Abstract
The mechanisms underlying the proapoptotic effect of the chemotherapeutic agent, cisplatin, are largely undefined. Understanding the mechanisms regulating cisplatin cytotoxicity may uncover strategies to enhance the efficacy of this important therapeutic agent. This study evaluates the role of activating transcription factor 3 (ATF3) as a mediator of cisplatin-induced cytotoxicity. Cytotoxic doses of cisplatin and carboplatin treatments consistently induced ATF3 expression in five tumor-derived cell lines. Characterization of this induction revealed a p53, BRCA1, and integrated stress response-independent mechanism, all previously implicated in stress-mediated ATF3 induction. Analysis of mitogen-activated protein kinase (MAPK) pathway involvement in ATF3 induction by cisplatin revealed a MAPK-dependent mechanism. Cisplatin treatment combined with specific inhibitors to each MAPK pathway (c-Jun N-terminal kinase, extracellular signal-regulated kinase, and p38) resulted in decreased ATF3 induction at the protein level. MAPK pathway inhibition led to decreased ATF3 messenger RNA expression and reduced cytotoxic effects of cisplatin as measured by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide cell viability assay. In A549 lung carcinoma cells, targeting ATF3 with specific small hairpin RNA also attenuated the cytotoxic effects of cisplatin. Similarly, ATF3-/- murine embryonic fibroblasts (MEFs) were shown to be less sensitive to cisplatin-induced cytotoxicity compared with ATF3+/+ MEFs. This study identifies cisplatin as a MAPK pathway-dependent inducer of ATF3, whose expression influences cisplatin's cytotoxic effects.
Introduction
cis-Diamminedichloroplatinum(II) (cisplatin) is among the most active antitumor agent used in human chemotherapy. Cisplatin and its derivative, carboplatin, are widely used agents in various tumor types including lung and ovarian cancers [1]. Acquired resistance and toxicities associated with treatment are major impediments inhibiting their efficacy [2]. Understanding the mechanisms regulating tumor cell cytotoxicity may uncover novel therapeutic strategies to enhance the efficacy of these platinum-based chemotherapeutics. Cisplatin and carboplatin are primarily considered as DNA-damaging anticancer drugs, forming different types of bifunctional adducts in reaction with cellular DNA [1]. Cisplatin and carboplatin become activated intracellularly by aquating one of two chloride-leaving groups and, subsequently, by covalently binding to DNA, forming DNA adducts [3]. Carboplatin is a less toxic compound with a more stable leaving group than chloride, which lowers toxicity and reduces nephrotoxicity [4]. Efficacious treatments of carboplatin generally require up to 20-fold higher doses than cisplatin and are tolerated because of its decreased toxicity [5]. The final cellular outcome of DNA adduct formation is generally apoptotic cell death, thought to occur through halting of cellular processes such as replication and transcription, leading to prolonged G2 phase cell cycle arrest and deregulation of signal transduction pathways involved in growth, differentiation, and stress responses [1,3]. Cellular mechanisms of resistance to platinum-based chemotherapeutics are multifactorial and contribute to severe limitation in their use in clinical practice. They include molecular events inhibiting drug-DNA interaction, such as reduction in cisplatin accumulation inside cancer cells or inactivation by thiol-containing species [2]. Other important mechanisms acting downstream of the initial reaction of cisplatin with DNA include an increase in adduct repair and a decrease in induction of apoptosis [2]. Although DNA is the primary target of cisplatin and carboplatin activity, gaps in our understanding of the process that translates cisplatin-induced DNA damage into its therapeutically beneficial process of apoptosis still remain. Two significant cellular pathways have been demonstrated to play key roles in platin-induced apoptosis/cytotoxicity, the mitogen-activated protein kinase (MAPK) cascades, and the tumor suppressor p53 [6,7]. An understanding of the mode of action is indeed desirable in refining therapeutic approaches that further enhance the antitumor activity of platinum-based chemotherapeutics.
Activating transcription factor 3 (ATF3) is a member of the basic region-leucine zipper proteins originally identified for their ability to bind the cAMP-responsive element (ATF/CRE) site (TGACGTCA) [8]. Whereas ATF3 messenger RNA (mRNA) and protein levels are not detectable under basal conditions in most cells, a large body of evidence shows that ATF3 is induced by a wide variety of stress-causing agents including hypoxia, metabolic stress, and DNA damage [9]. ATF3 is also induced in times of physiological stress such as liver regeneration [10], brain seizure [11], ischemia-reperfusion of the heart [12], nerve damage [13,14], and UV damage where it plays a role in maintaining genomic integrity [15]. ATF3 has been demonstrated to play a role in apoptosis and proliferation, two cellular processes critical for cancer progression [16–19]. ATF3 can either promote or suppress these processes. For example, overexpression of ATF3 in the sense orientation in colorectal cancer cells led to decreased focus formation in vitro and reduced the size of mouse tumor xenografts in vivo [16]. Divergence in function of ATF3 between a proapoptotic and an antiapoptotic factor in cancer models is likely dependent on both cellular model and state of malignancy [19,20]. ATF3 is also a member of the activating protein 1 transcription factors that consist of homodimers and heterodimers of the basic region-leucine zipper proteins that belong to the Jun (c-Jun, v-Jun, JunB, and JunD), Fos (c-Fos, v-Fos, FosB, Fra1, and Fra2), and the related activating transcription factor (ATF2, ATF3/LRF1, and B-ATF) subfamilies [21]. Activation of ATF3 by a wide array of stress signaling pathways has been demonstrated, including DNA repair pathway components p53 [22,23] and potentially BRCA1 [17,24], the integrated stress response (ISR) that is principally activated by hypoxia and metabolic stress [25], and the stress-induced MAPK cascades (SAPK/c-Jun N-terminal kinase [JNK], and p38) [26,27]. Of interest, p53 and the p38 MAPK pathway have also been shown to play roles in regulating cisplatin-induced cytotoxicity [28,29].
In this study, we evaluated the potential of cisplatin to induce ATF3 and determined the pathway regulating this induction. Furthermore, we determined the role of ATF3 as a mediator of the cytotoxic effects of cisplatin.
Materials and Methods
Tissue Culture
The A549, PC3, HCC1937, and MCF-7 cell lines were obtained from the American Type Culture Collection (Rockville, MD). Cell lines SKOV-3 and A2780-cp were kindly provided by Dr Barbara Vanderhyden, Ottawa Hospital Research Institute, Ottawa, Canada. The murine embryonic fibroblasts (MEFs) used in this study were derived from wild type and knockout mice from ATF3, ATF4, and ATF2 models kindly provided by D. Park (University of Ottawa, Ottawa, Ontario, Canada) and L. Glimcher (Harvard Medical School, Boston, MA). All cell lines were maintained in Dulbecco modified Eagle medium (Media Services, Ottawa Regional Cancer Centre) supplemented with 10% fetal bovine serum (Medicorp, Montreal, Canada) and 100 U/ml penicillin and 100 µg/ml streptomycin (GIBCO, Burlington, Ontario, Canada) of medium. Cells were exposed to cisplatin, carboplatin, and taxol (provided by the pharmacy at the Ottawa Hospital Regional Cancer Centre, Ottawa) alone or in combination with the p38 inhibitor SB203580 (Calbiochem, Gibbstown, NJ), JNK inhibitor, JNK inhibitor II (SP600125; Calbiochem), or extracellular signal-regulated kinase (ERK) inhibitor UO126 (Calbiochem) diluted in DMSO. Adenovirus p53wt and LacZ control were kindly provided by Dr Bruce McKay (Ottawa Hospital Research Institute).
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide Assay
In a 96-well flat-bottomed plate (Nunc, Naperville, IL), 5000 cells/150 µl of cell suspension were used to seed each well. The cells were incubated overnight to allow for cell attachment and recovery. Cells were treated with indicated drugs and incubated for 48 hours at 37°C. After treatment, 42 µl of a 5-mg/ml solution in PBS of the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) tetrazolium substrate (Sigma, St Louis, MO) was added to each well and incubated for ∼20 minutes at 37°C. The resulting violet formazan precipitate was solubilized by the addition of 82 µl of a 0.01 M HCl/10% SDS (Sigma) solution and was allowed to further incubate at 37°C overnight. The plates were then analyzed on a microplate reader (MRX; Dynex Technologies, West Sussex, United Kingdom) at 570 nm to determine the absorbance of the samples.
Flow Cytometry
Cells were plated at 1 x 106/10-cm dish and allowed to grow overnight and subsequently treated with cisplatin for 48 hours. Single-cell suspensions were labeled with 50 µg/ml propidium iodide (Sigma), and approximately 106 cells in 1 ml were analyzed by flow cytometry. Ten thousand cells were evaluated, and the percentage of cells in sub-2N phase was determined using a software (Modfit LT; VeritySoftware House, Topsham, ME).
Immunocytochemistry
MCF-7 and PC3 cells grown to 50% confluence were seeded on 2 x 15-cm plates/treatment/block with no treatment or with cisplatin (8 µg/ml) for 24 hours. Cells were washed twice in PBS, harvested in 10 ml of PBS per plate, and combined with 20 ml of 20% neutral-buffered formalin (Sigma). Cells were fixed at 4°C for 1 hour and spun down at 1600 rpm for 10 minutes at 4°C, and cells were washed once in PBS. Formalin-fixed cells were embedded in paraffin, cut into 5-µm sections, and allowed to dry at room temperature overnight. Sections were deparaffinized by washing in toluene (3 x 5 minutes) followed by absolute alcohol (2 x 1 minutes). Sections were washed with water (5 minutes) followed by Tris-buffered saline (TBS) and loaded on the intelliPAT FLX automated slide stainer (Davis Diagnostics, Brampton, Canada). The automated slide stainer was programmed with the following treatments: 3% H2O2 in TBS for 10 minutes, rinsed in TBS for 5 minutes, blocked with universal blocking agent Background Sniper (Biocare Medical, Brampton, Ontario, Canada) for 20 minutes at room temperature, incubated with ATF3 antibody (1:200 dilution in Da Vinci universal diluent [Biocare Medical]) for 1 hour at room temperature, rinsed with TBS for 5 minutes, incubated with universal mouse probe (Mach 4 Universal Polymer Detection Kit; Biocare Medical) for 5 minutes, rinsed with TBS for 5 minutes, incubated with Rabbit HRP Polymere (Mach 4 Universal Polymer Detection Kit; Biocare Medical) for 10 minutes at room temperature, rinsed with TBS, and developed for 5 minutes with DAB RTU (Betazoid DAB Chromatogen Kit; Biocare Medica) and rinsed with water. Slides were counterstained in hematoxylin for 1 minute, washed in running water, 0.2% HCl in 70% alcohol for five dips, washed in running water for 1 minute, dipped once in 2% aqueous saturated lithium carbonate, washed in running water for 5 minutes, dehydrated in absolute alcohol, cleared in toluene, and mounted on coverslips with Permount (Fisher Scientific, Mississauga, Canada).
Transfection
MCF-7 cells plated at 3 x 105 in six-well plates were transfected with 2 µg of each p38 constructs (p38 wild type [WT], p38 dominant-negative [DN], and p38 catalytically active [CA]; kindly provided by Dr Douglas Gray, Ottawa Hospital Research Institute) using a transfection reagent (FuGENE HD; Roche, Mississauga, Ontario, Canada) as per the manufacturer's protocol. After 24 hours, the medium was removed and replaced with medium containing cisplatin (10 µg/ml) treatment alone or in combination with SB203580 (10 µM), and cells were incubated for an additional 24 hours. Cells were then harvested and analyzed by Western blot analysis.
Adenovirus Infection
PC3 cells were plated at 2.5 x 105 cells per well (six-well dish) and infected with p53wt or LacZ control adenovirus (provided by Dr B. McKay; Ottawa Hospital Research Institute) at 25 plaque-forming units per milliliter per cell. After a 6-hour infection period, the medium was removed from the cells and replaced with medium containing cisplatin (10 µg/ml) or taxol (25 µM) for 24 hours. Cells were then harvested and analyzed by Western blot analysis as described below.
Design and Expression of Small Hairpin RNA
The two 19mer sequences targeting ATF3 mRNA are no. 1—5′-GCCAAAGAATATTCCATTT-3′ and no. 2—5′-GGGAGGGCCTGCAGTGATT-3′ to pSuper vector from Oligoengine small hairpin RNA (shRNA; no. 1: nucleotides 1524–1542 [GenBank accession number NM_001030287]; no. 2: nucleotides 1270–1289 [GenBank accession number NM_001030287]) target sequence. As controls, we used the green fluorescent protein (GFP)-targeted oligonucleotide 5′CATGCGTCCACTCTTCCTC-3′ with accession number NC_011521. These sequences were BLAST confirmed for specificity. The forward and reverse synthetic 60-nt oligonucleotides (Integrated DNA Technologies, Coralville, IA) were designed, annealed, and inserted into the BglII/HindIII sites of pSUPER.retro.puro vector, following the manufacturer's instructions (Oligoengine, Seattle, WA). These constructs express a 19mer targeting two independent locations within ATF3 mRNA or GFP (control shRNA) mRNA. A retroviral packaging cell line (RetroPack PT67; Clontech Laboratories, Mountain View, CA) was used for stable virus production according to the manufacturer's instructions. Briefly, packaging cells were transfected with ATF3-shRNA plasmid no. 1 or 2 or GFP-shRNA using FuGENE HD (Roche). After generation of stable clones and determination of viral titer, A549 cells were infected with viral supernatant using 4 µg/ml polybrene. Stable transfected clones expressing shRNA were selected using 3 µg/ml puromycin.
Western Blot Analysis
Cells plated at 0.7 x 106 per 60-mm dish were allowed to grow overnight and treated with the indicated drug for 24 hours. Protein samples were collected in RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM sodium chloride, 1 mM EDTA, 1% Triton X-100, 0.25% sodium deoxycholate, 0.1% SDS) containing 50 mM sodium fluoride, 1 mM sodium orthovanadate, 10 mM β-glycerophosphate, and 1x protease inhibitor cocktail (Sigma). Protein concentrations were assayed using a protein assay (Bio-Rad, Mississauga, Ontario, Canada) and a spectrophotometer (Biomate 3; Thermo Fisher Scientific, Waltham, MA). Protein extracts representing 60 µg were separated on a 12% SDS-PAGE gel and electrophoretically transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Billerica, MA). Membranes were blocked in 5% skim milk powder in Tris-buffered saline containing 10% Tween-20 (TBS-T) for 1 hour at room temperature followed by incubation with primary antibody diluted in 5% skim milk in TBS-T with shaking overnight at 4°C. Polyclonal antibody ATF3, ERK, and phospho-ERK (Tyr204) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal antiactin was purchased from Sigma-Aldrich, and monoclonal anti-p53 (Ab-6) was from Calbiochem (San Diego, CA). Polyclonal antibodies Jun, phospho-Jun (Ser73), p38, phospho-p38 (Thr180/Tyr182), and poly (ADP-ribose) polymerase (PARP) were purchased from Cell Signaling Technology (Beverly, MA). Polyclonal antibodies against heat shock protein 27 (hsp27) and phospho-hsp27 (Ser78) were purchased from Stessgen (Ann Arbor, MI). After washing in TBS-T, blots were incubated with the appropriate HRP-labeled secondary antibody for 1 hour at room temperature. Visualization of protein bands was performed using the SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL) exposed on a film (Kodak, Toronto, Canada) in a tabletop processor (SRX-101A; Konica Minolta, Mississauga, Canada).
RNA Isolation and Reverse Transcription-Polymerase Chain Reaction
MCF-7 cells plated at 0.8 x 106 cells per 10-cm dish were incubated at 37°C overnight. The next day, cells were treated with cisplatin, in the absence or presence of the three MAPK inhibitors used in this study, for 24 hours. Total RNA was extracted from cell samples using the RNeasy Kit (Qiagen, Mississauga, Canada). RNA concentrations were quantified using a spectrophotometer (ND-1000; NanoDrop Technologies, Inc, Wilmington, DE). One microgram of total RNA was reverse-transcribed to complementary DNA for quantitative real-time reverse transcription-polymerase chain reaction (RT-PCR) as previously described [30]. The AB 7500 real-time RT-PCR system (Applied Biosystems, Foster City, CA) was used to detect amplification. The real-time PCR was carried out in a total volume of 25 µl that contained 2.5 µl of synthesized complementary DNA (42 ng), 1.25 µl of TaqMan Gene Expression Assay Primer/Probe (20x; ATF3, HS00231069; Applied Biosystems), 12.5 µl of TaqMan Universal PCR Master Mix (2x; 4304437; Applied Biosystems), and 8.75 µl of RNase-free water for ATF3 expression. The endogenous control for ATF3 was the housekeeping gene, human GAPDH (20x; HS4333764-F; Applied Biosystems). Amplification conditions were 95°C for 5 minutes and 40 PCR cycles at 95°C for 15 seconds and at 60°C for 1 minute. Three independent experiments were performed to determine the mean and SD of gene expression.
Results
Cisplatin and Carboplatin Cytotoxicity Is Associated with ATF3 Induction
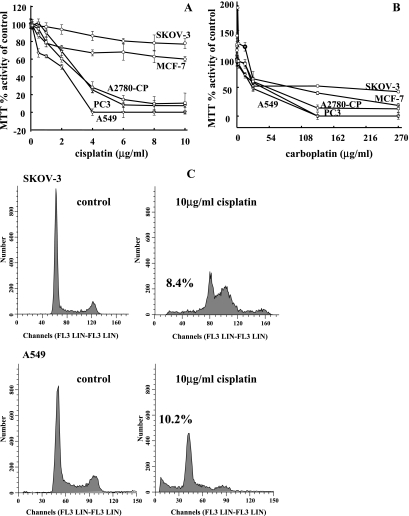
We analyzed the cytotoxic effects of cisplatin and carboplatin treatment on a number of human tumor cell lines, MCF-7 (breast adenocarcinoma), A549 (lung carcinoma), SKOV-3 (ovarian carcinoma), PC3 (prostate carcinoma), and A2780-cp (ovarian carcinoma), using the MTT cell viability assay and flow cytometry (Figure 1). In A549, PC3, and A2780-cp cell lines, higher doses of cisplatin lead to 100% cytotoxicity (Figure 1A), whereas the MCF-7 and SKOV-3 cell lines seemed more resistant as determined by the MTT cell viability assay. Resistance to the cytotoxic effects of cisplatin in the MCF-7 and SKOV-3 cell lines was evidenced by a plateau patterning at higher doses of the treatment, a pattern previously suggested to be related to deficiency in proapoptotic factors [2] (Figure 1A). Indeed, the MCF-7 cell line is caspase-3-deficient [31]. Therefore, the observed resistance to cisplatin in the MCF-7 cell line may be mediated by a blockage in downstream apoptotic pathways in which the platinum-based drug acts. Carboplatin also induced cell cytotoxicity in all cell lines where MCF-7, SKOV-3, and PC3 cell lines displayed the greatest resistance to the cytotoxic effects (Figure 1B). Interestingly, SKOV-3 and PC3 cell lines are functionally null for the tumor suppressor p53, which may contribute to their resistance. As expected, carboplatin was less cytotoxic at the same concentrations compared with cisplatin treatment. We further used flow cytometry analyses to determine the potential for apoptosis induction by the platinum-based chemotherapeutics in the A549 and SKOV-3 cell lines. Apoptosis was visualized as a sub-2N peak that identifies apoptotic bodies resulting from cellular fragmentation [32,33]. Cisplatin treatment (10 µg/ml for 48 hours) resulted in 10.19% and 8.42% of cells in the sub-2N fraction in A549 and SKOV-3 cell lines, respectively (Figure 1C). Differences between percentage of apoptotic cells observed between MTT assay and flow cytometry assay are largely due to the qualitative nature for the flow cytometry method [33]. Carboplatin treatment (270 µg/ml for 48 hours) resulted in 23.44% and 11.33% of cells in sub-2Nin A549 and SKOV-3 cell lines, respectively (data not shown).
Figure 1.
Cisplatin and carboplatin treatment induce cytotoxicity and apoptosis in a panel of human cancer cell lines. Cell lines SKOV-3, MCF-7, A2780-cp, PC3, and A549 were treated with cisplatin (A) or carboplatin (B) for 48 hours, and cell viability was assessed as measured by MTT activity. Data are represented as a percentage of MTT activity where untreated cells were taken to be 100%. Error bars are representative of six individual treated samples. (C) Flow cytometry analysis of SKOV-3 and A549 cell lines treated with cisplatin for 48 hours.
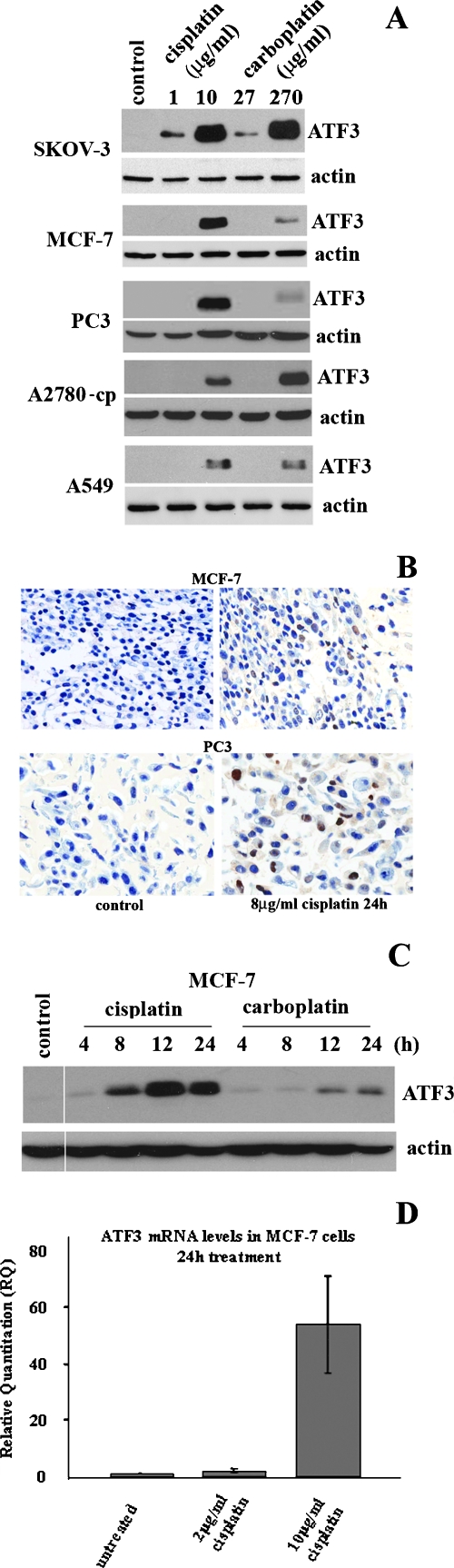
Previously, our laboratory had identified lovastatin, a potent inhibitor of mevalonate synthesis, as an inducer of the ISR pathway and subsequent mediator of lovastatin-induced apoptosis [34]. Downstream effectors of the ISR pathway activated by lovastatin included members of the activating transcription factor (ATF) family, ATF4 and ATF3. A role for the stress-inducible gene, ATF3, in tumorigenesis has been demonstrated and can act either as a tumor suppressor or as an oncogene depending on cell context. Because various stress pathways that induce ATF3 expression have also been shown to regulate cytotoxicity, we first evaluated the potential of cisplatin and carboplatin to affect ATF3 expression. Indeed, we found that ATF3 was significantly induced at the protein level when treated for 24 hours with cytotoxic concentrations of cisplatin (10 µg/ml) and carboplatin (270 µg/ml) in a panel of human cancer cell lines (Figure 2A). These higher concentrations of drug treatment induced significant cytotoxicity at 48-hour treatments as demonstrated by MTT assay analysis in all cell lines evaluated (Figure 1A). The noncytotoxic doses of each drug tested (cisplatin, 1 µg/ml; carboplatin, 2.7 µg/ml) produced either a weak or an undetectable induction of ATF3 (Figure 2A). Immunocytochemical analysis of ATF3 expression in cisplatin (8 µg/ml)-treated MCF-7 and PC3 cells for 24 hours showed that, although untreated cells did not express ATF3, a significant proportion of cisplatin-treated cells showed expression and nuclear localization of this transcription factor (Figure 2B). Time course analysis of ATF3 induction by cisplatin (10 µg/ml) and carboplatin (270 µg/ml) revealed maximal induction levels occurring at 12 and 24 hours in the MCF-7 cell line (Figure 2C). Furthermore, this cytotoxic cisplatin dose could specifically induce the levels of ATF3 mRNA (Figure 2D). In summary, ATF3 is highly induced at the protein and mRNA levels by cytotoxic doses of these agents in human cancer-derived cell lines.
Figure 2.
ATF3 is induced by cisplatin and carboplatin. (A) ATF3 protein expression levels after treatment with low and cytotoxic doses of cisplatin (1 and 10 µg/ml) and carboplatin (27 and 270 µg/ml) in SKOV-3, MCF-7, PC3, A2780-cp, and A549 cell lines. (B) Immunocytochemistry analysis of ATF3 expression (brown) in MCF-7 and PC3 cell lines in nontreated cells (control) and cisplatin treatment for 24 hours. Methylene blue stain is used as a nuclear counterstain. (C) Time course analysis of ATF3 expression in MCF-7 cells treated with cisplatin (10 µg/ml) or carboplatin (270 µg/ml) at the 4-, 8-, 12-, and 24-hour time points. (D) ATF3 mRNA quantified by RT-PCR in MCF-7 cells untreated, treated with 2 and 10 µg/ml cisplatin for 24 hours. Error bars are representative of quantified mRNA from three independent experiments. In all blots, actin is used as a loading control.
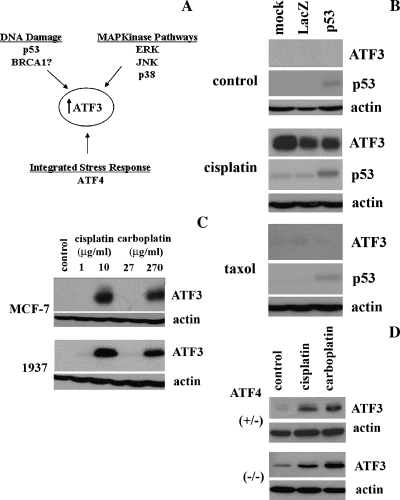
Induction of ATF3 by Cisplatin Is Independent of a p53, BRCA1, or ISR Mechanism
ATF3 mRNA and protein levels are readily induced by a wide range of stress causing agents [9]. The mechanism(s) of stress-induced ATF3 has been previously well documented (Figure 3A), and in this study, we evaluated these regulatory mechanisms with respect to cisplatin induction of ATF3. Because the tumor suppressor p53 had been previously implicated in ATF3 regulation [22,23], we determined its role in cisplatin induction of ATF3. ATF3 was induced by cisplatin in the p53 functionally null cell lines SKOV-3 and PC3, which suggested a p53-independent mechanism (Figure 1A). To further investigate a possible regulatory role, we virally expressed p53 in the PC3 cell line, treated the cells with cisplatin or taxol, a microtubule-depolarizing agent, and determined ATF3 expression levels. ATF3 induction by cisplatin (10 µg/ml for 24 hours) was unchanged between LacZ vector control and p53-containing viral infection under cisplatin treatment, further confirming a p53-independent mechanism (Figure 3B, middle panel). Taxol treatment (25 µM for 24 hours) had no effect on ATF3 expression levels (Figure 3B, bottom panel). We next looked at the possible involvement of the DNA damage response factor, BRCA1, in the regulation of ATF3 induction by platinum-based chemodrugs because previous reports suggested that BRCA1 could transcriptionally regulate ATF3 expression [17,24]. To determine whether BRCA1 played a role in ATF3 induction by cisplatin or carboplatin, ATF3 induction was contrasted in the breast adenocarcinoma cell lines expressing and null for BRCA1, MCF-7, and 1937, respectively. As previously demonstrated [35], we also confirmed the differential expression of BRCA1 in these cell lines and also showed that 1937 cells are sensitive to cisplatin and carboplatin at the higher cytotoxic doses used (data not shown). As shown in Figure 3C, no difference in ATF3 induction levels was observed between the two cell lines with either treatment, suggesting that induction of ATF3 by the chemodrugs is independent of BRCA1 expression. Next, we evaluated the role of the ISR pathway in mediating ATF3 induction because ATF3 is a downstream effector of the pathway [25]. We tested the ability of cisplatin and carboplatin to induce ATF3 expression in immortalized heterozygous or ATF4 null MEFs, the upstream inducer of ATF3 expression in the ISR pathway. Figure 3D demonstrates the absence of ATF4 had no affect on ATF3 induced by cisplatin and carboplatin, suggesting an ISR independent mechanism as well. Lower induction levels of ATF3 in the ATF4 MEFs compared with cell lines analyzed in Figure 2A is likely due to the lower doses of drug used (cisplatin, 1 µg/ml; carboplatin, 2.7 µg/ml) owing to the enhanced sensitivity of these MEFs to their cytotoxicity.
Figure 3.
ATF3 induction by cisplatin is independent of a p53, BRCA1, and ISR mechanism. (A) Schematic representation of the potential mechanisms involved in ATF3 induction by cisplatin. (B) ATF3 detection in PC3 cells transduced with no virus (mock), LacZ vector control (LacZ), or p53wt (p53) containing adenovirus for 6 hours after no treatment (control; top panel) or after treatment with cisplatin (10 µg/ml; middle panel) or taxol (25 µM; bottom panel) for 24 hours. (C) ATF3 expression detected in MCF-7 and 1937 (BRCA1 null) cells untreated (control) or treated with cisplatin (1 and 10 µg/ml) or carboplatin (27 and 270 µg/ml) for 24 hours. (D) ATF3 detection in ATF4-/- and ATF4+/- MEFs untreated (control) or treated with cisplatin (1 µg/ml) and carboplatin (27 µM) for 24 hours. In all blots, actin is used as a loading control.
MAPK Pathways Regulate ATF3 Induction by Cisplatin
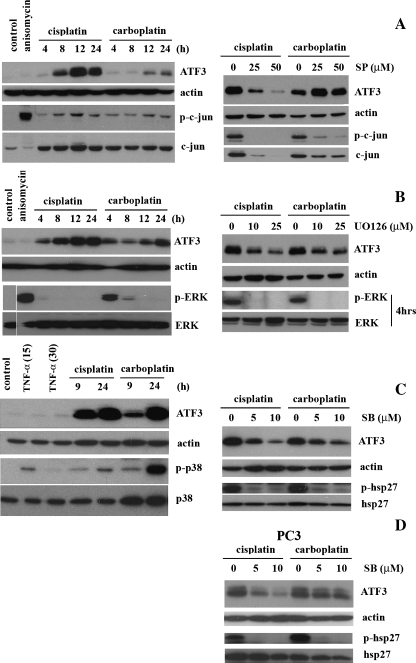
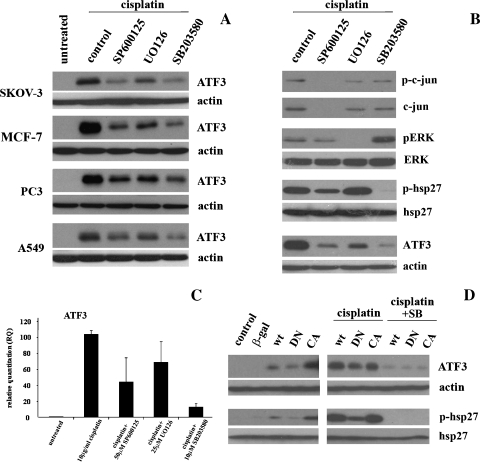
Recent characterization of ATF3 induction by anisomycin, an antibiotic that activates multiple signaling pathways, revealed a MAPK-dependent mechanism [27]; therefore, we investigated the individual MAPK pathways for potential regulation of ATF3 expression by platinum-based chemodrugs. First, we determined whether the pathways were activated under the drug treatments in the MCF-7 cell line. Time course analysis of JNK pathway activation after treatment with platinum-based chemodrugs as measured by the phosphorylation status of c-Jun, a downstream effector of the JNK pathway cascade, revealed a slight increase in phosphoc-Jun at 12 and 24 hours, which coincided with the maximal ATF3 induction by both cisplatin and carboplatin (Figure 4A, left panel). To determine the role of the JNK pathway in ATF3 induction by cisplatin (10 µg/ml) and carboplatin (270 µg/ml), MCF-7 cells were treated with cisplatin or carboplatin for 24 hours in the presence of a JNK-specific inhibitor, JNK inhibitor II SP600125 (SP), which inhibited this pathway as measured by phospho-c-Jun levels (Figure 4A, right panel). ATF3 induction levels were found to be reduced under cisplatin treatment in the presence of JNK inhibitor, but not with carboplatin, in MCF-7 cells (Figure 4A, right panel).
Figure 4.
ATF3 induction by cisplatin is mediated by MAPK pathways in MCF-7 cells. (A) MCF-7 cells without treatment (control) and those treated with anisomycin (20 µM) for 1 hour and cisplatin (10 µg/ml) or carboplatin (270 µg/ml) for the 4-, 8-, 12-, and 24-hour time points were analyzed by Western blot analysis for the detection of ATF3, actin, phospho-c-jun (p-c-jun), and total c-jun (left panel). MCF-7 cells similarly treated with cisplatin or carboplatin in the absence (0) or presence of JNK pathway inhibitor (SP; 25 and 50 µM) for 24 hours were analyzed for the detection of ATF3, actin, p-c-jun, and total c-jun (right panel). (B) MCF-7 cells without treatment (control) and those treated with anisomycin (20 µM) for 1 hour and cisplatin or carboplatin as previously mentioned for the 4-, 8-, 12-, and 24-hour time points were analyzed by Western blot analysis for the detection of ATF3, actin, phospho-ERK (p-ERK), and total ERK (left panel). MCF-7 cells treated with cisplatin or carboplatin in the absence (0) or presence of ERK pathway inhibitor (UO126; 10 and 25 µM) for 24 hours were analyzed for the detection of ATF3 and actin and at 4 hours for p-ERK and total ERK (right panel). (C) MCF-7 cells untreated (control), treated with tumor necrosis factor α (20 ng/ml) for 15 and 30 minutes, and cisplatin or carboplatin for the 9- and 24-hour time points were analyzed by Western blot analysis for the detection of ATF3, actin, phospho-p38 (p-p38), and total p38 (left panel). MCF-7 cells treated with cisplatin or carboplatin in the absence (0) or presence of p38 inhibitor, SB203580 (SB; 5 and 10 µM), for 24 hours were analyzed for the detection of ATF3, actin, phospho-hsp27 (p-hsp27), and total hsp27 (right panel). (D) PC3 cells treated with cisplatin (10 µg/ml) or carboplatin (270 µg/ml) in the absence (0) or presence of p38 inhibitor, SB203580 (SB; 5 and 10 µM), for 24 hours were analyzed for the detection of ATF3, actin, phospho-hsp27 (p-hsp27), and total hsp27.
Next, we evaluated the role of the ERK pathway in ATF3 induction by cisplatin and carboplatin. Time course treatment with chemodrugs revealed activation of the ERK pathway, as measured by the phosphorylation status of ERK, at 4 hours under cisplatin, and 4 and 8 hours under carboplatin treatment (Figure 4B, left panel). Phosphorylated ERK was not detected at 12 and 24 hours under either treatment (Figure 4A, left panel). To determine the role of the ERK pathway in ATF3 induction by cisplatin and carboplatin, MCF-7 cells were treated with cisplatin or carboplatin in the presence of the specific inhibitor to the ERK pathway, UO126, which was effective in blocking of the pathway as measured by phospho-ERK levels (Figure 4B, right panel). Treatment with ERK inhibitor in the presence of cisplatin and carboplatin revealed a dose-dependent decrease in ATF3 expression levels, suggesting a role for the pathway in mediating induction by the chemodrugs (Figure 4B, right panel).
Lastly, we determined the activation of the p38 pathway in MCF-7 cells after treatment with cisplatin or carboplatin at 9- and 24-hour time points. Indeed, it was observed that the pathway was active at these time points as measured by the phosphorylation status of p38, which correlated with ATF3 induction levels (Figure 4C, left panel). To investigate the role of p38 pathway in the induction of ATF3 by cisplatin and carboplatin, MCF-7 cells were treated with chemodrugs in the presence of the specific p38 inhibitor, SB203580 (SB), for 24 hours. This p38 inhibitor was shown to effectively block the pathway as measured by the phosphorylation status of hsp27, a downstream target of p38 (Figure 4C, right panel). ATF3 expression levels were also shown to be decreased by the inhibitor in a dose-dependent manner suggesting a regulatory role of the p38 pathway in chemodrug induction of ATF3 (Figure 4C, right panel3). Similarly, inhibition of the p38 pathway also resulted in the effective blockage of the ATF3 induction by cisplatin and carboplatin in the PC3 cell line (Figure 4D).
We similarly characterized the involvement of the MAPK pathways in cisplatin-induced ATF3 expression in the tumor-derived cells lines SKOV-3, MCF-7, PC3, and A459. These inhibitors against the three MAPK pathways evaluated had variable degrees of inhibition of ATF3 induction by cisplatin (10 µg/ml for 24 hours), implicating all three pathways in the mechanistic induction of ATF3 by cisplatin (Figure 5A). Interestingly, the pattern of reduced ATF3 induction by cisplatin in the presence of MAPK inhibitors was consistent between all four cancer cell lines, with the p38 pathway inhibitor showing the greatest inhibition of ATF3 induction and the ERK pathway inhibitor showing the least (Figure 5A). To determine whether the MAPK pathway inhibitors used in this study were specific, cisplatin-induced (10 µg/ml for 24 hours) activation of each pathway in the presence of these specific inhibitors was assessed. MCF-7 cells were treated with cisplatin alone, or in combination with JNK pathway inhibitor, SP600125 (50 µM), ERK inhibitor, UO126 (25 µM), or p38 pathway inhibitor, SB203580 (10 µM), and activation of the pathways was assessed by phosphorylation status of c-jun (JNK pathway), ERK, and hsp27 (p38 pathway; Figure 5B). Each pathway was inhibited by its respective inhibitor without significantly affecting the activation of the other pathways confirming their specificity (Figure 5B). We also demonstrate that the coadministration of all three MAPK inhibitors used in this study could downregulate 10 µg/ml cisplatin-induced ATF3 expression at the mRNA level in a pattern of inhibition that was similar to that observed at the protein level at 24 hours (Figure 5C).
Figure 5.
Induction of ATF3 by cisplatin is mediated by MAPK pathways in a panel of human cancer cells. (A) SKOV-3, MCF-7, PC3, and A549 cells untreated and treated with cisplatin (10 µg/ml) for 24 hours in the absence (control) or presence of MAPK pathway inhibitors (SP600125 [50 µM], UO126 [25 µM], and SB203580 [10 µM]) and analyzed by Western blot analysis for ATF3 and actin. (B) MCF-7 cells treated with cisplatin (10 µg/ml) for 24 hours in the absence (control) or presence of MAPK pathway inhibitors (SP600125 [50 µM], UO126 [25 µM], and SB203580 [10 µM]) and analyzed by Western blot analysis for p-c-jun, c-jun total, p-ERK, ERK total, p-hsp27, hsp27 total, ATF3, and actin. (C) ATF3 mRNA quantified by RT-PCR in MCF-7 cells untreated or treated with cisplatin (10 µg/ml) or cisplatin in the presence of SP (50 µM), UO126 (25 µM), and SB (10 µM) for 24 hours. Error bars are representative of three independent experiments. (D) MCF-7 cells transiently transfected with 2 µg of β-galactosidase (β-gal), p38-WT, p38-DN, and p38-CA, for 24 hours followed (left panel) by no treatment or by treatment with cisplatin (10 µg/ml) alone or in combination with p38 inhibitor, SB (10 µM), for an additional 24 hours (right panel) and analyzed by Western blot analysis for ATF3, actin, p-hsp27, and hsp27 total.
To further define the role of the p38 pathway in ATF3 induction by cisplatin, the MCF-7 cell line was transiently transfected with various p38 constructs (WT, DN, and CA) for 24 hours followed by no treatment (left panel) or 10 µg/ml cisplatin in the absence or presence of 10 µM SB203580 (right panel), for an additional 24 hours. Untransfected (control) and a β-galactosidase-expressing plasmid were used as negative controls (Figure 5D). In untreated samples, activation of the pathway as measured by phospho-hsp27 was highest in p38-CA-transfected cells and lowest in the p38-DN-transfected cells, confirming functionality of the constructs. ATF3 expression levels were also increased on p38-CA transfection, suggesting that activation of the p38 pathway is sufficient to induce ATF3 expression (Figure 5D, left panel). After cisplatin treatment, phospho-hsp27 was induced, at greater levels, which is consistent with activation of the pathway by the drug (Figure 4C, left panel), where p38-DN-transfected cells decreased expression of both p-hsp27 and ATF3 as compared with p38-WT- and p38-CA-transfected cells (Figure 5D, right panel). Cisplatin treatment in the presence of the p38 inhibitor resulted in the abolishment of phospho-hsp27 and ATF3 expressions irrespective of transfection status (Figure 5D, right panel). These data provide further evidence that ATF3 induction by cisplatin is mediated through MAPK pathway activation. Taken together, these results identify the MAPK pathways as regulators of ATF3 induction by platinum-based cytotoxic drugs.
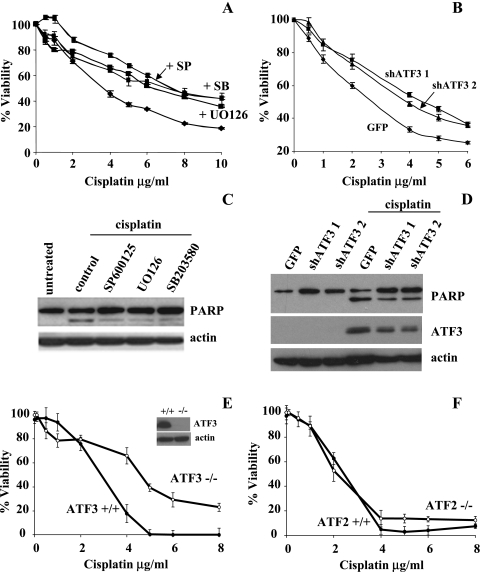
ATF3 Regulates, in Part, the Cytotoxic Effects of Cisplatin
Because ATF3 has been previously shown to play a proapoptotic role in cancer models, we investigated the role of ATF3 induction by cisplatin in regulating the cytotoxic effects of cisplatin. Treating A549 cells with increasing concentrations of cisplatin in the presence or absence of SP600125 (50 µM), U0126 (25 µM), or SB203580 (10 µM), the cytotoxic effects of cisplatin (48 h) was observed to be attenuated by approximately 20% as determined by the MTT cell viability assay (Figure 6A). Similar results were obtained in the PC3 cell line for the SB203580 inhibitor (data not shown). To further demonstrate ATF3's role as a factor in the cytotoxic effects of cisplatin, the stable expression of shRNA against two different sequences of the ATF3-mRNA and to green fluorescent protein (GFP) as a negative control was used in the A549 cell line. Cells expressing both shATF3 and treated with cisplatin showed similar attenuation (20%) of the cytotoxic effects of the drug in the shATF3 cell lines as compared with shGFP control as determined by MTT analysis (Figure 6B). The cleavage status of PARP, a marker of apoptosis, was determined in the A549 cell line after treatment with cisplatin (10 µg/ml for 24 hours) in the presence or absence of the MAPK inhibitors at the previously mentioned concentrations. Indeed, PARP cleavage induced by cisplatin was reduced in the presence of MAPK inhibitors to all three pathways (Figure 6C), correlating with the attenuated cytotoxicity observed in Figure 6A. Likewise, reduced PARP cleavage was also observed in the shATF3 (shATF31 and shATF32) compared with the GFP control cell line (Figure 6D).
Figure 6.
ATF3 expression mediates, in part, the cytotoxic effects of cisplatin. (A) A549 cells treated with cisplatin (0–10 µg/ml) in the presence or absence of 50 µM SP600125, 25 µM UO126, and 10 µM SB203580 for 48 hours were assessed for cell cytotoxicity as measured by MTT activity. (B) A549 cells stably expressing shRNA against two separate ATF3 mRNA regions (shATF3-1 and shATF3-2) and GFP (negative control) were treated with cisplatin for 48 hours and were analyzed for MTT activity. For A and B, data are presented as a percentage of MTT activity where untreated cells were taken to be 100%. Error bars are representative of six independently treated samples. (C) A549 cells without treatment (untreated) or treated with 10 µg/ml cisplatin for 24 hours in the presence of MAPK inhibitors of 50 µM SP600125, 25 µM UO126, and 10 µM SB203580 and analyzed by Western blot analysis for PARP and actin as a loading control. (D) GFP-, shATF3-1-, and shATF3-2-expressing cell lines treated with or without 10 µg/ml cisplatin for 24 hours and analyzed by Western blot analysis for PARP, ATF3, and actin expression. (E) ATF3-/- and ATF3+/+ MEFs treated with cisplatin (0–8 µg/ml) and analyzed for MTT activity. Western blot analysis for ATF3 and actin in ATF3-/- and ATF3+/+ MEFs treated with 10 µg/ml of cisplatin for 24 hours (inset). Actin was used as a loading control. (F) ATF2-/- and ATF2+/+ MEFs treated with cisplatin (0–8 µg/ml) and analyzed for MTT activity.
Lastly, we contrasted the cytotoxic effects of increasing cisplatin treatments on MEF cells expressing and knocked out for ATF3. ATF3+/+ MEFs were more sensitive to cytotoxic effects of cisplatin compared with ATF3-/- MEFs (Figure 6E). Cisplatin treatment induced ATF3 expression in the ATF3+/+ MEFs but not in the ATF3-/- MEFs (Figure 6E, inset). The differences in cytotoxic effects of cisplatin observed in ATF3+/+ and ATF3-/- MEFs were contrasted in MEFs (+/+) and (-/-) for the ATF2 family member. No differences in the effect of cisplatin cytotoxicity were observed between ATF2+/+ and ATF2-/- MEFs (Figure 6F). In all three models where ATF3 expression was targeted, there was approximately a two-fold increase in cisplatin dose required for 50% cytotoxicity. Taken together, this study provides evidence that the cytotoxic effects invoked by cisplatin can in part be correlated to the drug's ability to induce ATF3 expression as regulated by MAPK pathways.
Discussion
In summary, this study has identified the platinum-based chemocytotoxic drug, cisplatin, as an inducer of the stress-inducible gene, ATF3, at both the mRNA and protein levels. Through elimination of potential ATF3-regulatory mechanisms, namely, p53, BRCA1, and ISR, we identified ATF3 induction by cisplatin as regulated by the MAPK pathways JNK, ERK, and p38. Inhibition of the MAPK pathway with the p38 inhibitor SB203580 in cisplatin-treated cells resulted in the greatest decrease in ATF3 induction at the protein level in the human cancer cells analyzed. Increased ATF3 expression was associated with cisplatin-induced cytotoxicity as evidenced by attenuation of ATF3 expression and cytotoxicity with treatment of the three MAPK pathway inhibitors and in shATF3 knockdown cells compared with control. Targeting the activity of p38 with the expression of wild type, DN and CA expression constructs also modulated cisplatin-induced ATF3 expression as the DN version attenuated cisplatin-induced ATF3 expression. Furthermore, ATF3-/- MEFs were more resistant to cisplatin cytotoxicity compared with ATF3+/+ MEFs. Taken together, these results provide strong evidence that ATF3 is a regulator of platinum-based chemotherapeutic-induced cytotoxicity. Identifying ATF3 as a cisplatin-induced gene through MAPK pathway activation may have therapeutic relevance. Inducers of the MAPK pathways or other cell stress pathways that enhance ATF3 expression may augment the cytotoxic effects of cisplatin. For example, two agents that induce ATF3 expression through different mechanisms, salubrinal and proteosome inhibitors, have demonstrated synergistic cytotoxicity in myeloma cells and may represent a novel combinational therapeutic approach [36,37].
It is well characterized that the commencement of the anticancer effects of cisplatin involves DNA adduct formation resulting in apoptotic cell death if the DNA damage cannot be adequately repaired [1]. However, the specific mechanism(s) downstream of cisplatin-induced DNA damage, which leads to the apoptotic response, are poorly defined. In this study, we show that cisplatin and its derivative, carboplatin, could readily induce ATF3 expression. A role for ATF3 in tumorigenesis has been implicated through its ability to affect the transcription of a number of regulators of apoptosis and cell proliferation including CHOP and cyclin D1, respectively [38,39]. Depending on the cell type and the type and severity of the cell stressor, ATF3 has been implicated as both a proto-oncogene or tumor suppressor. For example, overexpression of ATF3 inhibited proliferation and induced cell cycle arrest in human cancer cells [17], whereas loss of ATF3 in a Ras-transformed model resulted in higher proliferation rates and increased G1- to S-phase transition efficiency [18]. Treating our panel of human cancer cell lines with a high and a low dose of cisplatin or carboplatin revealed an increase in ATF3 protein expression that was associated with the high cytotoxic doses of these drugs, implicating a role for ATF3 in regulating platin-induced cytotoxicity. Because ATF3 has been implicated as a biomarker for cell death in cancer models [16–19,40], based on these results, expression levels of ATF3 with respect to platin response in patients should be evaluated.
After the identification of ATF3 as a cisplatin-induced gene, the major objective of this study was to systematically identify the mechanism of induction of ATF3. A number of cellular stress pathways have been shown to regulate the expression of ATF3. The DNA damage response factors p53 and BRCA1 had previously been linked to ATF3 regulation. Likewise, ATF3 is a known downstream effector of the ISR induced by ER, hypoxia, and viral and metabolic stressors [25]. This study showed that BRCA1, p53, and ISR are not factors in ATF3 induction by cisplatin. Instead, the mechanism of induction of ATF3 by cisplatin was found to be largely MAPK pathway-dependent. Looking at the involvement of the ERK, JNK, and p38 pathways, we found that all three pathways, when inhibited, lead to decreased induction of ATF3 by cisplatin. Although the inhibition of ATF3 induction by cisplatin in the presence of MAPK pathway inhibitors was significant, it was not complete suggesting that other regulatory mechanism(s) may exist.
In conclusion, we determined the functional relevance of ATF3 expression in regulating cisplatin-induced cytotoxicity. Recent literature has implicated the MAPK pathways in the regulation of stress-induced ATF3 apoptosis. Similar to our results, that ribotoxic stress induction of ATF3 was shown to be mediated through the p38 pathway, and ATF3 expression was shown as a proapoptotic factor in HeLa cells [27]. Another recent study reported that cisplatin could induce ATF3 in T98G glioblastoma cells at both the protein and mRNA levels [26]. In contrast to our results, this report showed that the induction of ATF3 by cisplatin was antiapoptotic. Discrepancies between our results and others could stem from differences in cellular model or status of cell line malignancy both of which have previously been suggested to determine ATF3's role as an antiapoptotic or a proapoptotic factor [18,19].
The literature has previously reported that cisplatin treatment results in the activation of MAPKs [41]. Reports showing activation of the p38 pathway by cisplatin has been exclusively correlated with proapoptotic outcomes in a number of cell lines, whereas activation of the JNK and ERK pathways is correlated with both anti-death and pro-death outcomes [41]. Inhibition of the p38 pathway with specific inhibitors has been previously shown to increase resistance to cisplatin [42,43]. Likewise, reduced activation of the p38 pathway has been identified as a mechanism correlated with cisplatin resistance [2]. Although the activation of MAPK pathways by cisplatin treatment has been documented, the signaling pathways downstream of activation that determines cell fate are poorly understood. This study identifies ATF3, previously defined as a factor capable of influencing cellular fate, as a novel target of the MAPK pathways when activated by cisplatin treatment. This study suggests that ATF3 induction by cisplatin may identify a novel factor responsible for mediating the established link between cisplatin-induced MAPK pathway activation and cell cytotoxic outcomes. Whether ATF3 is directly activated by the MAPK pathway or is induced downstream of known MAPK pathway transcription factor targets remains to be determined. Defining the specific mechanism(s) responsible for the antitumor affects of cisplatin may lead to novel and improved therapeutic approaches.
Acknowledgments
The authors thank B. Vanderhyden, B. McKay, D. Park, and L. Glimcher for generously providing reagents used in this study.
Abbreviations
- ATF
activating transcription factor
- cisplatin
cis-diamminedichloroplatinum (II)
- ISR
integrated stress response
- MAPK
mitogen-activated protein kinase
- MEF
murine embryonic fibroblast
Footnotes
This study was supported by The Canadian Institute of Health Research (J.D.), the Ottawa Regional Cancer Foundation and the Motorcycle Ride for Dad (Prostate Cancer Fight Foundation) (J.D.) and Ortho-Biotech Canada.
References
- 1.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 2.Stewart DJ. Mechanisms of resistance to cisplatin and carboplatin. Crit Rev Oncol Hematol. 2007;63:12–31. doi: 10.1016/j.critrevonc.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 4.Knox RJ, Friedlos F, Lydall DA, Roberts JJ. Mechanism of cytotoxicity of anticancer platinum drugs: evidence that cis-diamminedichloroplatinum(II) and cis-diammine-(1,1-cyclobutanedicarboxylato)platinum(II) differ only in the kinetics of their interaction with DNA. Cancer Research. 1986;46:1972–1979. [PubMed] [Google Scholar]
- 5.Galanski M. Recent developments in the field of anticancer platinum complexes. Recent Pat Anticancer Drug Discov. 2006;1:285–295. doi: 10.2174/157489206777442287. [DOI] [PubMed] [Google Scholar]
- 6.Manic S, Gatti L, Carenini N, Fumagalli G, Zunino F, Perego P. Mechanisms controlling sensitivity to platinum complexes: role of p53 and DNA mismatch repair. Curr Cancer Drug Targets. 2003;3:21–29. doi: 10.2174/1568009033333727. [DOI] [PubMed] [Google Scholar]
- 7.Sedletska Y, Giraud-Panis MJ, Malinge JM. Cisplatin is a DNA-damaging antitumour compound triggering multifactorial biochemical responses in cancer cells: importance of apoptotic pathways. Curr Med Chem Anticancer Agents. 2005;5:251–265. doi: 10.2174/1568011053765967. [DOI] [PubMed] [Google Scholar]
- 8.Liang G, Wolfgang CD, Chen BP, Chen TH, Hai T. ATF3 gene. Genomic organization, promoter, and regulation. J Biol Chem. 1996;271:1695–1701. doi: 10.1074/jbc.271.3.1695. [DOI] [PubMed] [Google Scholar]
- 9.Hai T, Hartman MG. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene. 2001;273:1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- 10.Hsu JC, Laz T, Mohn KL, Taub R. Identification of LRF-1, a leucine-zipper protein that is rapidly and highly induced in regenerating liver. Proc Natl Acad Sci USA. 1991;88:3511–3515. doi: 10.1073/pnas.88.9.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen BP, Wolfgang CD, Hai T. Analysis of ATF3, a transcription factor induced by physiological stresses and modulated by gadd153/Chop10. Mol Cell Biol. 1996;16:1157–1168. doi: 10.1128/mcb.16.3.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin T, Sandhu G, Wolfgang CD, Burrier A, Webb RL, Rigel DF, Hai T, Whelan J. Tissue-specific pattern of stress kinase activation in ischemic/reperfused heart and kidney. J Biol Chem. 1997;272:19943–19950. doi: 10.1074/jbc.272.32.19943. [DOI] [PubMed] [Google Scholar]
- 13.Takeda M, Kato H, Takamiya A, Yoshida A, Kiyama H. Injury-specific expression of activating transcription factor-3 in retinal ganglion cells and its colocalized expression with phosphorylated c-Jun. Invest Ophthalmol Vis Sci. 2000;41:2412–2421. [PubMed] [Google Scholar]
- 14.Tsujino H, Kondo E, Fukuoka T, Dai Y, Tokunaga A, Miki K, Yonenobu K, Ochi T, Noguchi K. Activating transcription factor 3 (ATF3) induction by axotomy in sensory and motoneurons: a novel neuronal marker of nerve injury. Mol Cell Neurosci. 2000;15:170–182. doi: 10.1006/mcne.1999.0814. [DOI] [PubMed] [Google Scholar]
- 15.Turchi L, Fareh M, Aberdam E, Kitajima S, Simpson F, Wicking C, Aberdam D, Virolle T. ATF3 and p15PAF are novel gatekeepers of genomic integrity upon UV stress. Cell Death Differ. 2009;16:728–737. doi: 10.1038/cdd.2009.2. [DOI] [PubMed] [Google Scholar]
- 16.Bottone FG, Moon Y, Kim JS, Alston-Mills B, Ishibashi M, Eling TE. The anti-invasive activity of cyclooxygenase inhibitors is regulated by the transcription factor ATF3 (activating transcription factor 3) Mol Cancer Ther. 2005;4:693–703. doi: 10.1158/1535-7163.MCT-04-0337. [DOI] [PubMed] [Google Scholar]
- 17.Fan F, Jin S, Amundson SA, Tong T, Fan W, Zhao H, Zhu X, Mazzacurati L, Li X, Petrik KL, et al. ATF3 induction following DNA damage is regulated by distinct signaling pathways and over-expression of ATF3 protein suppresses cells growth. Oncogene. 2002;21:7488–7496. doi: 10.1038/sj.onc.1205896. [DOI] [PubMed] [Google Scholar]
- 18.Lu D, Wolfgang CD, Hai T. Activating transcription factor 3, a stressinducible gene, suppresses Ras-stimulated tumorigenesis. J Biol Chem. 2006;281:10473–10481. doi: 10.1074/jbc.M509278200. [DOI] [PubMed] [Google Scholar]
- 19.Yin X, Dewille JW, Hai T. A potential dichotomous role of ATF3, an adaptive-response gene, in cancer development. Oncogene. 2008;27:2118–2127. doi: 10.1038/sj.onc.1210861. [DOI] [PubMed] [Google Scholar]
- 20.Syed V, Mukherjee K, Lyons-Weiler J, Lau KM, Mashima T, Tsuruo T, Ho SM. Identification of ATF-3, caveolin-1, DLC-1, and NM23-H2 as putative antitumorigenic, progesterone-regulated genes for ovarian cancer cells by gene profiling. Oncogene. 2005;24:1774–1787. doi: 10.1038/sj.onc.1207991. [DOI] [PubMed] [Google Scholar]
- 21.Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 22.Buganim Y, Kalo E, Brosh R, Besserglick H, Nachmany I, Rais Y, Stambolsky P, Tang X, Milyavsky M, Shats I, et al. Mutant p53 protects cells from 12-O-tetradecanoylphorbol-13-acetate-induced death by attenuating activating transcription factor 3 induction. Cancer Res. 2006;66:10750–10759. doi: 10.1158/0008-5472.CAN-06-0916. [DOI] [PubMed] [Google Scholar]
- 23.Zhang C, Gao C, Kawauchi J, Hashimoto Y, Tsuchida N, Kitajima S. Transcriptional activation of the human stress-inducible transcriptional repressor ATF3 gene promoter by p53. Biochem Biophys Res Commun. 2002;297:1302–1310. doi: 10.1016/s0006-291x(02)02382-3. [DOI] [PubMed] [Google Scholar]
- 24.Harkin DP, Bean JM, Miklos D, Song YH, Truong VB, Englert C, Christians FC, Ellisen LW, Maheswaran S, Oliner JD, et al. Induction of GADD45 and JNK/SAPK-dependent apoptosis following inducible expression of BRCA1. Cell. 1999;97:575–586. doi: 10.1016/s0092-8674(00)80769-2. [DOI] [PubMed] [Google Scholar]
- 25.Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 26.Hamdi M, Popeijus HE, Carlotti F, Janssen JM, van der Burgt C, Cornelissen-Steijger P, van de Water B, Hoeben RC, Matsuo K, van Dam H. ATF3 and Fra1 have opposite functions in JNK- and ERK-dependent DNA damage responses. DNA Repair (Amst) 2008;7:487–496. doi: 10.1016/j.dnarep.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 27.Lu D, Chen J, Hai T. The regulation of ATF3 gene expression by mitogen-activated protein kinases. Biochem J. 2007;401:559–567. doi: 10.1042/BJ20061081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bragado P, Armesilla A, Silva A, Porras A. Apoptosis by cisplatin requires p53 mediated p38α MAPK activation through ROS generation. Apoptosis. 2007;12:1733–1742. doi: 10.1007/s10495-007-0082-8. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez-Prieto R, Rojas JM, Taya Y, Gutkind JS. A role for the p38 mitogen-activated protein kinase pathway in the transcriptional activation of p53 on genotoxic stress by chemotherapeutic agents. Cancer Res. 2000;60:2464–2472. [PubMed] [Google Scholar]
- 30.Kroupis C, Stathopoulou A, Zygalaki E, Ferekidou L, Talieri M, Lianidou ES. Development and applications of a real-time quantitative RT-PCR method (QRT-PCR) for BRCA1 mRNA. Clin Biochem. 2005;38:50–57. doi: 10.1016/j.clinbiochem.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 31.Janicke RU. MCF-7 breast carcinoma cells do not express caspase-3. Breast Cancer Res Treat. 2009;117:219–221. doi: 10.1007/s10549-008-0217-9. [DOI] [PubMed] [Google Scholar]
- 32.Dimitroulakos J, Ye LY, Benzaquen M, Moore MJ, Kamel-Reid S, Freedman MH, Yeger H, Penn LZ. Differential sensitivity of various pediatric cancers and squamous cell carcinomas to lovastatin-induced apoptosis: therapeutic implications. Clin Cancer Res. 2001;7:158–167. [PubMed] [Google Scholar]
- 33.Piacentini M, Fesus L, Melino G. Multiple cell cycle access to the apoptotic death programme in human neuroblastoma cells. FEBS Lett. 1993;320:150–154. doi: 10.1016/0014-5793(93)80081-5. [DOI] [PubMed] [Google Scholar]
- 34.Niknejad N, Morley M, Dimitroulakos J. Activation of the integrated stress response regulates lovastatin-induced apoptosis. J Biol Chem. 2007;282:29748–29756. doi: 10.1074/jbc.M705859200. [DOI] [PubMed] [Google Scholar]
- 35.Tassone P, Di Martino MT, Ventura M, Pietragalla A, Cucinotto I, Calimeri T, Bulotta A, Neri P, Caraglia M, Tagliaferri P. Loss of BRCA1 function increases the antitumor activity of cisplatin against human breast cancer xenografts in vivo. Cancer Biol Ther. 2009;8:648–653. doi: 10.4161/cbt.8.7.7968. [DOI] [PubMed] [Google Scholar]
- 36.Drexler HC. Synergistic apoptosis induction in leukemic cells by the phosphatase inhibitor salubrinal and proteasome inhibitors. PLoS One. 2009;4:e4161. doi: 10.1371/journal.pone.0004161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schewe DM, Aguirre-Ghiso JA. Inhibition of eIF2α dephosphorylation maximizes bortezomib efficiency and eliminates quiescent multiple myeloma cells surviving proteasome inhibitor therapy. Cancer Res. 2009;69:1545–1552. doi: 10.1158/0008-5472.CAN-08-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allan AL, Albanese C, Pestell RG, LaMarre J. Activating transcription factor 3 induces DNA synthesis and expression of cyclin D1 in hepatocytes. J Biol Chem. 2001;276:27272–27280. doi: 10.1074/jbc.M103196200. [DOI] [PubMed] [Google Scholar]
- 39.Wolfgang CD, Chen BP, Martindale JL, Holbrook NJ, Hai T. gadd153/Chop10, a potential target gene of the transcriptional repressor ATF3. Mol Cell Biol. 1997;17:6700–6707. doi: 10.1128/mcb.17.11.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyazaki K, Inoue S, Yamada K, Watanabe M, Liu Q, Watanabe T, Adachi MT, Tanaka Y, Kitajima S. Differential usage of alternate promoters of the human stress response gene ATF3 in stress response and cancer cells. Nucleic Acids Res. 2009;37:1438–1451. doi: 10.1093/nar/gkn1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brozovic A, Osmak M. Activation of mitogen-activated protein kinases by cisplatin and their role in cisplatin-resistance. Cancer Lett. 2007;251:1–16. doi: 10.1016/j.canlet.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 42.Losa JH, Parada Cobo C, Viniegra JG, Sanchez-Arevalo Lobo VJ, Ramon y Cajal S, Sanchez-Prieto R. Role of the p38 MAPK pathway in cisplatin-based therapy. Oncogene. 2003;22:3998–4006. doi: 10.1038/sj.onc.1206608. [DOI] [PubMed] [Google Scholar]
- 43.Olson JM, Hallahan AR. p38 MAP kinase: a convergence point in cancer therapy. Trends Mol Med. 2004;10:125–129. doi: 10.1016/j.molmed.2004.01.007. [DOI] [PubMed] [Google Scholar]