein kinase (DNA-PK).
DNA-PK activity requires the DSBs-binding protein Ku
In 1991, I set up my laboratory to study the control of transcription… and to try to discover the physiological functions of DNA-PK. In work I carried out together with my first graduate student, Tanya Gottlieb, we discovered by DNase I footprinting experiments that purified preparations of DNA-PK contained an activity that bound specifically to double-stranded DNA termini. Moreover, building on the observation that DNA-PK was activated strongly by linear but not by super-coiled plasmid DNA, we reached the conclusion that DNA-PK binds to and is activated by DNA DSBs [16]. At that time, one of the very few proteins reported to bind to DNA ends was Ku, a human auto-immune antigen whose function was not known and which is made up of two tightly-associated polypeptides of around 70 and 80 kDa (Ku70 and Ku80, respectively). Strikingly, we discovered that Ku was present in our DNA-PK preparations and that optimal DNA-PK activity required Ku together with a very large polypeptide, which we named DNA-PK catalytic subunit (DNA-PKcs, [16]). Upon discussing our data with a colleague, I learned that William (Bill) Dynan (now at the Medical College of Georgia, USA) had recently given a seminar where he had described experiments showing that the DNA-PK holoenzyme comprised of Ku and a separate catalytic subunit. After I had contacted Bill, we set out to publish our work co-ordinately; our work appeared in press in January 1993, with Bill’s paper appearing one month earlier [16, 17]. Collectively, these data gave rise to a model in which Ku first binds DNA ends and then recruits DNA-PKcs, thus activating DNA-PK catalytic activity towards substrate proteins, with most effective phosphorylation taking place when the protein target is also bound to the same DNA molecule as DNA-PK itself.
DNA-PKcs and Ku function in DNA DSB repair
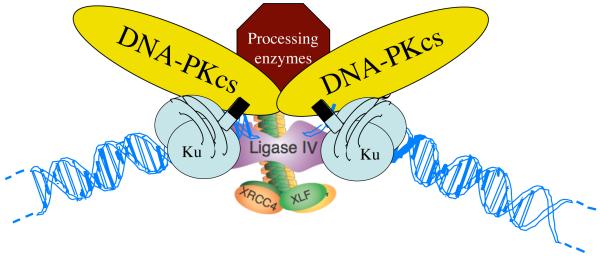
The above work suggested that DNA-PK might bind to DSBs in vivo, and raised the possibility that it might function in DNA repair and/or recombination. In early 1994, I was fortunate to enter into discussions with Penny Jeggo from the University of Sussex, UK, who for several years had been identifying and characterizing mutant mammalian cell lines that were hypersensitive to IR and were defective in DSB repair by NHEJ (for example, [18]). Furthermore, such cells had been shown to be deficient in V(D)J recombination, the site-specific recombination process that helps to generate the mature genes for immunoglobulin and T-cell receptor proteins in B- and T-lymphocytes, respectively (for example, [19]). Specifically, V(D)J recombination had been shown to occur via a cut-and-ligation process that involves DNA DSBs as intermediates, with the specific radiosensitive cell lines being defective in the ligation step. In light of these findings and our data on DNA-PKcs and Ku, Nick Finnie and Tanya Gottlieb in my laboratory collaborated with Penny and colleagues together with the group of Fred Alt (Harvard University, USA) to explore whether the IR-sensitivity and V(D)J recombination defects of such cells might be caused by mutations in DNA-PK components. To cut a long story short, our work and related work carried out by a series of other laboratories revealed that some of the mutant cells lacked functional Ku, while others had DNA-PKcs defects ([20-26]; for reviews, see [27-29]). In parallel, the labs of Susan Lees-Miller and Jane Allalunis Turner collaboratively discovered that a DNA-PKcs mutation caused the radiosensitivity of a cell line derived from a human cancer [30]. In addition to identifying the first mammalian NHEJ components, these studies led to the resolution of a long-standing question in the immunology field by our demonstration that a DNA-PKcs mutation causes the phenotype of the radiosensitive and immune-deficient severe-combined immune-deficient (Scid) mouse [31]. Over the ensuing years, my group and others have identified and characterized other components of the NHEJ apparatus, which include DNA ligase IV, XRCC4 [32-34] and XLF/Cernunnos [35, 36], thereby leading to a model for how this apparatus assembles at DSB sites to promote their repair (Figure 2).
Figure 2.
Model for assembly of the NHEJ apparatus at DSB sites (see main text for details).
Notably, while NHEJ factors were initially identified and characterized in higher eukaryotic systems, yeast genetic studies failed to identify NHEJ components. Although this was initially believed to reflect simpler eukaryotes relying solely on HR-based mechanisms for DSB repair, Simon Boulton and Soo-Hwang Teo in my laboratory noted that there were genes in the S. cerevisiae genome that could encode for Ku subunits or a counterpart of mammalian DNA Ligase IV. Consequently, through work carried out by Simon, Soo-Hwang and researchers in several other laboratories, we now know that, with the notable exception of DNA-PKcs, both S. cerevisiae and S. pombe carry out NHEJ by mechanisms closely related to those that operate in human cells (for review, see [37]).
DNA-PK, ATM and ATR respond to DNA damage by analogous mechanisms
By probing human cDNA libraries with synthetic oligonucleotides corresponding to peptide sequences obtained by Arie Admon (then in Tjian’s laboratory in Berkeley, USA) from purified DNA-PKcs, I isolated a partial cDNA for DNA-PKcs. Subsequent work by Kathy Hartley and David Gell in my laboratory isolated cDNA clones spanning the entire DNA-PKcs cDNA, thus revealing DNA-PKcs to be a ~470 kDa polypeptide [38]. Strikingly, this revealed the existence of a kinase domain in the carboxyl-terminal region of DNA-PKcs that was unlike those of most protein serine/threonine kinases. Instead, this domain was most similar in sequence to the kinase domains of a relatively small set of proteins, characterized members of which had been shown to mediate phosphorylation of inositol phospholipids (this family of enzymes is often referred to as the PI 3-kinase family because of the activity of one of its best known members: phosphatidyl inositol 3-kinase). Although this initially suggested to us that DNA-PK might function as a phospho-lipid kinase, collaborative work between Graeme Smith in my laboratory and Nullin Divecha (Babraham Institute, Cambridge, UK) established that DNA-PKcs was a protein kinase and did not have detectable activity against any phospholipids that we tested [38]. Notably, sequence comparisons indicated that the PI 3-kinase family of enzymes could be subdivided into two distinct subgroups: one containing mammalian PI 3-kinase and other lipid kinases; the other containing DNA-PKcs and a series of other large (>250 kDa) proteins… which suggested to us that these other kinases might share functional features with the DNA-PKcs/Ku system [39]. Indeed, we now know that this is the case: other proteins in the DNA-PKcs subgroup are protein kinases; and moreover, some of these – like DNA-PKcs itself – play important roles in the DDR (for reviews, see [29, 40, 41]).
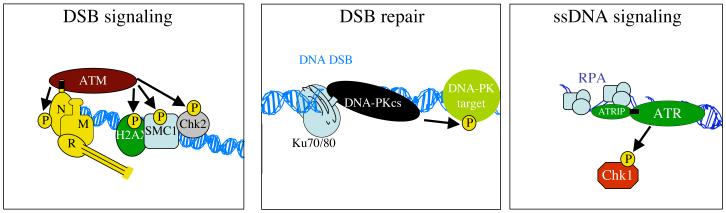
To date, the most extensively characterised of the DNA-PKcs-related proteins in human cells is ATM, which was identified in 1995 as the protein whose defect causes the human autosomal recessive syndrome ataxia telangiectasia (A-T) [42, 43]. The most critical features of this disease are an unsteady posture (ataxia) that progressively worsens over time due to degeneration of neuronal Purkinje cells in the cerebellum, heightened predisposition to cancer and radiosensitivity. At the cellular level, A-T is characterised by hypersensitivity to IR and other DNA DSB-generating agents, defects in both the G1/S and G2/M checkpoint responses, defective inhibition of DNA replication in response to DSB formation and an inability to effectively up-regulate p53 in response to IR due to defective DSB signalling [44]. Strikingly, we and other groups established that ATM is recruited to chromosomal DSBs but, rather than this being mediated by Ku, it is mediated by the MRE11-RAD50-NBS1 (MRN) protein complex [45-47]. Furthermore, in parallel with work from the labs of Tony Hunter and Paul Russell, Jacob Falck and Julia Coates in my group established that this recruitment involves (together with other important interactions) direct interactions between ATM and the carboxyl-terminal region of NBS1 [48, 49]. In addition, we found that this NBS1 region has homology to the carboxyl-terminal region of Ku80 that is needed for DNA-PKcs recruitment to DSB sites [49]; a region previously shown by David Gell in my laboratory and the group of Penny Jeggo to interact with DNA-PKcs and be required for DNA-PK activity in vivo [50, 51]. In further parallels with the Ku/DNA-PKcs system, the above work, together with other studies (for example, [52]) established that ATM recruitment to DSB sites promotes ATM activation via auto-phosphorylation [53] and helps ATM to effectively phosphorylate some of its protein targets. The phosphorylation of such targets, which include p53 and the protein kinase CHK2, then brings about activation of various cellular events, including cell-cycle checkpoint delays [54-56]. Thus, like DNA-PKcs, ATM appears to be a DNA-damage responsive kinase. However, while DNA-PK mainly controls DSB repair by NHEJ, ATM’s best-characterised functions are in promoting cell-cycle delay mechanisms. Nevertheless, recent collaborative work from the labs of Jeggo and Markus Lobrich (Darmstadt University, Germany) has revealed overlap between DNA-PK and ATM functions by showing that a subset of DSBs are repaired by mechanisms that rely on both DNA-PK and ATM [57].
Another DDR protein whose kinase domain is related in sequence to those of DNA-PKcs and ATM is ATR [58-60]; for review see [61]. Like DNA-PK and ATM, ATR is also recruited to sites of DNA damage, although in this case recruitment is to single-stranded DNA bound by replication protein A (RPA). This recruitment occurs by a mechanism that requires the ATR-interacting protein ATRIP [62, 63], and which involves the carboxyl-terminal region of ATRIP that has some homology to the Ku80 and NBS1 carboxyl-terminal regions [49]. It should be noted, however, that several other regions of ATRIP are important for ATR function [64]. Activated ATR mediates phosphorylation of various downstream target proteins, the best characterised of these being the protein kinase CHK1, which promotes, amongst other things, slowing of cell-cycle progression and activation of the HR pathway of DSB repair [65-67]. Because of its key roles in controlling such processes, which play particularly critical roles during S-phase, ATR is essential for cellular viability. Indeed, homozygous inactivation of ATR in mice results in early embryonic lethality associated with loss of cellular proliferative potential, chromosomal fragmentation and extensive apoptosis. While lack of ATR function is presumably incompatible with viability in humans, it is notable that hypomorphic ATR mutations are found in patients with Seckel syndrome, which is characterized by growth retardation, microcephaly, mental retardation and DNA-damage hypersensitivity [68]. Collectively, these studies have established that, while DNA-PKcs, ATM and ATR are activated by analogous mechanisms that employ interactions with distinct accessory proteins that bind sites of damaged DNA, their major downstream protein targets and biological functions are largely distinct (Figure 3).
Figure 3.
Models depicting how ATM, DNA-PKcs and ATR are recruited to sites of DNA damage by analogous mechanisms but possess different functions.
DSB repair and DNA-damage signaling are cell-cycle regulated
While the DNA structures activating ATM and ATR are distinct, both DSBs and single-stranded DNA (ssDNA) regions are generated when cells are treated with IR. In light of this, in 2004 Ali Jazayeri and Jacob Falck in my laboratory decided to study (with the assistance of Julia Coates) potential relationships between ATM- and ATR-mediated signalling after IR treatment. Consistent with previous studies, our initial analyses (carried out in collaboration with the groups of Jiri Lukas and Jiri Bartek in Copenhagen, Denmark) revealed that ATM activation ensues extremely rapidly after IR exposure as measured by ATM and MRN recruitment to DSB sites and the phosphorylation of ATM-target proteins such as NBS1 and CHK2. By contrast, we found that ATR/ATRIP recruitment to sites of DNA damage and phosphorylation of ATR targets such as CHK1 took place slightly more slowly, and that this correlated with a time delay between DSB generation and ssDNA production, presumably brought about by nuclease enzymes [69]. Moreover, Ali and Jacob made the striking discovery that efficient ssDNA generation and ATR activation required functional MRN and ATM kinase activity, thus revealing that, after IR or laser micro-irradiation of cells, prompt ATR activation requires prior activation of ATM-dependent signalling events. Similar observations were also made by the groups of Nick Lakin, Oskar Fernandez-Capetillo and David Cortez at around the same time [70-72].
Notably, our studies also revealed that, while ATM activation took place in all cells within an asynchronous population, effective ATR activation only occurred in a subset of such cells. Further analyses revealed that this is because, following laser micro-irradiation or IR exposure, ssDNA production and ATR activation take place effectively in S or G2 cells but not in G0 or G1 cells [69]. Subsequently, work by Alex Sartori and Julia Coates in my group established that ATR activation is promoted by the human CtIP protein, which achieves this function by working in conjunction with MRN to bring about resection of DNA ends in the 5′ to 3′ direction to generate ssDNA that binds RPA and then activates ATR [73]. Most recently, Pablo Huertas here established that cell-cycle regulation of DSB resection, and thereby ATR activation, is governed by cyclin-dependent kinases (CDK) phosphorylating a highly evolutionarily conserved motif present within CtIP and in its S. cerevisiae counterpart Sae2,[74] and Huertas et al. in press. Our current work is aimed at trying to understand precisely how this phosphorylation – and additional phosphorylations carried out by ATM and ATR (and their yeast counterparts) – brings about activation of CtIP/Sae2 and the MRN complex.
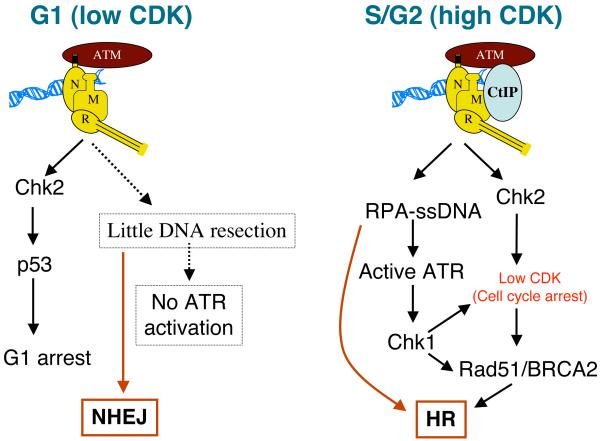
Why would a cell want to respond to DSBs differently in S and G2 than it does in G0 or G1? While we do not know the full answer to this question, it seems that a major rationale lies at the level of DSB-repair-pathway choice. As mentioned previously, there are two main ways to repair DSBs: NHEJ and HR. One crucial difference between the two pathways is that HR requires DSB resection, while NHEJ does not. Another key difference is that NHEJ is able to operate throughout all phases of the cell cycle, while HR is mainly restricted to S and G2, mainly because the partner DNA molecule used as a repair template is generally the sister chromatid – a structure that is not present in G0 or G1 cells. Collectively, the available data hence suggest a model for how and why DSBs are dealt with differently in different cell-cycle stages (Figure 4). In G0 or G1, DSBs are not extensively resected, allowing ATM activation and NHEJ to operate effectively, while ATR activation and DSB repair by HR are largely suppressed. By contrast, in S/G2 cells, DSBs are resected, thereby allowing ATR activation and HR. Notably, this resection and events based on the ensuing ssDNA production take place effectively only after ATM activation has occurred and also require the activities of MRN and CtIP. It is therefore tempting to speculate that responses to DSBs in S/G2 take place as a carefully orchestrated cascade, where the instigation of resection only occurs when cells have already instigated rapid ATM-mediated processes, such as slowing of cell cycle progression and, perhaps, up-regulation of latent DNA-repair-promoting factors. Once resection is progressing, this then provides the ssDNA substrate for HR and also leads to ATR activation that, amongst other things, might also act to promote HR. Consistent with this idea, CHK1 has been shown to facilitate HR, at least in part by mediating phosphorylation of the key HR protein, RAD51 [67].
Figure 4.
Model for how and why cells resect DSBs efficiently in the S and G2 phases of the cell cycle but not in G0 and G1 (see main text for details).
What might happen if the above control mechanisms went awry? To address this question, Pablo Huertas in my group recently mutated the CDK-target site in Sae2 that mediates cell-cycle control of resection in S. cerevisiae. Critically, he found that mutating the site to a non-phosphorylatable alanine residue prevented effective resection in S and G2, thus curtailing HR and causing hyper-sensitivity towards DSB-generating agents in S and G2 cells but not in G1 cells [74]. By contrast, mutating the site to a glutamic acid residue to mimic constitutive phosphorylation allowed HR but impaired the accuracy (and to a lesser degree efficiency) of NHEJ, presumably because of resection occurring in G1. In accord with this idea, Pablo found that the glutamic-acid substitution caused hypersensitivity towards DSB-generating agents in G1 cells but not in S/G2 cells [74]. Most recently, Pablo has observed similar effects when he mutated the analogous CDK-target site in human CtIP. Moreover, this work showed that deregulating HR by mimicking constitutive CtIP phosphorylation leads to high levels of gross-chromosomal rearrangements in irradiated cells, which at least in part appear to result from G1 cells engaging in aberrant HR events with ectopic loci (Huertas et al. in press). Collectively, the available data therefore indicate that, while there may be more than one way to repair a DSB, these pathways do not really act in a redundant manner with one another. Instead, the HR and NHEJ pathways are complementary, with each being suited to mediate repair under different circumstances. It is tempting to speculate that there may be many other instances where multiple pathways – initially seemingly carrying out the same DDR function – in fact primarily operate under different circumstances. As we will see below, such relationships between DDR pathways have a significant bearing on the use of DNA-damaging agents and DDR inhibitors in the treatment of cancer.
Therapeutic potential of DDR inhibition
It has long been clear that the field of DDR research is of huge relevance to cancer. First, aside from surgery, DNA-damaging agents – in the form of radiotherapy and many chemotherapies – are numerically by far the most effective and broadly used treatments for cancer. Second, the effectiveness of such treatments and the side-effects caused by them reflect the inability of cancer cells and normal cells, respectively, to repair therapy-induced DNA lesions. Conversely, when such treatments fail and cancer recurs, this is generally due to a fraction of the cancer cells having repaired the therapy-induced DNA lesions. Third, the majority of cancer-causing environmental agents known appear to exert their carcinogenic effects primarily through generating DNA damage. Fourth, although heightened predisposition to cancer in certain individuals can arise from various genetic causes, in many cases this results from inherited defects in the detection, signalling and/or repair of DNA damage, particularly DSBs [6, 75].
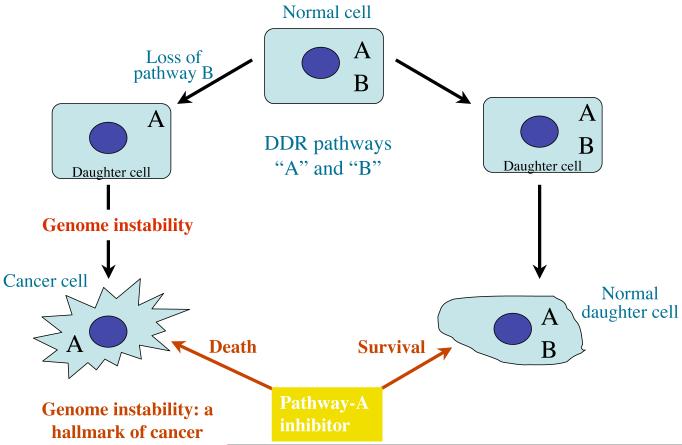
While the above issues have been apparent for many years, in 1994-1995 various things came to my attention that caused me to conclude that DDR-inhibitory drugs could have great potential in cancer therapy. First, our work on DNA-PK, ATM, ATR and PARP (which will be explained in more detail below) showed that it might be possible to develop highly specific inhibitors of these enzymes. For instance, our realization that DNA-PK was related to PI 3-kinase lead us to test various small-molecule drug-like compounds that were initially developed as potential PI 3-kinase inhibitors. Thus, Graeme Smith in my group established that the “PI 3-kinase inhibitors” wortmannin and LY294002 were actually also very effective against DNA-PK [38]. Subsequent work by us and others (for example, [76-78]) established that compounds like these also worked against ATM and ATR, and were able to sensitize cells to the cytotoxic effects of various DNA-damaging agents. Another factor that suggested to me that there was anti-cancer potential for DDR-inhibitory drugs was the insight that, in many instances, cancer cells should be more sensitive to DDR-inhibitors than normal cells. For example, cancer cells tend to replicate more frequently than most normal cells, meaning that they will generally have less time to repair damage before executing a key cell-cycle transition such as entry into S-phase or mitosis. Furthermore, unlike non-dividing cells, cancer cells traverse S-phase, when many forms of DNA damage are converted into highly toxic DSBs at the replication fork. In addition, cancer cells are invariably deficient in one or more aspects of the DDR. A prime example of this is provided by p53 deficiency, which occurs in a large proportion of cancers and which leads to defective DNA-damage induced cell-cycle delays/arrests [79]. Although there has been much debate about why and how cancer cells acquire such DDR defects, a popular current model is that these defects are selected for to allow the cancer cells (and their progenitors) to continue proliferating in the presence of chronic DNA-damage and/or replicative stress [80, 81]. Finally – and perhaps most crucially – there has been growing evidence over the past dozen-or-so years that DDR deficiencies tend to make cancer cells particularly highly reliant on the DDR pathways that they still retain. As a consequence, one could envision that certain DDR-inhibitory drugs might be much more cytotoxic towards certain cancer cells than normal cells (Figure 5).
Figure 5.
Model depicting how certain components of the DDR are often lost during the evolution of cancer. In the hypothetical situation shown, there are two DDR pathways “A” and “B” that are both able to promote cell survival in response to a particular form of DNA damage. While normal cells possess both pathways, pathway “B” is shown to be lost during tumourigenesis. Consequently, administration of a drug targeting pathway “A” would be expected to be much more cytotoxic to cancer cells than the normal cells, thus providing a potential therapeutic window for the use of this drug in treating a cancer patient.
Establishing KuDOS Pharmaceuticals Ltd
In 1996, with the above ideas in mind, I approached Richard Jennings of the University of Cambridge and my main sponsors, Cancer Research UK (then Cancer Research Campaign) with the concept of establishing a biotech company focused on making DDR inhibitors for use in cancer therapy. After much debate with Sue Foden and Guy Heathers at CRT (Cancer Research UK Technology), we decided that this was worth a try and, in December 1997, KuDOS Pharmaceuticals Ltd was founded (the name was suggested by my wife, Teresa Clarke because “it is all based on what Ku does”). With advice from Sue, Guy and a consultant, Nick Holladay, I wrote the KuDOS Business Plan… and after much effort, in May 1999 we obtained venture-capital funding, allowing the company to establish its operations on the Cambridge Milton Road Science Park. By employing the complementary expertise of myself (Chief Scientific Officer), Barrie Ward (Chief Executive Officer), Graeme Smith (Head of Drug Evaluation) and Niall Martin (Head of Drug Discovery), KuDOS established high throughput small-molecule-library-based screening and drug evaluation capabilities. As described below, these capabilities and their subsequent broadening into areas including medicinal and combinatorial chemistry allowed KuDOS to develop highly potent and selective inhibitors of key DDR enzymes… that are currently in various stages of pre-clinical and clinical development. Still based in Cambridge, KuDOS now benefits from the extensive, complementary resources and capabilities of AstraZeneca, which acquired KuDOS in late 2005.
Development of DNA-PK, ATM and PARP inhibitors
One initial goal for KuDOS was to show that it was possible to develop specific inhibitors of DNA-PK, ATM and/or ATR. A major obstacle to doing this was the fact that these enzymes are very large polypeptides that work in conjunction with yet other proteins (several of which were actually not known of at the time of KuDOS’ inception). Consequently, we and various others found it impossible to generate active, recombinant versions of these enzymes; and moreover, when expressed alone, the kinase domains of these enzymes were unstable, insoluble or inactive. To circumvent these issues, KuDOS used expertise that Graeme Smith had acquired when he was in my academic laboratory to develop efficient ways to purify the enzymes from human cell extracts; and together with the establishment of highly sensitive kinase assays by Graeme and Niall Martin, this allowed effective – albeit very laborious –compound screening. As a result of this work and many years of iterative medicinal chemistry, combinatorial chemistry and compound evaluation, KuDOS was able to develop highly potent and very selective inhibitors of DNA-PK and ATM. Notably, in addition to such compounds and their derivatives having promise as anti-cancer agents, they have proved to be very useful reagents with which to characterise ATM- and DNA-PK-dependent processes (for example, [57, 82-86]).
Another major focus for KuDOS over the past decade has been to develop inhibitors of Poly (ADP-ribose) polymerase (PARP-1). The production of protein-linked poly ADP-ribose chains is a long-established aspect of the DDR, this being carried out by PARP-1 and, to a lesser extent, PARP-2 (for reviews, see [87, 88]). Although PARP-1 and PARP-2 have been implicated in controlling other nuclear events such as transcription, their best-characterised functions are in promoting DNA base-excision repair (BER) and the repair of DNA single-strand breaks (SSBs). This is believed to be mediated by PARP1/2 binding to SSBs or DNA nicks that are produced by agents such as reactive oxygen species and which also occur as intermediates in the BER process [89]. Such DNA SSB-binding then triggers PARP1/2 enzymatic activity, leading to the production of large, branched chains of poly (ADP-ribose) on PARP-1 itself and on various other proteins; and this in turn appears to help the recruitment of other proteins that facilitate BER and SSB repair. Consequently, lack of PARP-1 or PARP-2 causes persistence of endogenously-arising SSBs, and hyper-sensitises cells to IR and drugs such as DNA-alkylating agents [90, 91]. Because of these issues, KuDOS and others have carried out biochemical screens of chemical libraries for PARP-1 inhibitors, and have then evolved the ensuing “hits” through medicinal and combinatorial chemistry to produce highly potent and selective compounds. As anticipated, such inhibitors have been found to sensitise cancer cells to IR and towards DNA-damaging agents that include alkylating drugs, topoisomerase I inhibitors and platinum compounds (for example, [92-95]). On the basis of these findings, PARP-1/2 inhibitors developed by several companies have entered into clinical trials (for reviews, see [86, 96, 97]).
PARP inhibitors are selectively cytotoxic to BRCA1/2-deficient cells
As described in preceding sections, as they lack particular DDR pathways, cancer cells might be more susceptible than normal cells to the inhibition of certain other DDR pathways (Figure 5). My colleagues and I at KuDOS were, therefore, very interested in trying to identify situations where such “therapeutic windows” might exist for the various KuDOS DDR inhibitors. Although there are many examples of DDR loss in cancer cells, one that captured our specific attention was the loss of BRCA1 or BRCA2 function in hereditary breast and ovarian cancer. In these conditions, a woman inherits one inactive copy of BRCA1/2, meaning that she has an extremely high risk of developing breast or ovarian malignancies. Notably, while normal cells in such a cancer patient possess one wild-type copy of the BRCA1/2 gene, rendering them BRCA1/2 proficient, the tumour cells turn out to have invariably lost the function of the other BRCA1/2 allele, making them dysfunctional for BRCA1/2-dependent events. Crucially, in 1999-2001, work from various laboratories – in particular the group of Maria Jasin (Sloan Kettering Cancer Centre, New York) – revealed that BRCA1/2 defects lead to markedly impaired DSB repair by HR (for example, [98, 99]). In light of these findings, in mid 2002, I approached Alan Ashworth (Breakthrough Breast Cancer Centre, London, UK), who is an expert in studying BRAC1/2 functions, with a proposal: that we collaboratively assess the impact of DNA-PK, ATM and PARP-1 inhibitors in cells proficient or deficient in BRCA1 or BRCA2. It turned out that Alan was very receptive to this proposal, as he had been thinking along similar lines himself.
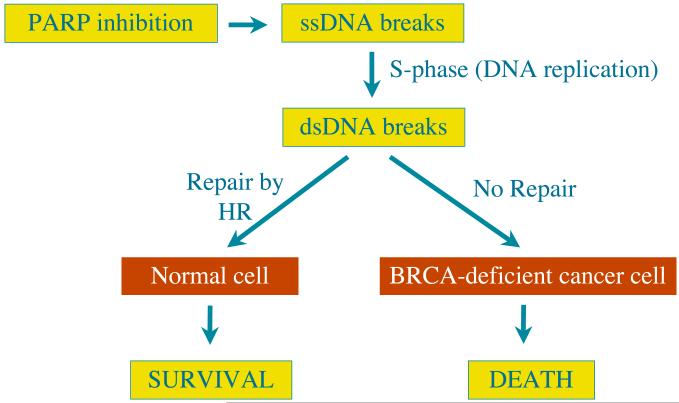
To cut a long story short, work carried out collaboratively by Alan’s group in London and the groups of Niall Martin, Mark O’Connor and Graeme Smith at KuDOS revealed that BRCA−/− cells are strikingly hyper-sensitive to PARP inhibition, being killed at concentrations of KuDOS PARP inhibitors that are several hundred-fold lower than are needed to kill BRCA+/+ or BRCA+/− cells [100]. In parallel, work carried out collaboratively by the groups of Thomas Helleday (University of Sheffield, UK and University of Stockholm, Sweden) and Nicola Curtin (University of Newcastle-Upon-Tyne, UK) reached the same conclusions [101]. Taken together with subsequent studies revealing that other HR-deficient cells are hyper-sensitive to PARP inhibition [102], we were thus lead to the model that PARP inhibition slows the repair of spontaneously-arising SSBs, which are then converted into DSBs during S-phase by replication (Figure 6). Notably, such DSBs are not very toxic to normal cells, because – as we have seen in previous sections – they can efficiently and accurately repair these lesions by HR-dependent processes. By contrast, this type of repair is not effective in BRCA1/2-defective cells, leading to cell death by apoptosis or mitotic catastrophe.
Figure. 6.
Model depicting how PARP inhibition is well tolerated by normal cells that are proficient in HR but is highly cytotoxic to HR-deficient cells, such as those defective in BRCA1 or BRCA2 (see main text for details).
PARP inhibitors in the treatment of hereditary breast and ovarian cancers
On the basis of the above findings and subsequent data generated through the use of mouse cancer models (for example [103-105]), the orally-active KuDOS PARP inhibitory compound, KU-0059436 (now termed AZD2281), was selected for a Phase I clinical trial in human patients. Initial evaluation, which was on advanced patients with various types of progressive solid tumours that had proved resistant to previous therapies, revealed that AZD2281 is generally well tolerated, with reversible and generally relatively mild toxicities being observed only at doses sufficient to largely ablate PARP enzymatic activity. As a result of these observations and indications of anti-tumour activity in several patients, including some of whom possessed BRCA1/2 mutations [106], the Phase I trial was expanded by recruiting a further cohort of ovarian cancer patients with BRCA1/2 mutations. Strikingly, in addition to there being responses in the other patient sub-groups, a substantial proportion of the BRCA-deficient ovarian cancer patients displayed stable disease or tumour regression, with several patients remaining being treated even after formal closure of the trial [107]. Encouraged by such findings, as of January 2009, AZD2281 is being evaluated in three Phase II trials as a stand-alone agent against BRCA-defective breast or ovarian cancers. It is also being assessed in a further seven Phase I trials against various tumour types in combination with DNA-damaging agents (http://www.cancer.gov/search/ResultsClinicalTrialsAdvanced.aspx?protocolsearchid=5678174).
Conclusions and future prospects
I have described how work in my laboratory and elsewhere has led to the identification and characterisation of proteins that function in the detection, signalling and repair of DNA DSBs. Although this and other work has taken us to a fairly sophisticated understanding of the DDR, much remains to be learned. In my view, key issues for future studies will be to determine exactly how DDR proteins work at the molecular level, how they interact with one another and how their activities are controlled by such interactions and by post-translational modifications. As has been the case so far in the DDR field, biochemical studies will surely play major roles in shaping such developments. Another prime aspect of future studies will be to try to gain a more “holistic” picture of the DDR, which we now know more resembles a complex meshwork of activities rather than operating as a series of distinct, largely linear pathways. Such understandings will presumably arise through integration of data provided by biochemical, molecular and genetic studies, as well as information generated through more global “omic” approaches (for example, [108, 109]). Finally, I speculate that we are currently just seeing the “tip of the iceberg” in regards to the therapeutic potential of DDR modulating drugs… and that the next five-to-ten years will witness many more DDR-regulatory drugs progressing through clinical trials. Hopefully, these and related developments will translate into significant benefits for cancer patients and their families.
Acknowledgements
The work that I have described from my laboratory was carried out by a group of dedicated, enormously talented and highly interactive group of individuals that I have been fortunate to recruit over the past seventeen years. I apologise to colleagues in my lab and elsewhere whose work I did not mention because of constraints on the length of this manuscript (and possibly deficiencies in my memory). I also express thanks to Kate Dry for assistance with this manuscript and for reworking the figures. Thanks also to my many other colleagues for their advice and encouragement over the years, particularly Peter Rigby, Ron Laskey, John Gurdon, Robert Tjian, Penny Jeggo, Tomas Lindahl and Barrie Ward. Finally, I thank Cancer Research UK for their strong long-term support of my laboratory, and also the other agencies who have funded my laboratory and members within it over the years, including the Association for International Cancer Research, the AT Children’s Project, the AT Medical Research Trust, the Biotechnology and Biological Sciences Research Council, the European Molecular Biology Organization, the European Union, the Medical Research Council and the Wellcome Trust.
References
- 1.Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol. 2000;65:127–133. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. ASM Press; Washington, DC: 2006. [Google Scholar]
- 3.Mills KD, Ferguson DO, Alt FW. The role of DNA breaks in genomic instability and tumorigenesis. Immunol Rev. 2003;194:77–95. doi: 10.1034/j.1600-065x.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 4.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 5.Lobrich M, Jeggo PA. The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat Rev Cancer. 2007;7:861–869. doi: 10.1038/nrc2248. [DOI] [PubMed] [Google Scholar]
- 6.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 7.Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu Rev Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 8.Zhou B-BS, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 9.Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297:547–551. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- 10.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 11.San Filippo J, Sung P, Klein H. Mechanism of Eukaryotic Homologous Recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 12.Walker AI, Hunt T, Jackson RJ, Anderson CW. Double-stranded DNA induces the phosphorylation of several proteins including the 90 000 mol. wt. heat-shock protein in animal cell extracts. EMBO J. 1985;4:139–145. doi: 10.1002/j.1460-2075.1985.tb02328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson S, Gottlieb T, Hartley K. Phosphorylation of transcription factor SP1 by the DNA-dependent protein kinase. Advances in Second Messenger and Phosphoprotein Research. 1993;28:279–286. [PubMed] [Google Scholar]
- 14.Lees-Miller SP, Chen Y-R, Anderson CW. Human cells contain a DNA-activated protein kinase that phosphorylates simian virus 40 T antigen, mouse p53, and the human Ku autoantigen. Mol. Cell. Biol. 1990;10:6472–6481. doi: 10.1128/mcb.10.12.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter T, Vancurova I, Sun I, Lou W, Deleon S. A DNA-activated protein kinase from HeLa cell nuclei. Mol. Cell. Biol. 1990;10:6460–6471. doi: 10.1128/mcb.10.12.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 17.Dvir A, Peterson SR, Knuth MW, Lu H, Dynan WS. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc. Natl. Acad. Sci. USA. 1992;89:11920–11924. doi: 10.1073/pnas.89.24.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeggo PA, Caldecott K, Pidsley S, Banks GR. Sensitivity of Chinese hamster ovary mutants defective in DNA double strand break repair to topoisomerase II inhibitors. Cancer Res. 1989;49:7057–7063. [PubMed] [Google Scholar]
- 19.Taccioli GE, Rathbun G, Oltz E, Stamato T, Jeggo PA, Alt FW. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 20.Taccioli GE, Gottlieb TM, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann AR, Alt FW, Jackson SP, Jeggo PA. Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 21.Finnie NJ, Gottlieb TM, Blunt T, Jeggo PA, Jackson SP. DNA-dependent protein kinase activity is absent in xrs-6 cells: implications for site-specific recombination and DNA double-strand break repair. Proc. Natl. Acad. Sci. USA. 1995;92:320–324. doi: 10.1073/pnas.92.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Getts RC, Stamato TD. Absence of a Ku-like DNA end-binding activity in the xrs double-strand DNA repair-deficient mutant. J. Biol. Chem. 1994;269:15981–15984. [PubMed] [Google Scholar]
- 23.Rathmell WK, Chu G. Involvement of the Ku autoantigen in the cellular response to DNA double-strand breaks. Proc. Natl. Acad. Sci. USA. 1994;91:7623–7627. doi: 10.1073/pnas.91.16.7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boubnov NV, Hall KT, Wills Z, Lee SE, He DM, Benjamin DM, Pulaski CR, Band H, Reeves W, Hendrickson EA, Weaver DT. Complementation of the ionizing radiation-sensitivity, DNA end-binding, and V(D)J recombination defects of double-strand break repair mutants by the p86 Ku autoantigen. Proc. Natl. Acad. Sci. USA. 1995;92:890–894. doi: 10.1073/pnas.92.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peterson SR, Kurimasa A, Oshimura M, Dynan WS, Bradbury EM, Chen DJ. Loss of the catalytic subunit of the DNA-dependent protein kinase in DNA double-strand break repair mutant mammalian cells. Proc. Natl. Acad. Sci. USA. 1995;92:3171–3174. doi: 10.1073/pnas.92.8.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smider V, Rathmell WK, Lieber MR, Chu G. Restoration of X-ray resistance and V(D)J recombination in mutant cells by Ku cDNA. Science. 1994;266:288–291. doi: 10.1126/science.7939667. [DOI] [PubMed] [Google Scholar]
- 27.Jeggo PA, Taccioli GE, Jackson SP. Menage à trois: double-strand break repair, V(D)J recombination and DNA-PK. BioEssays. 1995;17:949–957. doi: 10.1002/bies.950171108. [DOI] [PubMed] [Google Scholar]
- 28.Lieber MR, Grawunder U, Wu X, Yaneva M. Tying loose ends: roles of Ku and DNA-dependent protein kinase in the repair of double-strand breaks. Curr. Opin. Genet. Dev. 1997;7:99–104. doi: 10.1016/s0959-437x(97)80116-5. [DOI] [PubMed] [Google Scholar]
- 29.Smith GCM, Jackson SP. The DNA-dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 30.Lees-Miller SP, Godbout R, Chan DW, Weinfeld M, Day RS, Barron GM, Allalunis-Turner J. Absence of p350 subunit of DNA activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 31.Blunt T, Finnie NJ, Taccioli GE, Smith GCM, Demengeot J, Gottlieb TM, Mizuta R, Varghese AJ, Alt FW, Jeggo PA, Jackson SP. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine SCID mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 32.Li Z, Otevrel T, Gao Y, Cheng H-L, Seed B, Stamato TD, Taccioli GE, Alt FW. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell. 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- 33.Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 34.Critchlow SE, Bowater RP, Jackson SP. Mammalian DNA double-strand break repair protein XRCC4 interacts with DNA ligase IV. Curr. Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 35.Ahnesorg P, Smith P, Jackson SP. XLF Interacts with the XRCC4-DNA Ligase IV Complex to Promote DNA Nonhomologous End-Joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 36.Buck D, Malivert L, de Chasseval R, Barraud A, Fondaneche MC, Sanal O, Plebani A, Stephan JL, Hufnagel M, le Deist F, Fischer A, Durandy A, de Villartay JP, Revy P. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 37.Critchlow SE, Jackson SP. DNA end joining: from yeast to man. Trends Biochem. Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- 38.Hartley KO, Gell D, Smith GCM, Zhang H, Divecha N, Connelly MA, Admon A, Lees-Miller SP, Anderson CW, Jackson SP. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 39.Durocher D, Jackson SP. DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr. Opin. Cell Biol. 2001;13:225–231. doi: 10.1016/s0955-0674(00)00201-5. [DOI] [PubMed] [Google Scholar]
- 40.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 41.Keith CT, Schreiber SL. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- 42.Savitsky K, Barshira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, Ashkenazi M, Pecker I, Frydman M, Harnik R, Patanjali SR, Simmons A, Clines GA, Sartiel A, Gatti RA, Chessa L, Sanal O, Lavin MF, Jaspers NGJ, Malcolm A, Taylor R, Arlett CF, Miki T, Weissman SM, Lovett M, Collins FS, Shiloh Y. A single ataxia telangiectasia gene with a product similar to PI3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 43.Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- 44.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nature Reviews Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 45.Lee JH, Paull TT. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- 46.Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. Embo J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee JH, Paull TT. ATM Activation by DNA Double-Strand Breaks Through the Mre11-Rad50-Nbs1 Complex. Science. 2005 doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 48.You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 50.Gell D, Jackson SP. Mapping of protein–protein interactions within the DNA-dependent protein kinase complex. Nucleic Acids Res. 1999;27:3494–3502. doi: 10.1093/nar/27.17.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singleton BK, Taccioli GE, Rottinghaus S, Jeggo PA. Interaction of the C-terminus of Ku80 with the DNA-dependent protein kinase catalytic subunit. Mol. Cell. Biol. 1999;19:3267–3277. doi: 10.1128/mcb.19.5.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dupre A, Boyer-Chatenet L, Gautier J. Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nat Struct Mol Biol. 2006 doi: 10.1038/nsmb1090. [DOI] [PubMed] [Google Scholar]
- 53.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 54.Banin S, Moyal L, Shieh S-Y, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 55.Canman CE, Lim D-S, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 56.Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc. Natl. Acad. Sci. USA. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riballo E, Kuhne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, Parker AR, Jackson SP, Gennery A, Jeggo PA, Lobrich M. A Pathway of Double-Strand Break Rejoining Dependent upon ATM, Artemis, and Proteins Locating to gamma-H2AX Foci. Mol Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 58.Cimprich KA, Shin TB, Keith CT, Schreiber SL. cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc. Natl. Acad. Sci. USA. 1996;93:2850–2855. doi: 10.1073/pnas.93.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cliby WA, Roberts CJ, Cimprich KA, Stringer CM, Lamb JR, Schreiber SL, Friend SH. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wright JA, Keegan KS, Herendeen DR, Bentley NJ, Carr AM, Hoekstra MF, Concannon P. Protein kinase mutants of human ATR increase sensitivity to UV and ionizing radiation and abrogate cell cycle checkpoint control. Proc. Natl. Acad. Sci. USA. 1998;95:7445–7450. doi: 10.1073/pnas.95.13.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008 doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cortez D, Guntuku S, Qin J, Elledge SJ. ATR and ATRIP: partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- 63.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 64.Ball HL, Ehrhardt MR, Mordes DA, Glick GG, Chazin WJ, Cortez D. Function of a Conserved Checkpoint Recruitment Domain in ATRIP Proteins. Mol Cell Biol. 2007 doi: 10.1128/MCB.02238-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo Z, Kumagai A, Wang SX, Dunphy WG. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Q, Guntuku S, Cui X-S, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, Donehower LA, Elledge SJ. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 67.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 68.O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 69.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2005 doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 70.Adams KE, Medhurst AL, Dart DA, Lakin ND. Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene. 2006 doi: 10.1038/sj.onc.1209426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cuadrado M, Martinez-Pastor B, Murga M, Toledo LI, Gutierrez-Martinez P, Lopez E, Fernandez-Capetillo O. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J Exp Med. 2006;203:297–303. doi: 10.1084/jem.20051923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem. 2006 doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–692. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 76.Izzard RA, Jackson SP, Smith GCM. Competitive and noncompetitive inhibition of the DNA-dependent protein kinase. Cancer Res. 1999;59:2581–2586. [PubMed] [Google Scholar]
- 77.Sarkaria JN, Tibbetts RS, Busby EC, Kennedy AP, Hill DE, Abraham RT. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res. 1998;58:4375–4382. [PubMed] [Google Scholar]
- 78.Boulton S, Kyle S, Yalçintepe L, Durkacz BW. Wortmannin Is a potent inhibitor of DNA double strand break but not single strand break repair in chinese hamster ovary cells. Carcinogenesis. 1996;17:2285–2290. doi: 10.1093/carcin/17.11.2285. [DOI] [PubMed] [Google Scholar]
- 79.Rodier F, Campisi J, Bhaumik D. Two faces of p53: aging and tumor suppression. Nucleic Acids Res. 2007;35:7475–7484. doi: 10.1093/nar/gkm744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 81.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, Maestro R, Pelicci PG, d’Adda di Fagagna F. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 82.Veuger SJ, Curtin NJ, Richardson CJ, Smith GC, Durkacz BW. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res. 2003;63:6008–6015. [PubMed] [Google Scholar]
- 83.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 84.Hollick JJ, Rigoreau LJ, Cano-Soumillac C, Cockcroft X, Curtin NJ, Frigerio M, Golding BT, Guiard S, Hardcastle IR, Hickson I, Hummersone MG, Menear KA, Martin NM, Matthews I, Newell DR, Ord R, Richardson CJ, Smith GC, Griffin RJ. Pyranone, Thiopyranone, and Pyridone Inhibitors of Phosphatidylinositol 3-Kinase Related Kinases. Structure-Activity Relationships for DNA-Dependent Protein Kinase Inhibition, and Identification of the First Potent and Selective Inhibitor of the Ataxia Telangiectasia Mutated Kinase. J Med Chem. 2007 doi: 10.1021/jm061121y. [DOI] [PubMed] [Google Scholar]
- 85.Zhao Y, Thomas HD, Batey MA, Cowell IG, Richardson CJ, Griffin RJ, Calvert AH, Newell DR, Smith GC, Curtin NJ. Preclinical Evaluation of a Potent Novel DNA-Dependent Protein Kinase Inhibitor NU7441. Cancer Res. 2006;66:5354–5362. doi: 10.1158/0008-5472.CAN-05-4275. [DOI] [PubMed] [Google Scholar]
- 86.O’Connor MJ, Martin NM, Smith GC. Targeted cancer therapies based on the inhibition of DNA strand break repair. Oncogene. 2007;26:7816–7824. doi: 10.1038/sj.onc.1210879. [DOI] [PubMed] [Google Scholar]
- 87.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 88.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 89.de Murcia G, Ménissier-de Murcia J. Poly(ADP-ribose) polymerase: a molecular nick-sensor. Trends Biochem. Sci. 1994;19:250. doi: 10.1016/0968-0004(94)90280-1. [DOI] [PubMed] [Google Scholar]
- 90.de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, Walztinger C, Chambon P, de Murcia G. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl. Acad. Sci. USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schreiber V, Amé JC, Dollé P, Schultz I, Rinaldi B, Fraulob V, Ménissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- 92.Curtin NJ, Wang LZ, Yiakouvaki A, Kyle S, Arris CA, Canan-Koch S, Webber SE, Durkacz BW, Calvert HA, Hostomsky Z, Newell DR. Novel poly(ADP-ribose) polymerase-1 inhibitor, AG14361, restores sensitivity to temozolomide in mismatch repair-deficient cells. Clin Cancer Res. 2004;10:881–889. doi: 10.1158/1078-0432.ccr-1144-3. [DOI] [PubMed] [Google Scholar]
- 93.Calabrese CR, Almassy R, Barton S, Batey MA, Calvert AH, Canan-Koch S, Durkacz BW, Hostomsky Z, Kumpf RA, Kyle S, Li J, Maegley K, Newell DR, Notarianni E, Stratford IJ, Skalitzky D, Thomas HD, Wang LZ, Webber SE, Williams KJ, Curtin NJ. Anticancer chemosensitization and radiosensitization by the novel poly(ADP-ribose) polymerase-1 inhibitor AG14361. J Natl Cancer Inst. 2004;96:56–67. doi: 10.1093/jnci/djh005. [DOI] [PubMed] [Google Scholar]
- 94.Miknyoczki SJ, Jones-Bolin S, Pritchard S, Hunter K, Zhao H, Wan W, Ator M, Bihovsky R, Hudkins R, Chatterjee S, Klein-Szanto A, Dionne C, Ruggeri B. Chemopotentiation of temozolomide, irinotecan, and cisplatin activity by CEP-6800, a poly(ADP-ribose) polymerase inhibitor. Mol Cancer Ther. 2003;2:371–382. [PubMed] [Google Scholar]
- 95.Chalmers A, Johnston P, Woodcock M, Joiner M, Marples B. PARP-1, PARP-2, and the cellular response to low doses of ionizing radiation. Int J Radiat Oncol Biol Phys. 2004;58:410–419. doi: 10.1016/j.ijrobp.2003.09.053. [DOI] [PubMed] [Google Scholar]
- 96.Ratnam K, Low JA. Current development of clinical inhibitors of poly(ADP-ribose) polymerase in oncology. Clin Cancer Res. 2007;13:1383–1388. doi: 10.1158/1078-0432.CCR-06-2260. [DOI] [PubMed] [Google Scholar]
- 97.Curtin N. Therapeutic potential of drugs to modulate DNA repair in cancer. Expert Opin Ther Targets. 2007;11:783–799. doi: 10.1517/14728222.11.6.783. [DOI] [PubMed] [Google Scholar]
- 98.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol. Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 99.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 100.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 101.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 102.McCabe N, Turner NC, Lord CJ, Kluzek K, Bialkowska A, Swift S, Giavara S, O’Connor M,J, Tutt AN, Zdzienicka MZ, Smith GC, Ashworth A. Deficiency in the Repair of DNA Damage by Homologous Recombination and Sensitivity to Poly(ADP-Ribose) Polymerase Inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 103.Hay T, Jenkins H, Sansom OJ, Martin NM, Smith GC, Clarke AR. Efficient deletion of normal Brca2-deficient intestinal epithelium by poly(ADP-ribose) polymerase inhibition models potential prophylactic therapy. Cancer Res. 2005;65:10145–10148. doi: 10.1158/0008-5472.CAN-05-1186. [DOI] [PubMed] [Google Scholar]
- 104.Evers B, Drost R, Schut E, de Bruin M, van der Burg E, Derksen PW, Holstege H, Liu X, van Drunen E, Beverloo HB, Smith GC, Martin NM, Lau A, O’Connor MJ, Jonkers J. Selective Inhibition of BRCA2-Deficient Mammary Tumor Cell Growth by AZD2281 and Cisplatin. Clin Cancer Res. 2008;14:3916–3925. doi: 10.1158/1078-0432.CCR-07-4953. [DOI] [PubMed] [Google Scholar]
- 105.Rottenberg S, Jaspers JE, Kersbergen A, van der Burg E, Nygren AO, Zander SA, Derksen PW, de Bruin M, Zevenhoven J, Lau A, Boulter R, Cranston A, O’Connor MJ, Martin NM, Borst P, Jonkers J. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci U S A. 2008;105:17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yap TA, Boss DS, Fong PC, Roelvink M, Tutt A, Carmichael J, O’Connor MJ, Kaye SB, Schellens JH, de Bono JS. First in human phase I pharmacokinetic (PK) and pharmacodynamic (PD) study of KU-0059436 (Ku), a small molecule inhibitor of poly ADP-ribose polymerase (PARP) in cancer patients (p), including BRCA1/2 mutation carriers. J Clin Oncol (Meeting Abstracts) 2007;25:3529. [Google Scholar]
- 107.Fong PC, Boss DS, Carden CP, Roelvink M, De Greve J, Gourley CM, Carmichael J, De Bono JS, Schellens JH, Kaye SB. AZD2281 (KU-0059436), a PARP (poly ADP-ribose polymerase) inhibitor with single agent anticancer activity in patients with BRCA deficient ovarian cancer: Results from a phase I study. J Clin Oncol (Meeting Abstracts) 2008;26:5510. [Google Scholar]
- 108.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 109.Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]