Abstract
Traditional robots1 rely on computing to coordinate sensing and actuating components and to store internal representations of their goals and environment. Any implementation of single-molecule based robotics must overcome the limited ability of individual molecules to store complex programs and, for example, use architectures that obtain complex behaviors from the interaction of simple robots with their environment2-4. Previous research in DNA walkers5 focused on transitioning from non-autonomous systems6,7 to directed but brief motion on one-dimensional tracks8-11. Herein, we obtain elementary robotic behaviors from the interaction between a random walker incorporating deoxyribozymes12 and a precisely defined environment. Using single-molecule microscopies we demonstrate that such walkers achieve directionality by sensing and modifying their environment, following trails of recognition elements (“bread crumbs”) laid out on a two-dimensional DNA origami landscape13. These molecular robots autonomously carry out sequences of actions such as “start”, “follow”, “turn”, and “stop”, thus laying the foundation for the synthesis of more complex robotic behaviors at the molecular level by incorporating additional layers of control mechanisms. For example, interactions between multiple molecular robots could lead to collective behavior14,15, while the ability to read and transform secondary cues on the landscape could provide a mechanism for Turing-universal algorithmic behavior2,16,17.
Our walkers, called molecular spiders, comprise an inert body (streptavidin) and three catalytic “legs”. Legs are adapted from DNA enzyme 8-17 that binds and cleaves oligodeoxynucleotide (henceforth “oligonucleotide”) substrates with a single ribose moiety (Fig. 1a,b) into two shorter products that have lower affinities for the enzyme18. A spider’s interactions with a layer of immobilized substrate and/or product sites can be modeled using a simple ‘memory’ principle19: Each leg moves independently from sites to accessible neighboring sites, but if a leg is on a site not visited before, it will stay longer on average. Restated in a biochemically more intuitive manner: A deoxyribozyme on a site that was previously converted to a product will dissociate faster, whereas it will stick longer on the substrates and eventually cleave them. Because spiders have multiple legs, a single dissociated leg will quickly reattach to nearby product or substrate. It follows that the body of a spider positioned at the interface between products and substrates will move toward the substrate region, because after cleaving, each leg will explore neighboring sites until it finds another substrate. On a linear track of substrates this mechanism predicts a deviation from an otherwise random walk process, yielding directional movement as the substrates are cleaved. Unlike previously engineered “burnt bridge” mechanisms6-9,11 and those found in nature20, which render revisiting the same path impossible, spiders will perform Brownian walks on product tracks until they again encounter substrate.
Figure 1. Deoxyribozyme based molecular walker and origami prescriptive landscape schematics.
a, The NICK3.4A3+1 spider consists of a streptavidin core that displays a 20 base ssDNA that positions the spider at the start (green), and three deoxyribozyme legs. b, The 8-17 deoxyribozyme cleaves its substrate at an RNA base creating two shorter products (seven and eleven bases). Dissociation from these products allows legs to associate with the next substrate. c, Spider actions: after release by a 27-base ssDNA trigger, the spider follows the substrate track, turns, and continues to a stop site (red). d, Schematic of the DNA origami landscape with positions A-E labeled; track EABD is shown. e, A representative origami landscape shows the start position (green), the substrate track (brown), stop and control sites (red), and a topographical marker (blue),
In analogy to the reactive planning used in simple robots4, the sensor-actuator feedback afforded when legs sense and modify nearby oligonucleotides allows us to design prescriptive landscapes that direct the spiders’ motion along a predefined path (Fig. 1c,d). Prescriptive landscapes were constructed using the DNA origami scaffolding technique13. The scaffold consists of a 7249-nucleotide single-stranded DNA folded by 202 distinct staple strands into a rectangular shape roughly 65×90×2 nm in size and with 6-nm feature resolution (Fig. 1e). Each staple can be extended on its 5′ end with probes that recruit substrates, products, goal and control strands21.
We designed pseudo-one dimensional tracks on origami of about spider width (three adjacent rows of substrates, Fig. 1d). Tracks are coded by a sequence of points (A, B, C, D, E; i.e., on an ABD landscape the spider starts at A, and passes through B before ending at D). Staples were modified to position: (1) A START oligonucleotide, used to position a spider at the start of the experiment, that is complementary to a TRIGGER oligonucleotide used to release the spider22 (the “start” action); (2) Substrate TRACK probes to capture the 5′ extension on substrates forming the TRACK (directing the “follow” and “turn” actions); (3) STOP probes complementary to the 5′ extension on STOP strands (non-chimeric and uncleavable analogs of the substrate) that do not influence directional movement but trap spiders to prevent them from walking backwards after completing the track (the “stop” action); (4) CONTROL probes (identical to the STOP, but disconnected from the track), used to assess the extent to which free-floating spiders are captured directly from solution; and (5) MARKER oligonucleotides based on inert dumbbell hairpins, aiding in origami classification within atomic force microscopy (AFM) images (Fig. 1e). To position spiders at START sites, we replaced one of the four catalytic legs of the NICK-4.4A12 spider with a tethering oligonucleotide (Supplementary Figs 1-4 and Supplementary Information) partially complementary to the START oligonucleotide.
To estimate the efficiency of spider motion directed by the TRACK, we defined and tested four paths with no (EAC), one (ABD), or two (EABD, EABC) turns (Fig. 2 and Supplementary Figs 8, 11, 14, 17). Our basic procedure consisted of: (1) Assembling the origami; (2) attaching the spider to the START site; (3) adding TRACK, STOP, and CONTROL strands to complete the landscape; and (4) initiating an experiment by releasing the spider through addition of TRIGGER and 1 mM Zn2+ cofactor23 (Supplementary Figs 6, 25, and Supplementary Information). We sampled the origami solution before and after spider release, and imaged individual samples by AFM to determine the locations of spiders. We scored only “face-up” origami (substrates projected away from mica) to avoid artifacts and developed procedures to minimize readout bias (see Supplementary Information for details).
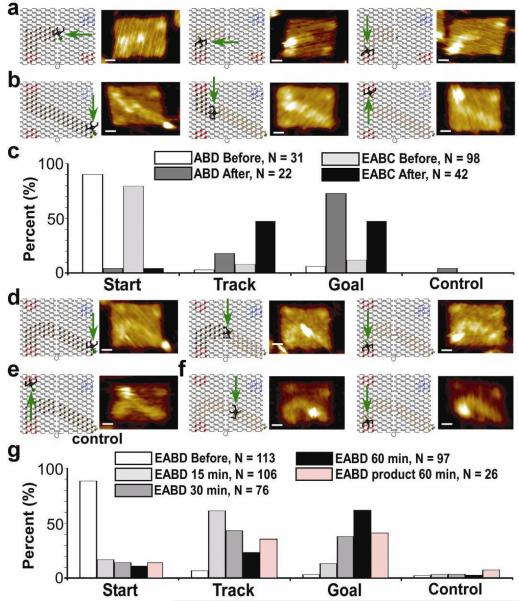
Figure 2. Results of spider movement along three tracks with schematics and AFM images of the spider at the start, on the track, and at the stop site.
a, ABD track. b, EABC track. c, Graph of ABD and EABC spider statistics before and 30 minutes after release. d, EABD track. e, EABD track with spider on control. f, EABD product-only track. g, Graph of the EABD spider statistics before, and 15, 30 and 60 min after release, and 60 min after release on the EABD product-only track. All AFM images are 144 x 99.7 nm, the scale bar is 20 nm. Legend text indicates the number of origami with a single spider that were counted for the given sample.
In all samples imaged before spider release, 30-40% of the assembled origami carry at least one spider, 80-95% of which are singly occupied, and of these 80-90% bound their spider at the START position (Supplementary Table 1 and Supplementary Figs 9, 10, 12, 13, 15, 16, 18, and 19). Upon adding trigger, all four landscapes with substrate tracks showed that the fraction of spiders at the START diminishes with a concomitant increase in spiders observed on the STOP sites (Fig. 2c,g and Supplementary Fig. 23). A spider’s ability to reach the STOP sites decreased with increased TRACK length and with decreased time of incubation in solution. In time-lapse experiments on a long path (EABD, spanning ~ 90 nm) we observed a gradual increase of up to 70% of spiders on STOP sites within 60 min (Fig. 2c,g). A short path (ABD, ~ 48 nm) was completed to the same extent within 30 min.
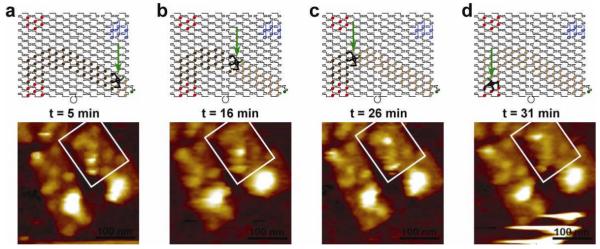
We captured one series of AFM images of a spider moving along an origami track (Fig. 3). The rate of spider movement (~90 nm over 30 min, with approximately 6 nm per three parallel cleavage events) was consistent with the processive cleavage rates (~1 min−1) of spiders on a 2D surface as obtained by SPR (Supplementary Fig. 6). More systematic sequential imaging proved difficult due to mica’s inhibitory effects on the spider.
Figure 3. AFM movie of spider movement.
a, b, c, d, Schematics and AFM images of the spider moving along the EABD track at 5 min (a), 16 min (b), 26 min, (c) and 31 min (d) after trigger was added. AFM images are 300 × 300 nm and the scale bar is 100 nm.
We can eliminate deviations from the proposed mechanism of spider motion as major contributors to these results. First, to test that spiders can indeed traverse product tracks by means of unbiased random walks, we challenged spiders with EABD origami in which the substrate was replaced by product on the TRACK. Spiders still reached the STOP sites albeit more slowly (Fig. 2f,g), as expected from purely Brownian spider movement even if individual steps are somewhat faster19. Second, we wished to confirm that spiders don’t often ‘jump’; if all three legs simultaneously dissociate before any leg reattaches, a spider could completely dissociate from the origami and subsequently reattach elsewhere at random. Evidence against frequent jumping (or an excess of spiders in solution during the initial assembly stage) comes from the low level of spider occupancy at CONTROL sites in both substrate and product track experiments (Fig. 2c,e,g) and the stable proportions of unoccupied and multiply-occupied origami (Supplementary Table 1; both before and after the addition of trigger, 5-10% of origami displayed more than one spider on its track). In contrast, when spiders were released on ABD landscapes with no TRACK strands, after 30 min we observed an equal distribution between STOP and CONTROL sites (Supplementary Fig. 24 and Supplementary Table 2), as expected for a process that involves spider dissociation from and random rebinding to the origami. In independent ensemble experiments using surface plasmon resonance, we observed that up to 15% of spiders may dissociate from a non-origami 2D product-covered surface within 60 min under flow conditions (Supplementary Fig. 5). On similar substrate-covered surfaces, spiders show an average processivity of ~200 substrates before being removed by flow (Supplementary Figs 5 and 6). Together, these results rule out that spiders move predominantly by jumping; there is insufficient jumping even on product tracks to explain the 50-70% occupation of the STOP sites after walks on ABD, EABC, and EABD substrate tracks.
For a more facile real-time observation of the movement of individual spiders, we applied particle tracking by super-resolution total internal reflection fluorescence (TIRF) video microscopy24. Four biotin molecules were attached to the underside of the origami for immobilization on the avidin-coated quartz slide. Spiders were covalently labeled with on average 2.3 Cy3 fluorophores, and STOP sites were labeled with 6 Cy5 fluorophores. The labeling allowed us to monitor changes in spider position relative to the STOP site by two-color fluorescent particle tracking25,26. In a typical experiment, spider-loaded tracks were incubated with TRIGGER and immobilized on the slide (Supplementary Fig. 26), then Zn2+ was added to promote spider movement via substrate cleavage. Recognizing that the 8-17 activity depends on buffer conditions23, we obtained the best results from SSC or HEPES with increased Zn2+ concentrations but without Mg2+ (Supplementary Figs 6 and 25).
Our resolution was not sufficient to reliably detect turns, so we focused on EAC landscapes. Individual particle traces showed a distribution of behaviors that may result from variations across molecules, idiosyncrasies of the sample preparation, the stochastic nature of the observed process, photobleaching, and/or instrument measurement error (Fig. 4a,b, Supplementary Figs 29-31, Supplementary Information and Supplementary Table 3). Despite this variability, moving traces commonly had net displacements between 60 and 140 nm and their mean velocity varied between 1 and 6 nm/min, within error consistent with track length (~90 nm) and deoxyribozyme cleavage rate (~1 min−1/leg), respectively.
Figure 4. Spiders imaged on origami tracks in real-time using super-resolution TIRF microscopy.

a, Position-time trajectory of a selected spider (EAC 2, Cy3-labeled) on the EAC substrate track. The position as a function of time is represented by color-coded dots (see Supplementary Information for details). A small green dot represents the START and a large red oval represents the Cy5-labeled STOP site. ZnSO4 was added at time zero. b, Displacement of the spider trajectory in panel a from its initial position as a function of time. The green line represents displacement calculated using averaged position measurements of 1 min intervals, and the black line represents the displacement from a rolling 4-min average (see Supplementary Information). c, Ensemble root mean square displacement (RMSD) of exemplary spiders on the EAC substrate track in the presence (red, corresponding to the 15 Tier 1 Spiders in Supplementary Fig. 29) and absence (black, 7 spiders) of Zn2+, with the corresponding displacements used to calculate each ensemble RMSD for each buffer condition (similarly colored line graphs). d, Ensemble RMSD for spiders on EAC tracks satisfying simple filtering criteria. Curves are shown for spiders on EAC substrate track (red, 85 spiders), EAC product track with TRIGGER introduced to the sample 10-15 min before imaging (blue, 18 spiders), and EAC product track with TRIGGER introduced 30-60 min before imaging (black, 29 spiders). EAC substrate and 10-15 min trigger product RMSD plots are fit to a power law function, and the EAC 30-60 min trigger product RMSD is fit to a straight line. Individual displacements are shown with colors corresponding to the respective ensemble RMSD plots. All Figure 4 data were obtained in SSC buffer.
To confirm that our particle traces reflect genuine spider movement, we performed tests with and without Zn2+ and/or TRIGGER, both on substrate and product tracks. In each case, RMSD plots varied in a way consistent with the expected corresponding behavior of spiders on origami tracks, despite the inherent noise associated with single particle tracking over tens-of-nanometer length scales and tens-of-minute time scales (Fig. 4c,d). For instance, RMSD plots indicated substantially more movement on substrate tracks in the presence of Zn2+ and trigger than in their individual absence (Fig. 4c, Supplementary Figs 30-32 and Supplementary Table 4). On product tracks, results were consistent with an unbiased random walk with no dependence on Zn2+. When product tracks were pre-incubated with TRIGGER 30-60 min prior to addition of Zn2+ and onset of imaging (as were substrate tracks), little or no movement was observed (Fig. 4d), consistent with spiders having been released and having diffused toward or to the STOP sites prior to imaging. In contrast, when TRIGGER and Zn2+ were both added shortly prior to imaging, substantial movement was observed (Fig. 4d), consistent with our AFM results for spiders on product tracks (Fig. 2f,g) and with Monte Carlo simulations of spider movement (Supplementary Information and Supplementary Fig. 32).
Our single-molecule experiments provided results consistent with random DNA-based walkers guided by their landscapes for as far as 100 nm, for up to 50 cleavage steps, at speeds of roughly 3 nm/min. Still, there are mechanistic limitations: (1) The distance over which a spider can move is confined by dissociation or backtracking, with an increase in processivity achievable only at the cost of a slower velocity12; (2) the current mechanism consumes substrate, which must be recharged to sustain directed movement; (3) spiders are subject to the stochastic uncertainty as to whether each individual robot can accomplish its task (cf., “faulty” behavior in robotics and “yield” in chemistry); and (4) our walkers are not as fast, efficient, or powerful as protein based walkers with solution phase fuels27. As candidates for molecular robots, however, they offer the advantages of programmability5,10,28-30, predictable biophysics5, and designable landscapes13. The ability to obtain programmed behavior from the interaction of simple molecular robots with a complex modifiable environment suggests that exploiting stochastic local rules and programming the environment are effective ways to minimize the limitations that molecular construction places on the complexity of robotic behavior at the nanoscale.
Supplementary Material
Acknowledgements
This research was supported by the NSF CBC and EMT grants to all authors, fellowships and grants from the Searle Foundation, the Lymphoma and Leukemia Society and the Juvenile Diabetes Research Foundation to MNS, awards from ARO, AFOSR, ONR, NIH and a Sloan Research Fellowship to HY, an NSF Graduate Fellowship to ND, and Molecular Biophysics and Microfluidics in Biomedical Sciences Training Fellowships from the NIH to AJB and NM.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Siegwart R, Nourbakhsh IR. Introduction to Autonomous Mobile Robots. MIT Press; Cambridge, MA: 2004. [Google Scholar]
- 2.Turing AM. On computable numbers, with an application to the Entscheidungsproblem. Proc. London Math. Soc. Series. 1936;2:230–265. [Google Scholar]
- 3.Braitenberg V. Vehicles: Experiments in Synthetic Psychology. MIT Press; Cambridge, MA: 1984. [Google Scholar]
- 4.Brooks RA. Intelligence without representation. Artif. Intell. 1991;47:139–159. [Google Scholar]
- 5.Bath J, Turberfield A. DNA nanomachines. Nat. Nanotechnol. 2007;2:275–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- 6.Sherman WB, Seeman NC. A precisely controlled DNA biped walking device. Nano Lett. 2004;4:1203–1208. [Google Scholar]
- 7.Shin JS, Pierce NA. A synthetic DNA walker for molecular transport. J. Am. Chem. Soc. 2004;126:10834–10835. doi: 10.1021/ja047543j. [DOI] [PubMed] [Google Scholar]
- 8.Bath J, Green SJ, Turberfield AJ. A free-running DNA motor powered by a nicking enzyme. Angew. Chem. Int. Edn. 2005;44:4358–4361. doi: 10.1002/anie.200501262. [DOI] [PubMed] [Google Scholar]
- 9.Tian Y, He Y, Chen Y, Yin P, Mao C. A DNAzyme that walks processively and autonomously along a one-dimensional track. Angew. Chem. Int. Edn. 2005;44:4355–4358. doi: 10.1002/anie.200500703. [DOI] [PubMed] [Google Scholar]
- 10.Yin P, Choi H, Calvert CR, Pierce NA. Programming biomolecular self-assembly pathways. Nature. 2008;451:318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 11.Omabegho T, Sha R, Seeman NC. A Bipedal DNA Brownian Motor with Coordinated Legs. Science. 2009;324:67–71. doi: 10.1126/science.1170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pei R, et al. Behavior of Polycatalytic Assemblies in a Substrate-Displaying Matrix. J. Am. Chem. Soc. 2006;128:12693–12699. doi: 10.1021/ja058394n. [DOI] [PubMed] [Google Scholar]
- 13.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:298–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 14.Bonabeau E, Dorigo M, Theraulaz G. Swarm Intelligence: From Natural to Artificial Systems. Oxford University Press; New York, NY: 1999. [Google Scholar]
- 15.Rus D, Butler Z, Kotay K, Vona M. Self-reconfiguring robots. Commun. ACM. 2002;45:39–45. [Google Scholar]
- 16.Von Neumann J. In: Theory of Self-Reproducing Automata. Burks AW, editor. University of Illinois Press; Urbana, IL: 1966. [Google Scholar]
- 17.Bennett CH. The thermodynamics of computation—a review. Int. J. Theor. Phys. 1982;21:905–940. [Google Scholar]
- 18.Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antal T, Krapivsky PL. Molecular spiders with memory. Phys. Rev. E. 2007;76:021121–021129. doi: 10.1103/PhysRevE.76.021121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saffarian S, Collier IE, Marmer BL, Elson EL, Goldberg G. Interstitial Collagenase is a Brownian Ratchet Driven by Proteolysis of Collagen. Science. 2004;306:108–111. doi: 10.1126/science.1099179. [DOI] [PubMed] [Google Scholar]
- 21.Ke Y, Lindsay S, Chang Y, Liu Y, Yan H. Self-Assembled Water-Soluble Nucleic Acid Probe Tiles for Label-Free RNA Hybridization Assays. Science. 2008;319:180–183. doi: 10.1126/science.1150082. [DOI] [PubMed] [Google Scholar]
- 22.Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 23.Li J, Zheng W, Kwon AH, Lu Y. In vitro selection and characterization of a highly efficient Zn (II)-dependent RNA-cleaving deoxyribozyme. Nucleic Acids Res. 2000;28:481–488. doi: 10.1093/nar/28.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walter NG, Huang C-Y, Manzo AJ, Sobhy MA. Do-it-yourself guide: How to use the modern single-molecule toolkit. Nat. Methods. 2008;5:475–489. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Churchman LS, Okten Z, Rock RS, Dawson JF, Spudich JA. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proc. Natl. Acad. Sci. USA. 2005;102:1419–1423. doi: 10.1073/pnas.0409487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yildiz A, Selvin PR. Fluorescence imaging with one nanometer accuracy: application to molecular motors. Acc. Chem. Res. 2005;38:574–582. doi: 10.1021/ar040136s. [DOI] [PubMed] [Google Scholar]
- 27.Hess H. Toward Devices Powered by Biomolecular Motors. Science. 2006;312:860–861. doi: 10.1126/science.1126399. [DOI] [PubMed] [Google Scholar]
- 28.Adleman L. Molecular computation of solutions to combinatorial problems. Science. 1994;266:1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- 29.Stojanovic MN, Stefanovic D. A deoxyribozyme-based molecular automaton. Nat. Biotechnol. 2003;21:1069–1074. doi: 10.1038/nbt862. [DOI] [PubMed] [Google Scholar]
- 30.Seelig G, Soloveichik D, Zhang DY, Winfree E. Enzyme-Free Nucleic Acid Logic Circuits. Science. 2006;314:1585–1588. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.