Abstract
TMPRSS2-ERG gene fusions occur in 50% of prostate cancers and result in the overexpression of a chimeric fusion transcript that encodes a truncated ERG product. Previous attempts to detect truncated ERG products have been hindered by a lack of specific antibodies. Here, we characterize a rabbit anti-ERG monoclonal antibody (clone EPR 3864; Epitomics, Burlingame, CA) using immunoblot analysis on prostate cancer cell lines, synthetic TMPRSS2-ERG constructs, chromatin immunoprecipitation, and immunofluorescence. We correlated ERG protein expression with the presence of ERG gene rearrangements in prostate cancer tissues using a combined immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) analysis. We independently evaluated two patient cohorts and observed ERG expression confined to prostate cancer cells and high-grade prostatic intraepithelial neoplasia associated with ERG-positive cancer, as well as vessels and lymphocytes (where ERG has a known biologic role). Image analysis of 131 cases demonstrated nearly 100% sensitivity for detecting ERG rearrangement prostate cancer, with only 2 (1.5%) of 131 cases demonstrating strong ERG protein expression without any known ERG gene fusion. The combined pathology evaluation of 207 patient tumors for ERG protein expression had 95.7% sensitivity and 96.5% specificity for determining ERG rearrangement prostate cancer. In conclusion, this study qualifies a specific anti-ERG antibody and demonstrates exquisite association between ERG gene rearrangement and truncated ERG protein product expression. Given the ease of performing IHC versus FISH, ERG protein expression may be useful for molecularly subtyping prostate cancer based on ERG rearrangement status and suggests clinical utility in prostate needle biopsy evaluation.
Introduction
Vanaja et al. [1] first reported the overexpression of the oncogene ERG (v-ets erythroblastosis virus E26 oncogene homolog [avian], chromosome 21q22.3) at the transcript level in 50% of clinically localized and metastatic prostate cancer samples. Shortly after, Tomlins et al. [2] demonstrated that the basis for this overexpression was due to a recurrent gene rearrangement involving the 5′ untranslated region of the androgen-regulated TMPRSS2 gene with ETS family members, either ERG or ETV1 (ets variant 1, chromosome 7p21.3). Numerous independent studies confirmed the existence of ETS gene rearrangements in prostate cancer, with TMPRSS2-ERG gene fusion being the most common variant, seen in approximately 50% of all prostate-specific antigen screened prostate cancers detected in the United States [3–5]. Subsequent works have demonstrated that ERG can be rearranged and fused with SLC45A3 [6,7] or NDRG1 [7,8], accounting for approximately 5% of the ERG-overexpressing prostate cancers. The gene fusion event occurs early in prostate cancer development, with approximately 15% of the precursor lesion, high-grade prostatic intraepithelial neoplasia (HG PIN), demonstrating ERG rearrangement [9–11], however, only in immediate association with similarly ERG-rearranged cancer. Gene fusions are not observed in benign prostate epithelial glands, atrophy, or stroma as demonstrated by Perner et al. [11] and additional studies using fluorescence in situ hybridization (FISH) to determine ERG rearrangement status [4,5]. TMPRSS2-ERG gene fusions result in a truncated ERG protein product, which has been difficult to characterize given a lack of specific anti-ERG antibodies for in vitro and in situ applications. Here, we qualify a novel rabbit anti-ERG monoclonal antibody (clone EPR 3864; Epitomics, Burlingame, CA) and demonstrate exquisite concordance between ERG protein expression and the presence of ERG gene rearrangements in prostate cancer using a combined IHC and FISH analysis. Given the ease of performing IHC compared with FISH, our results demonstrating uniform ERG protein expression in most tumor cells with ERG gene fusions, but not adjacent benign prostate tissue or stroma, suggest diagnostic utility.
Materials and Methods
Cohort Description and Tissue Microarray Construction
The clinical cohorts studied consisted of 131 men from Weill Cornell Medical College (WCMC) and 79 men from the University of Michigan (UM) who underwent radical prostatectomy for clinically localized prostate cancer as a monotherapy. The clinical demographics for the both cohorts are presented in Table 1. Four tissue microarrays (TMAs; three from WCMC and one from UM) were used for the study, representing tumors and benign prostate tissue samples. The TMAs were constructed from formalin-fixed paraffin-embedded tissue blocks from radical prostatectomy specimens. Review of pathological findings and selection of tissue samples for the TMAs were performed by the study pathologists. The TMAs from WCMC were composed of three representative TMA cores of the primary tumor consisting of the tumor with the highest Gleason pattern (three 0.6-mm cores) from 131 patients. Secondary tumors were sampled when present. In addition, normal prostatic tissue was selected in a subset of cases. The TMA from UM consisted of one TMA core sampled from 79 patients. Cases were selected in part based on previous assessment of ETS rearrangement status by FISH as described [6,12]. All patients provided written informed consent, and this study was approved by the institutional review boards at WCMC and at the UM Medical School, respectively.
Table 1.
Clinical and Pathological Demographics of Cohorts from WCMC and UM.
| WCMC Patients (n = 131) | UM Patients (n = 79) | ||
| Age, median (range), years | 61 (42–75) | 58 (45–77) | |
| Clinical stage | T1c | 115 (88%) | 62 (78%) |
| T2 | 16 (12%) | 17 (22%) | |
| Preoperative PSA, median (range), ng/ml | 6.1 (1.1–24.2) | 6.4 (0.5-18.4) | |
| Prostatectomy Gleason score | 6 | 19 (14%) | 6 (8%) |
| 7 | 98 (75%) | 70 (90%) | |
| 8–9 | 14 (11%) | 2 (3%) | |
| Tumor stage | pT2a-c | 92 (70%) | 64 (81%) |
| pT3a | 30 (23%) | 13 (16%) | |
| pT3b | 6 (5%) | 1 (1%) | |
| pT4 | 3 (2%) | 1 (1%) | |
| Surgical margin status | No | 111 (85%) | 61 (77%) |
| Yes | 20 (15%) | 18 (23%) | |
| PSA biochemical recurrence | No | 121 (92%) | 70 (89%) |
| Yes | 10 (8%) | 9 (11%) | |
| Vascular invasion status | No | 122 (93%) | N/A |
| Yes | 9 (7%) | ||
| Lymph node status | Negative | 130 (99%) | 78 (99%) |
| Positive | 1 (1%) | 1 (1%) | |
N/A indicates not available.
Assessment of Gene Rearrangement Status Using Two-color Interphase FISH
Four-micrometer-thick TMA sections were used for interphase FISH analysis. Rearrangement status for individual cohorts was determined independently at WCMC and UM by the study pathologists using a dual-color break-apart interphase FISH assay as described previously [2,13]. Briefly, two differentially labeled probes were designed to span the telomeric and centromeric neighboring regions of each locus. ERG status was evaluated for all 207 cases on four TMAs. We have previously evaluated 88 patients from two TMAs from the WCMC cohort for three other ETS genes, namely, ETV1, ETV4 and ETV5, and four known five prime fusion partners, namely, TMPRSS2, SLC45A3, NDRG1, and Herv-K22q11.23 (Svensson and Rubin, unpublished observations). The following centromeric/telomeric BAC clones were used to assess for rearrangement status in the WCMC cohort: ERG (RP11-24A11 and RP11-372O17), ETV1 (RP11-661L15 and RP11-79G16), ETV5 (RP11-480B15 and RP11-822O23), ETV4 (CTP-3215I16 and RP11-147C10), TMPRSS2 (RP11-35C4 and RP11-120C17), SLC45A3 (RP11-249H15 and RP11-131E5), NDRG1 (RP11-185E14 and RP11-1145H17), and Herv-K22q11.23 (RP11-61N10 and RP11-71G19). RP11-95I21 (5′ to ERG) and RP11-476D17 (3′ to ERG) were used in the UM cohort.
For all FISH experiments using break-apart probes, a nucleus without a rearrangement demonstrates two pairs of juxtaposed red and green signals (mostly forming two yellow signals). A nucleus with rearrangement through insertion shows the split of one red-green (yellow) signal pair, resulting in a single red and green signal for the rearranged allele, and a still combined (yellow) signal for the nonrearranged allele in each nucleus. Finally, a nucleus with a rearrangement through deletion shows one juxtaposed red-green signal pair (yellow) for the nonrearranged allele and a single red signal for the allele involved in the rearrangement. For each case, at least 100 cancer nuclei were assessed. Benign epithelial and stromal cells served as internal controls for most TMA cores evaluated.
Evaluation of ERG Protein Expression by IHC
Immunohistochemical (IHC) analyses on paraffin-embedded formalin-fixed tumor tissue sections were carried out using the automated DiscoveryXT staining platform from Ventana Medical Systems. The primary rabbit monoclonal antibody was obtained from Epitomics (San Diego, CA). Antigen recovery was conducted using heat retrieval and CC1 standard, a high pH Tris/borate/EDTA buffer (VMSI, catalog no. 950-124). Slides were incubated with 1:100 of the ERG primary antibody for 1 hour at room temperature. Primary antibody was detected using the ChromoMap DAB detection kit (VMSI, catalog no. 760-159) and UltraMap anti-Rb HRP (VMSI, catalog no. 760-4315). The anti-Rb HRP secondary antibody was applied for 16 minutes at room temperature. Slides were counterstained with Hematoxylin II (VMSI, catalog no. 790-2208) for 8 minutes followed by Bluing Reagent (VMSI, catalog no. 760-2037) for 4 minutes at 37°C. Subjective evaluation of ERG protein expression in both the WCMC and UM cohorts was determined for each tumor core using a four-tier grading system: negative (0), weakly positive (1+), moderately positive (2+), and strongly positive (3+). In addition, benign prostate glands, benign prostatic hyperplasia, and HGPIN (isolated and associated with adenocarcinoma) were evaluated. Altogether, 207 cases from WCMC and UM were assessable both for ERG rearrangement by FISH and for ERG protein expression by the study pathologists.
A subset of cases (WCMC cohort) were also scanned using the Ariol Platform (Genetix Corp, San Jose, CA) for objective measurements of ERG protein expression. This microscope system scans the entire TMA slide at 20x magnification. The image analysis component was run with the Multistain assay on the Ariol 3.2.125 software version. The color and shape characteristics (“classifiers”) were set by the investigator to properly identify cells with positive staining. The software applies the color classifiers to identify regions of positive nuclear staining, excluding objects that are either too light or too dark. The shape classifiers are applied to the remaining objects to further screen out objects that do not resemble cells of interest. After determining that endothelial cells and lymphocytes consistently express ERG protein, endothelial cell and lymphocyte masking was applied to each slide to avoid measuring ERG expression from the endothelial compartment. A total of 131 cases were assessed for ERG protein expression using the Ariol Platform.
Cell Line Immunoblot Analyses
Lysates from benign (RWPE and PrEC) and cancerous (LNCaP, VCaP, DU145) prostatic cell lines were transferred to polyvinylidene fluoride membranes and probed with rabbit monoclonal anti-ERG (clone EPR3864; Epitomics) at 1:1000 dilution. Membranes were stripped and reprobed with mouse monoclonal anti-β-actin (clone AC-74; Sigma, St Louis,MO) at 1:20,000 dilution for loading control.
Antibody Epitope Mapping
DNA expression vectors encoding ERG deletion constructs with 3′ 3xFLAG tags were purchased from GeneART (Regensburg, Germany). HEK293 cells were transiently transfected with complementary DNA expression vectors 48 hours before lysis. Immunoblot analysis was as above, except rabbit monoclonal anti-FLAG (no. F7425; Sigma) at 1:1000 dilution was used for loading control.
Immunofluorescence
Formalin-fixed paraffin-embedded prostate tissue sections were soaked in xylene for 1 hour to remove paraffin, and antigen retrieval was performed using Target Retrieval Solution (Dako). Slides were blocked in PBS-T containing 5% normal donkey serum for 1 hour. Slides were incubated overnight at 4°C with rabbit monoclonal anti- ERG (clone EPR3864, 1:200 dilution; Epitomics), washed, and incubated with donkey antirabbit Alexa 488 (1:1000 dilution; Invitrogen, Carlsbad, CA) for 1 hour. Slides were mounted using Vectashield mounting medium containing 4′,6-diamidino-2-phenylindole-2HCl (Vector Laboratories, Burlingame, CA) after washing with PBS-T and PBS. Confocal images were taken with a Zeiss LSM510 META (Carl Zeiss, San Francisco, CA) imaging system.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was carried out on stable RWPE-ERG or RWPE-GUS (control) cells [14] as previously described [15] using 5 µg of rabbit monoclonal anti-ERG (clone EPR3864; Epitomics) or rabbit immunoglobulin G (no. sc-2027; Santa Cruz Biotechnology, Santa Cruz, CA). The final ChIP yield was 10 to 30 ng for each antibody. Quantitative polymerase chain reaction (PCR) after ChIP was performed for ERG target genes PLAU, MMP3, and the TMPRSS2 enhancer and for control gene KIAA0066 as described [15].
Statistical Analysis
The objective ERG protein expression level by Ariol system was defined as the ratio of “ERG nuclear area” to “analyzed tissue area.” Because ERG protein expressions were collected from three representative TMA cores and multiple focus for each patient, the average of ratios from three cores for each focus was calculated. When multiple foci were available from one patient, we prioritized the focus with ERG rearrangement (FISH) or maximum protein expression for each patient. Once the focus was selected for each patient, the corresponding subjective pathologist evaluation of ERG protein expression by IHC was used. The distribution of objective ERG protein expression (continuous scale) stratified by the study pathologists (categorical) or by ERG rearrangement status (categorical) was assessed by descriptive statistic and box plots (box showing the first, median, and third quartiles; and whiskers showing the 10th and 90th percentiles). Wilcoxon ranksum test was used to evaluate the association between a continuous variable and a categorical variable (two-group). The Bonferroni adjustment was used for multiple tests between groups. To compare the predicted accuracy of ERG protein expression (both categorical and continuous) to ERG rearrangement status (criterion standard), sensitivity, specificity, and positive (PPV) and negative (NPV) predictive values were calculated. In addition, receiver operating characteristic (ROC) curve was plotted, and the area under the curve (AUC) was assessed. κ Statistic was calculated to determine the agreement between subjective ERG protein expression and automated image analysis ERG protein expression. Associations for ERG expression status and clinical and pathologic features were also explored by χ2 or Fisher exact test where applicable. Analyses were performed in SAS Version 9.2 (SAS Institute, Inc., Cary, NC).
Results
Characterization of Anti-ERG Antibody for Assessing ERG Gene Fusion Prostate Cancer
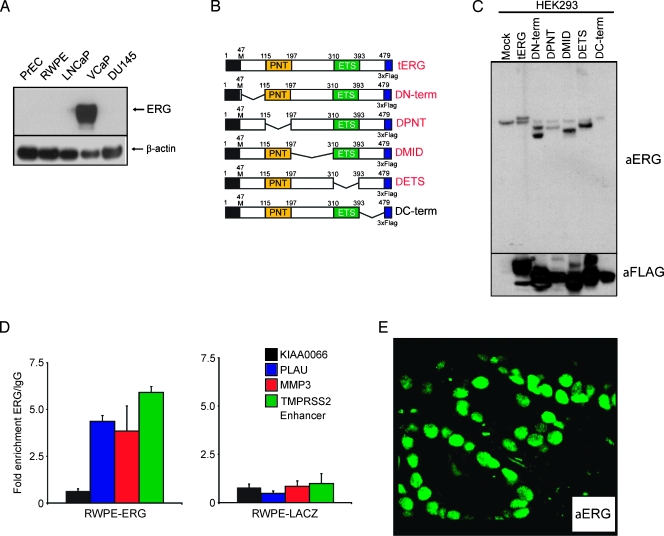
Because almost all reported TMPRSS2-ERG transcript isoforms encode for a truncated ERG protein rather than a chimeric protein, an antibody-based detection strategy would need to target the 3′ end of ERG. We previously evaluated multiple commercially available anti- ERG antibodies but could not identify any that were both sensitive and specific for ERG rearrangement prostate cancer through in vitro and in situ experiments (S.A. Tomlins, R. Mehra, and A.M. Chinnaiyan, unpublished observations). Here we sought to characterize and qualify a novel rabbit monoclonal anti-ERG antibody. First, we assessed cell lysates from benign (PrEC and RWPE) and cancerous (LNCaP, VCaP, DU145) prostatic epithelial cell lines for ERG expression using immunoblot analysis with the rabbit monoclonal anti-ERG antibody. Importantly, VCaP harbors a TMPRSS2-ERG gene fusion resulting in marked overexpression of ERG, whereas LNCaP harbors a rearrangement of the entire ETV1 locus, resulting in marked overexpression of ETV1, and no ETS gene rearrangements have been identified in DU145. As expected, the antibody showed marked overexpression of ERG in VCaP cells, with minimal background signal in the other cell lines and no cross-reactivity with ETV1 in LNCaP cells (Figure 1A).
Figure 1.
Characterization of a monoclonal anti-ERG antibody for detecting TMPRSS2-ERG gene fusion expression products. (A) Immunoblot detection of endogenous TMPRSS2-ERG gene fusion product (predicted molecular weight, 49.0 kDa) using anti-ERG monoclonal antibody. Cell lysates from benign (PrEC and RWPE) and cancerous (LNCaP, VCaP, and DU145) prostatic epithelial cell lines were immunoblotted for ERG expression using the rabbit monoclonal anti-ERG antibody. VCaP harbors a TMPRSS2-ERG gene fusion resulting in marked overexpression of ERG, LNCaP harbors a rearrangement of the entire ETV1 locus, resulting in marked overexpression of ETV1, and no ETS gene rearrangements have been identified in DU145. β-Actin was used as a loading control. (B) Schematic diagram showing different ERG deletion constructs used to identify the anti-ERG epitope. Constructs detected by the antibody are shown in red. (C) Western blot analysis of HEK293 cells transiently mock-transfected (showing endogenous ERG expression) or transfected with the constructs in panel B, and blotted with anti-ERG or anti-FLAG as loading control. (D) ChIP-quantitative PCR assays from stable RWPE-ERG and RWPE-GUS (control) cells, for ERG target genes PLAU, MMP3, and the TMPRSS2 enhancer, and the negative control gene KIAA0066. Means±SE are shown. Experiments were run in triplicate. (E) Immunofluorescence with anti-ERG on a formalin-fixed paraffin-embedded prostate tissue section with a TMPRSS2:ERG-positive tumor demonstrates strong nuclear staining in neoplastic cells. Endothelial cells in the stromal compartment also demonstrate expression of ERG.
Next, we mapped the antibody binding site on ERG through immunoblot analysis on a series of expression vectors representing portions of the most prevalent gene fusion protein (TMPRSS2 exon 1 to ERG1 exon 2) by making large tilling deletions of the N-terminus (ΔN-term, deletion of 47–115, 44.6 kDa), pointed domain (ΔPNT, deletion of 115–197, 43.4 kDa), the middle (ΔMID, deletion of 197–310, 41.6 kDa), the ETS domain (ΔETS, deletion of 310–393, 43.7 kDa), or the C-terminus (ΔC-term, deletion of 393–479, 43.6 kDa) of ERG. Triple FLAG antigen sequences were fused to the C-terminus of each construct, which were transiently transfected into HEK293 cells. As shown in Figure 1C, the antibody recognized all constructs except ΔC-term, consistent with the antibody epitope being found in the C-terminal amino acids 393–479 of the ERG protein, which is retained in all known ERG gene fusion isoforms. This immunogenic region is also highly specific to ERG relative to other ETS transcription factors.
We also assessed whether the anti-ERG antibody could recognize ERG in ChIP experiments using stable RWPE-ERG or RWPE-GUS (control) cells [15]. The anti-ERG antibody was able to specifically enrich known ERG target gene promoters including PLAU, MMP3, and the TMPRSS2 enhancer in the RWPE-ERG cells (two-tailed t test, P < .05 for all three targets), whereas no significant enrichment of these targets was observed in RWPE-GUS cells. Similarly, the negative control gene KIAA0066 was not specifically enriched by anti-ERG in either cell line (Figure 1D). Finally, to characterize the antibody for in situ experiments, we performed immunofluorescence on formalin-fixed paraffin-embedded tissue sections with TMPRSS2:ERG-positive prostate cancer (previously assessed by FISH), with almost all tumor nuclei in cancerous foci showing strong nuclear protein expression (Figure 1E). In the same field, one can also appreciate endothelial cell expression of ERG.
ERG Protein Expression Is Highly Concordant with ERG Rearrangement
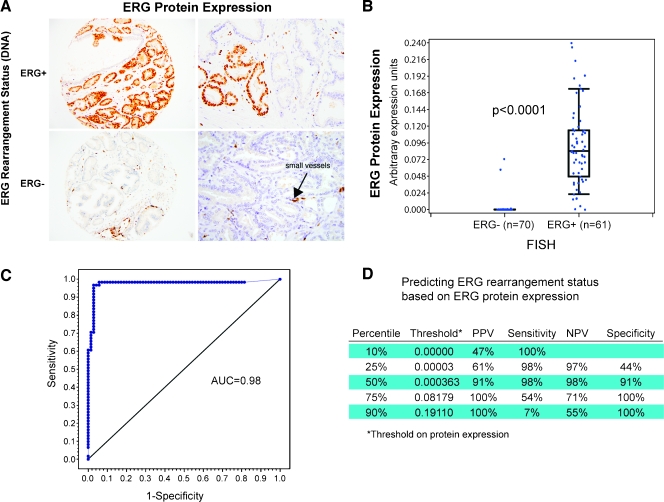
Previous studies demonstrated that ERG rearrangement prostate cancer (ERG+) demonstrate high levels of ERG messenger RNA transcript [2,13], the current study extends this observation to ERG protein expression. Using TMAs from 131 cases, ERG protein expression was evaluated by the study pathologists and by using the automated Ariol imaging system. ERG protein expression was confined to neoplastic cells (Figure 2A), vessels, and lymphocytes (Figure W1). The expression was strongly concentrated in the nuclei, but weak cytoplasmic expression could be appreciated in cases with strong nuclear ERG protein expression. None of the benign prostate glands in any of the samples reviewed demonstrated ERG protein expression. Using arbitrary expression units, the ERG protein expression was strongly associated with ERG rearrangement status in 131 cases from the WCMC cohort (median = 0.0 in ERG- vs 0.08 in ERG+, P < .0001) using the Wilcoxon rank-sum test (Figure 2B).
Figure 2.
ERG rearrangement by break-apart FISH is highly correlated with ERG protein expression by IHC. (A) ERG protein expression in tumors with and without the TMPRSS2-ERG gene fusion. The endothelial cells of small vessels show positive endogenous ERG expression in the context of surrounding ERG-negative cancer glands (left 20x, right 40x). (B) The box plot demonstrates a highly significant association between the automated image evaluation of ERG protein expression and the ERG gene rearrangement status for 128 cases (P < .0001, Wilcoxon rank-sum test). Of 70 ERG rearrangement-negative cases, 2 demonstrate ERG protein expression. (C) Using a threshold of 0.4 as the cut point for determining ERG status, we observed excellent ROC curve performance with an AUC of 0.98 using ERG rearrangement by FISH as the criterion standard. (D) A summary of the performance of ERG protein expression to predict ERG rearrangement status is presented for different thresholds.
We identified two cases demonstrating ERG protein expression (Figure W2) without any detectable ERG rearrangement as determined by FISH and reverse transcription-PCR. Conversely, three cases with ERG rearrangement by FISH had nearly no ERG protein expression. The sensitivity and specificity for using ERG protein expression on a continuous scale to predict ERG rearrangement status are presented in the ROC plot (Figure 2C). The AUC was 0.98. The PPV, sensitivity, NPV, and specificity based on variable ERG protein expression thresholds demonstrate that even low levels of expression are highly suggestive of ERG rearrangement (Figure 2D). By applying increasing thresholds to the protein expression levels, increased specificity and decreased sensitivity are observed.
We next explored the correlation between the subjective evaluation by the study pathologists for ERG protein expression for 128 cases from WCMC (three cases were excluded by the pathologists) and by automated imaging. The correlation demonstrated nearly perfect agreement [16] with a κ statistic of 0.84 (0.75–0.94) (Figure W3). Interestingly, no ERG protein negative cases (n = 66) evaluated by the study pathologists demonstrated ERG rearrangement by FISH. Two cases with no known ERG rearrangement were scored as having moderate ERG protein expression. Importantly, these results were independently confirmed in a second cohort with 79 patients from the UM (Table 2). The combined evaluations from both cohorts consisting of 207 patient tumors that could be evaluated by the pathologists demonstrated a sensitivity and specificity of 95.7% and 96.5%, respectively. Interestingly, between the two cohorts, there were six cases harboring either ETV1 (n = 4 or 1.9%), ETV4 (n = 1 or 0.48%), or ETV5 (n = 1 or 0.48%) rearrangements (all ERG rearrangement-negative). None of these cases demonstrated ERG protein expression (Figure W4), supporting the in vitro specificity of the antibody described above.
Table 2.
Pathological Evaluation of ERG Protein Expression Versus ERG Rearrangement Status as Determined by FISH in Cohorts from the WCMC and the UM Cohorts.
| ERG Status | Patient Cohort | |||||
| Pathologist evaluation | Positive | Negative | Combined | Sensitivity | 95.7% | |
| Positive | 88 | 4 | Specificity | 96.5% | ||
| Negative | 4 | 111 | PPV | 95.7% | ||
| NPV | 96.5% | |||||
| Fisher = 6.07e - 48 | ||||||
| Pathologist evaluation | Positive | Negative | WCMC | Sensitivity | 100.0% | |
| Positive | 60 | 2 | Specificity | 97.1% | ||
| Negative | 0 | 66 | PPV | 96.8% | ||
| NPV | 100.0% | |||||
| Fisher = 1.01e - 34 | ||||||
| Pathologist evaluation | Positive | Negative | UM | Sensitivity | 87.5% | |
| Positive | 28 | 2 | Specificity | 95.7% | ||
| Negative | 4 | 45 | PPV | 93.3% | ||
| NPV | 91.8% | |||||
| Fisher = 7.05e - 15 | ||||||
ERG FISH: negative versus positive (insertion + deletion); IHC: negative versus (weak + moderate + strong). Note that for the WCMC cohort, 128 cases were evaluated.
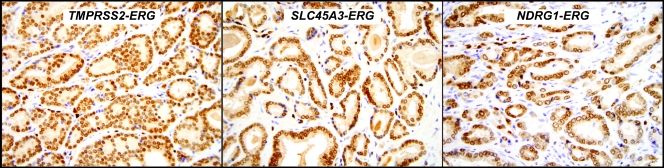
In the WCMC cohort, 47% (60/128) demonstrated ERG rearrangement by FISH with the majority harboring the TMPRSS2-ERG gene fusion (n = 57) and the remaining cases demonstrating either the SLC45A3-ERG (n = 2) or the NDRG1-ERG (n = 1) gene fusions. We did not appreciate any differences in protein expression in these other types of ERG rearranged cancers, although the numbers are too small to make any conclusive comments (Figure 3). The UM cohort was selected based on previous assessment of ETS status, and the prevalence 40% (32/79) of ERG rearranged cases is thus not representative of our prostatectomy series.
Figure 3.
ERG-rearranged cases express high levels of truncated ERG protein regardless of 5′ fusion partner. Representative examples of prostate cancers harboring ERG rearrangements with different 5′ partners but showing similar ERG protein expression by IHC.
ERG rearrangement heterogeneity has been well documented between discrete tumor nodules within the same prostate gland [17,18]. In this cohort, we had an example of two geographically discrete tumors from the same patient, with one ERG rearrangement positive and the other negative. As expected, ERG protein was strongly expressed in the ERG-rearranged case, and no expression was observed in the fusion-negative tumor (Figure W5). These findings are consistent with observations that multifocal localized prostate cancer is clonally distinct and that ERG rearrangement (through FISH or protein expression) can be used as a clonal marker.
ERG Protein Expression Is Present in High-grade PIN Associated with ERG-Expressing Carcinoma
We also had several areas with HG PIN from cases harboring ERG rearrangement. We observed similar ERG expression status in HG PIN and associated adjacent prostate cancer (Figure 4), consistent with previous FISH-based studies [17–19]. Interestingly, one can appreciate a distinct demarcation where HG PIN begins morphologically (Figure 4A, arrowhead), and in this ERG rearrangement-positive cancer, the ERG protein expression clearly identifies the initiation of HG PIN (Figure 4B, arrowhead).
Figure 4.
ERG protein expression in high-grade prostatic intraepithelial neoplasia (HG PIN) adjacent to ERG-expressing cancer. (A) H&E stain demonstrates small prostatic cancerous glands on the left and a larger atypical gland on the right with a subset of epithelial cells with neoplastic features consistent with HG PIN. (B) ERG protein expression by IHC demonstrates strong expression in both cancer and HG PIN. The arrowheads indicate a discrete demarcation between HG PIN and histologically benign luminal epithelial cells (40x).
Expression in Vessels/Endothelial Cells and Lymphocytes
As described above, the endothelial cells of small vessels in benign and cancerous prostate tissue demonstrated moderate to strong ERG protein expression, regardless of tumor ERG rearrangement status. In addition, lymphocytes in both benign and cancerous tissue also demonstrated ERG protein expression (Figure W1). However, ERG rearrangements were never detected by FISH in endothelial cells or lymphocytes.
Lack of Correlation with Clinical or Pathology Parameters
In the WCMC cohort (n = 128) and UM (n = 79) cohorts, we did not observe any associations between ERG rearrangement status and clinical or pathologic parameters (data not shown) and did not have sufficient follow-up to explore for clinical outcome.
Discussion
The current study demonstrates that recurrent ERG gene fusions lead to the over expression of a truncated ERG protein. When ERG protein is expressed in tumor cells, most of the tumor nuclei are positive consistent with earlier observations by FISH [2,11,13]. A range of ERG expression levels was observed in these clinically localized prostate cancer cases; however, this was in contrast to benign glands, which never showed staining, consistent with previous FISH results. We did not appreciate ERG protein expression differences based on the mechanism of ERG rearrangement (i.e., through deletion or insertion; data not shown). We also did not observe differences in ERG protein expression based on the involved 5′ partners of ERG, including TMPRSS2, SLC45A3, and NDRG1. Likewise, we saw no cross-reactivity with cases harboring ETV1, ETV4, or ETV5 rearrangements. Although we observed highly consistent results for ERG rearrangements by FISH and for ERG expression by IHC, we did not observe perfect concordance. In addition to technical factors, such discordant cases may be due to 1) ERG rearrangement but decreased transcript/protein expression (e.g., due to decreased androgen signaling); 2) insertion of the entire ERG locus into a genomically active region, which does not result in an ERG rearrangement detectable by split-signal FISH but results in transcript/protein overexpression (similar to the mechanism for ETV1 rearrangement in LNCaP [5]; and 3) activation of ERG overexpression by a non-rearrangement-based mechanism. The combined FISH/IHC strategy used herein will allow for identification of such cases in future studies.
In the current study, we observed ERG protein expression in vessels surrounding prostate cancer, consistent with staining of endogenous wild-type ERG, which is known to be expressed during angiogenesis (reviewed by Sato [20]). Vlaeminck-Guillem et al. [21] first described ERG gene expression during murine embryogenesis in mesodermal tissues, including the endothelial, precartilaginous, and urogenital areas. Importantly, ERG rearrangements by FISH were never observed in endothelial cells or lymphocytes, consistent with the observed staining representing endogenous wild-type ERG expression. Supporting this, expression in vessels and lymphocytes was not related to ERG rearrangement status of the tumor and was not cancer-specific, as ERG expressions in both vessels and lymphocytes adjacent to benign prostate tissue and stroma were observed (Figure W1). This is consistent with original observations made by Gavrilov et al. [22] describing the protein expression of ERG in small-caliber vessels adjacent to prostate cancer using IHC. The ERG protein expression of vessels and lymphocytes can be conveniently used as an internal staining control, regardless of ERG rearrangement status in cancer.
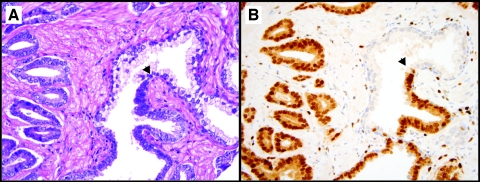
Because prostate needle biopsies can be readily evaluated for ERG protein expression (Figure 5), ERG staining may have immediate clinical utility in cases with atypical small acinar proliferation or HG PIN. Given the specificity of ERG staining for cancer observed herein, ERG staining in an atypical focus would be highly suggestive of cancer and evidence to support expedited rebiopsy. In addition to highly specific staining for cancer, we also observed a distinct demarcation between ERG-expressing HG PIN and adjacent benign glands (Figure 4). Importantly, HG PIN with ERG staining was always located adjacent to cancer with similar levels of ERG protein expression, consistent with previous FISH results demonstrating that ERG-rearranged HG PIN is nearly always found in close association with ERG-rearranged cancer [9–11]. Thus, ERG expression in isolated HG PIN would be highly suspicious for unsampled adjacent cancer or for the rapid progression to invasive disease. The utility of this finding will need to be evaluated in future clinical studies.
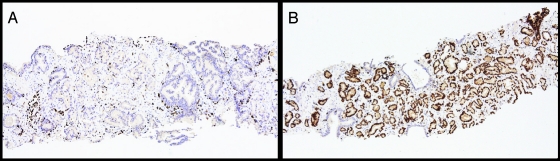
Figure 5.
ERG protein expression in prostate cancer biopsy samples. A prostate needle biopsy without ERG protein expression (A) and one with intense ERG protein expression (B) demonstrates the potential of determining the ERG rearrangement status using a prostate needle biopsy.
An antibody-based detection strategy for ERG rearrangement status may also have utility in the setting of castrate-resistant metastatic prostate cancer. We anticipate that ERG expression should be maintained in castrate-resistant cancers with ERG rearrangements because most of these tumors have selected for mechanisms to restore androgen signaling [4] and continue to express high levels of the gene fusion transcript [2,23]. Intriguingly, phase 1/2 results in men with castrate-resistant cancers treated with the novel antiandrogen abiraterone acetate demonstrated higher maximal prostate-specific antigen response in men with ERG rearrangement-positive versus -negative tumors, suggesting the potential utility of antibody- and/or FISH-based assessments of ERG rearrangement status for patient stratification. Importantly, FISH only demonstrates the presence of an ERG rearrangement, whereas continued ERG protein expression indicates both ERG rearrangement and continued androgen signaling (because androgen signaling-independent cancers with ERG rearrangements express low or no detectable TMPRSS2-ERG transcript) [24,25]. Antibody-based detection may also facilitate screening for ERG rearrangements in other clinical cohorts for molecular stratification, with possible confirmation by FISH (because IHC does not show perfect concordance with FISH and cannot determine fusion mechanism).
This current study is not the first to explore ERG expression in the prostate. Gavrilov et al. [22] described the protein expression of several ETS family members in prostate tissue samples using IHC. They reported nuclear expression of Elf-1 and Fli-1 in 16 and 20 of 25 high-grade PCA, respectively. Interestingly, ERG expression was only observed in 44% (11/25) of PCA samples tested, of which 7 had a Gleason score of 7 or higher. Vanaja et al. [1] reported ERG overexpression by quantitative PCR in 50% of both hormone-naive and metastatic prostate cancer samples. More recently, Petrovics et al. [26] identified ERG as the most frequently overexpressed oncogene in the transcriptome by using a combination of expression array analysis and quantitative real-time reverse transcription-PCR on 114 PCA samples isolated with laser capture microdissection.
In summary, we present the characterization of a specific rabbit monoclonal anti-ERG antibody that shows very high concordance with FISH, the current criterion standard for determining ERG rearrangement status in prostate cancer. In this study, we observed ERG staining exclusively in prostate cancer and HG PIN, consistent with FISH results, suggesting immediate potential clinical utility. In addition, this antibody will likely facilitate functional studies of ERG gene fusions and molecular subtyping of prostate cancer.
Supplementary Material
Acknowledgments
The authors thank Robert Kim of the WCMC Translational Research Program of the Department of Pathology and Laboratory Medicine and Yifang Liu for technical expertise and Ashley Santa Cruz at Ventana/Roche for optimizing the final protocol for ERG IHC. The authors also thank Javed Siddiqui of the UM Prostate SPORE Tissue/Informatics Core and Nalla Palanisamy for managing previous FISH probe preparations at UM.
Abbreviations
- ERG
v-ets erythroblastosis virus E26 oncogene homolog (avian)
- TMA
tissue microarray
- FISH
fluorescence in situ hybridization
- ChIP
chromatin immunoprecipitation
Footnotes
This study was supported by National Cancer Institute (NCI) grants CA125612 (F.D. and M.A.R.), U01CA111275-06 Early Detection Research Network (S.A.T., M.A.R., and A.M.C.), and R01CA132874 (A.M.C.) and Prostate SPORE P50CA69568 (A.M.C. to M.A.R.). S.A.T. is supported by a Young Investigator Award from the Prostate Cancer Foundation. A.M.C. is supported by the Doris Duke Charitable Foundation Clinical Scientist Award, Burroughs Welcome Foundation Award in Clinical Translational Research, and the Prostate Cancer Foundation. A.M.C. is an American Cancer Society Research Professor. F.D. is supported by a Developmental Project Award from Dana-Farber Harvard Cancer Center Prostate Cancer SPORE.
This article refers to supplementary materials, which are designated by Figures W1 to W5 and are available online at www.neoplasia.com.
References
- 1.Vanaja DK, Cheville JC, Iturria SJ, Young CY. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res. 2003;63(14):3877–3882. [PubMed] [Google Scholar]
- 2.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 3.Mosquera JM, Mehra R, Regan MM, Perner S, Genega EM, Bueti G, Shah RB, Gaston S, Tomlins SA, Wei JT, et al. Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin Cancer Res. 2009;15(14):4706–4711. doi: 10.1158/1078-0432.CCR-08-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomlins SA, Bjartell A, Chinnaiyan AM, Jenster G, Nam RK, Rubin MA, Schalken JA. ETS gene fusions in prostate cancer: from discovery to daily clinical practice. Eur Urol. 2009;56(2):275–286. doi: 10.1016/j.eururo.2009.04.036. [DOI] [PubMed] [Google Scholar]
- 5.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8(7):497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han B, Mehra R, Dhanasekaran SM, Yu J, Menon A, Lonigro RJ, Wang X, Gong Y, Wang L, Shankar S, et al. A fluorescence in situ hybridization screen for E26 transformation-specific aberrations: identification of DDX5-ETV4 fusion protein in prostate cancer. Cancer Res. 2008;68(18):7629–7637. doi: 10.1158/0008-5472.CAN-08-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esgueva R, Perner S, J LaFargue C, Scheble V, Stephan C, Lein M, Fritzsche FR, Dietel M, Kristiansen G, Rubin MA. Prevalence of TMPRSS2-ERG and SLC45A3-ERG gene fusions in a large prostatectomy cohort. Mod Pathol. 2010;23(4):539–546. doi: 10.1038/modpathol.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pflueger D, Rickman DS, Sboner A, Perner S, LaFargue CJ, Svensson MA, Moss BJ, Kitabayashi N, Pan Y, de la Taille A, et al. N-myc downstream regulated gene 1 (NDRG1) is fused to ERG in prostate cancer. Neoplasia. 2009;11(8):804–811. doi: 10.1593/neo.09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT, et al. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet. 2009;41(5):619–624. doi: 10.1038/ng.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han B, Mehra R, Lonigro RJ, Wang L, Suleman K, Menon A, Palanisamy N, Tomlins SA, Chinnaiyan AM, Shah RB. Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol. 2009;22(8):1083–1093. doi: 10.1038/modpathol.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perner S, Mosquera JM, Demichelis F, Hofer MD, Paris PL, Simko J, Collins C, Bismar TA, Chinnaiyan AM, De Marzo AM, et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31(6): 882–888. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 12.Mehra R, Tomlins SA, Shen R, Nadeem O, Wang L, Wei JT, Pienta KJ, Ghosh D, Rubin MA, Chinnaiyan AM, et al. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod Pathol. 2007;20(5):538–544. doi: 10.1038/modpathol.3800769. [DOI] [PubMed] [Google Scholar]
- 13.Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, Tchinda J, Tomlins SA, Hofer MD, Pienta KG, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66(17):8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 14.Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, Cao Q, Prensner JR, Rubin MA, Shah RB, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10(2):177–188. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu J, Cao Q, Mehra R, Laxman B, Yu J, Tomlins SA, Creighton CJ, Dhanasekaran SM, Shen R, Chen G, et al. Integrative genomics analysis reveals silencing of β-adrenergic signaling by polycomb in prostate cancer. Cancer Cell. 2007;12(5):419–431. doi: 10.1016/j.ccr.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 16.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–174. [PubMed] [Google Scholar]
- 17.Mehra R, Han B, Tomlins SA, Wang L, Menon A, Wasco MJ, Shen R, Montie JE, Chinnaiyan AM, Shah RB. Heterogeneity of TMPRSS2 gene rearrangements in multifocal prostate adenocarcinoma: molecular evidence for an independent group of diseases. Cancer Res. 2007;67(17):7991–7995. doi: 10.1158/0008-5472.CAN-07-2043. [DOI] [PubMed] [Google Scholar]
- 18.Barry M, Perner S, Demichelis F, Rubin MA. TMPRSS2-ERG fusion heterogeneity in multifocal prostate cancer: clinical and biologic implications. Urology. 2007;70(4):630–633. doi: 10.1016/j.urology.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mosquera JM, Perner S, Genega EM, Sanda M, Hofer MD, Mertz KD, Paris PL, Simko J, Bismar TA, Ayala G, et al. Characterization of TMPRSS2-ERG fusion high-grade prostatic intraepithelial neoplasia and potential clinical implications. Clin Cancer Res. 2008;14(11):3380–3385. doi: 10.1158/1078-0432.CCR-07-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sato Y. Role of ETS family transcription factors in vascular development and angiogenesis. Cell Struct Funct. 2001;26(1):19–24. doi: 10.1247/csf.26.19. [DOI] [PubMed] [Google Scholar]
- 21.Vlaeminck-Guillem V, Carrere S, Dewitte F, Stehelin D, Desbiens X, Duterque-Coquillaud M. The Ets family member Erg gene is expressed in mesodermal tissues and neural crests at fundamental steps during mouse embryogenesis. Mech Dev. 2000;91(1–2):331–335. doi: 10.1016/s0925-4773(99)00272-5. [DOI] [PubMed] [Google Scholar]
- 22.Gavrilov D, Kenzior O, Evans M, Calaluce R, Folk WR. Expression of urokinase plasminogen activator and receptor in conjunction with the ets family and AP-1 complex transcription factors in high grade prostate cancers. Eur J Cancer. 2001;37(8):1033–1040. doi: 10.1016/s0959-8049(01)00077-6. [DOI] [PubMed] [Google Scholar]
- 23.Attard G, Swennenhuis JF, Olmos D, Reid AH, Vickers E, A'Hern R, Levink R, Coumans F, Moreira J, Riisnaes R, et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 2009;69(7):2912–2918. doi: 10.1158/0008-5472.CAN-08-3667. [DOI] [PubMed] [Google Scholar]
- 24.Hermans KG, van Marion R, van Dekken H, Jenster G, van Weerden WM, Trapman J. TMPRSS2:ERG fusion by translocation or interstitial deletion is highly relevant in androgen-dependent prostate cancer, but is bypassed in late-stage androgen receptor-negative prostate cancer. Cancer Res. 2006;66(22):10658–10663. doi: 10.1158/0008-5472.CAN-06-1871. [DOI] [PubMed] [Google Scholar]
- 25.Mertz KD, Setlur SR, Dhanasekaran SM, Demichelis F, Perner S, Tomlins S, Tchinda J, Laxman B, Vessella RL, Beroukhim R, et al. Molecular characterization of TMPRSS2-ERG gene fusion in the NCI-H660 prostate cancer cell line: a new perspective for an old model. Neoplasia. 2007;9(3):200–206. doi: 10.1593/neo.07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrovics G, Liu A, Shaheduzzaman S, Furasato B, Sun C, Chen Y, Nau M, Ravindranath L, Chen Y, Dobi A, et al. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24(23):3847–3852. doi: 10.1038/sj.onc.1208518. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.