Abstract
Background
Self-complementary RNA transcripts form a double-stranded RNA (dsRNA) that triggers a sequence-specific mRNA degradation, in a process known as RNA interference (RNAi), leading to gene silencing. In vascular plants, RNAi molecules trafficking occur between cells and systemically throughout the plant. RNAi signals can spread systemically throughout a plant, even across graft junctions from transgenic to non-transgenic stocks. There is also a great interest in applying RNAi to pathogenic fungi. Specific inhibition of gene expression by RNAi has been shown to be suitable for a multitude of phytopathogenic filamentous fungi. However, double-stranded (ds)RNA/small interfering (si)RNA silencing effect has not been observed in vivo.
Results
This study demonstrates for the first time the in vivo interference phenomenon in the pathogenic fungus Fusarium verticillioides, in which expression of an individual fungal transgene was specifically abolished by inoculating mycelial cells in transgenic tobacco plants engineered to express siRNAs from a dsRNA corresponding to the particular transgene.
Conclusion
The results provide a powerful tool for further studies on molecular plant-microbe and symbiotic interactions. From a biotechnological perspective, silencing of fungal genes by generating siRNAs in the host provides a novel strategy for the development of broad fungi-resistance strategies in plants and other organisms.
Background
The genetic interference phenomenon [RNA interference (RNAi)] was described in Caenorhabditis elegans in which double-stranded RNA (dsRNA) induces individual sequence-specific posttranscriptional gene silencing. The spreading silencing effect has been demonstrated, in which the interference is observed in a broad region of the animal after the injection of dsRNA into the extracellular body cavity. Fire et al. [1] demonstrated that RNAi abolished expression of targeted genes in C. elegans by injecting dsRNA. Subsequently, others have shown that the effect also occurs when C. elegans is fed the bacterium Escherichia coli, which transcribes the recombinant dsRNA [2]. It also results from simply soaking the animals in dsRNA preparations [3]. Data obtained from in vivo and in vitro studies are now being used to engineer resistance against parasitic nematodes in transgenic plants [4-6].
Cellular boundaries play a pivotal role in this integration by maintaining a level of cell autonomy while enabling communication between cells for coordinated gene expression and metabolism. In vascular plants, trafficking of RNAi molecules occurs between cells and systemically throughout the plant [7,8]. Such RNA trafficking breaks the boundaries of our traditional thinking of RNAs as functioning solely within the cells in which they are produced, and ushers in a new frontier of plant biology [9]. Intercellular and systemic movement occurs via plasmodesmata, which provide the continuity of cytoplasm and endoplasmic reticulum between adjacent cells and the phloem. Analyses of vascular exudates from oilseed rape (Brassica napus) showed that phloem sap contained a large number of small (sm)RNAs, predominantly of 21 and 24 nucleotides in length [10]. In addition, RNAi signals can spread systemically throughout a plant, even across graft junctions from transgenic stocks to non-transgenic scions [11,12]. Moreover, recent experiments described by Tomilov et al. [13] demonstrated that the movement of RNAi molecules between a parasite and its host plants. The gus silencing signal generated by lettuce roots was functional in its parasite Triphysaria versicolor, translocating across the haustorium interface. In nematodes gene silencing may also be triggered by a diet composed of transgene-encoded RNAi plants [4,14]. The same phenomenon has been observed in herbivorous insects fed on a plant engineered to express dsRNAs targeting vital insect genes [15,16].
Plants have been genetically manipulated by introducing constructs encoding self-complementary hairpin RNA (hpRNA) to efficiently silence genes [17-19]. The expressed transcripts form a dsRNA that triggers a sequence-specific messenger (m)RNA degradation (RNAi). Briefly, the dsRNA is recognized by DICER-like enzymes, which cleaves the molecule into a series of 21-23 bp duplexes [called small interfering (si)RNAs], that complexes with a ribonucleoprotein complex (RISC). The duplex is unwound to give single stranded siRNA leading to activation of the RISC, which searches for homologous mRNA transcripts by a base-pairing mechanism, leading to mRNA degradation. In addition, siRNA can affect the chromatin structure of targeted genes, resulting in transcriptional inhibition [20].
There is a great interest in applying RNAi to pathogenic fungi. Specific inhibition of gene expression by RNAi has been shown to be suitable for a multitude of phytopathogenic filamentous fungi, such as Magnaporthe oryzae [21], Sclerotinia sclerotiorum [22], Phytophthora sojae [23], Aspergillus nidulans [24], A. fumigatus [25-28], A. oryzae [29], Bipolaris oryzae [30], Colletotrichum lagenarium [31], Coprinus cinereus [32,33], Fusarium solani [34], Mucor circinelloides [35], which were transformed with plasmid constructs to express self-complementary hairpin RNA molecules [36]. It has also been shown that simply adding synthetic siRNA molecules to the culture medium can result in specific suppression of the corresponding target gene in Aspergillus nidulans [37]. In addition, the reporter gfp transgene and the endogenous genes coding for hydrophobins and a peroxiredoxin were silenced in Moniliophthora perniciosa transfected with in vitro synthesized specific dsRNA [38]. However, the dsRNA/siRNA silencing effect has not been observed in vivo. Here we show that the gus gene expression can be specifically silenced in Fusarium verticillioides (= F. moniliforme) interacting with a transgenic plant engineered with a gus gene-inferring cassette [hairpin (hp)GUS].
Results and Discussion
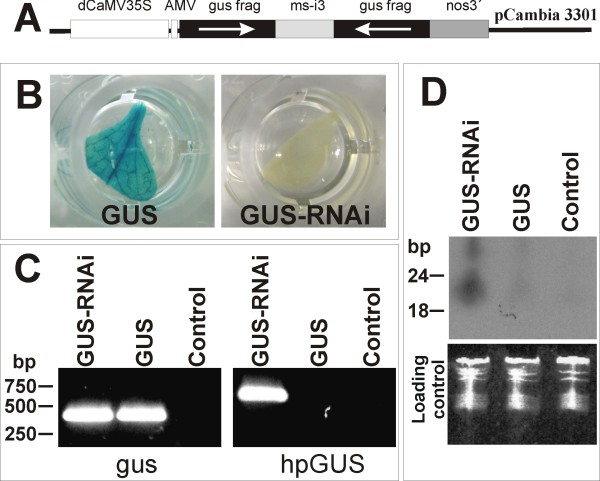
In this study, we hypothesized that a plant expressing dsRNA could result in in vivo trans-specific suppression of the corresponding target gene in fungal cells attached to plant tissues. Tobacco plants expressing the gus gene were re-transformed with an interfering intron-hairpin construct (Figure 1A). None of the re-transformed lines (GUS-RNAi lines) presented an observed gus expression in the leaves (Figure 1B) and no significant phenotypical differences were observed in the transgenic lines when compared with wild-type tobacco. Polymerase chain reaction (PCR) analysis confirmed the presence of both transgenes in the GUS-RNAi lines (Figure 1C). Northern analysis was carried out to detect the siRNA in the transgenic tobacco lines showed siRNA bands of expected size range only in the GUS-RNAi lines (Figure 1D). No signal was observed in either gus-expressing or non-transgenic plants. The constitutive expression of the gus dsRNA in these transgenic tobacco lines provides gus siRNA molecules for absorption by F. verticillioides feeding cells and subsequent RNAi of the constitutive gus transgene in fungal mycelium and conidial cells. In addition, uptake of unprocessed dsRNA and others RNAi molecules could be also occurs.
Figure 1.
Tobacco GUS-expressing line was re-transformed to express a double-stranded RNA for silencing the gus gene. (A) General scheme of the intron-spliced hairpin (hp)RNA vector (pC1302GUSi) constructed to promote gus gene silencing in GUS+ transformed tobacco lines. In order to generate the gus inferring cassette (hpGUS) a 627-bp fragment from gus gene (gus frag) was directionally cloned in order to generate sense and antisense arms, flanking the malate synthase gene intron 3 from Arabidopsis thaliana (ms-i3). (B) A GUS expressing tobacco line (GUS) was re-transformed with pC1302GUSi and regenerated plants (GUS-RNAi lines) did not show observable gus gene expression. (C) transgenic plants were analysed by polymerase chain reaction n order to detect both gus transgene in transformed plants (GUS) and the gus inferring cassette (hpGUS) in the GUS-RNAi lines. (D) Detection of small interfering RNA in a GUS expressing plants, GUS-RNAi line and control. Ethidium bromide-stained RNA serves as the loading control. Control in C and D is a non-transformed plant. Molecular size markers are indicated on the left in C and D.
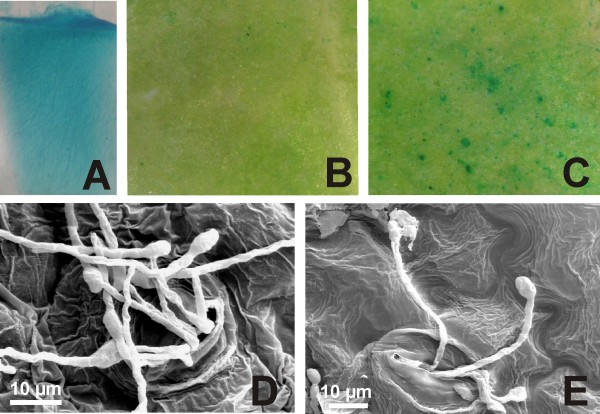
F. verticillioides fungal strain S68 (a nia-mutant) was transformed to express the GUS and nitrate reductase coding genes. Only colonies harbouring the nitrate reductase gene (nia+) were able to grow on MM plates and all co-transformants showed mitotic stability in respect of the GUS+ phenotype. One line over-expressing the gus gene was selected to inoculate both GUS-RNAi and non-transformed tobacco lines (Figure 2A). Leaves detached from in vitro plants (n = 12 per line; repeated twice) were inoculated with F. verticillioides culture, applied to the adaxial surface. After 11 days the agar plug was removed and leaves containing penetrative food-absorbing structures that remained attached to leaf surface were analysed for gus gene expression. When the F. verticillioides expressing the gus transgene was inoculated on leaves of GUS-RNAi lines, expressing dsRNA corresponding to the gus gene, a small number of fungal structures exhibiting the gus gene expression (7 ± 3 blue spots/cm2) were observed by GUS protein assay (Figure 2B). Non-transgenic tobacco leaves were unable to cause an evident gus gene silencing in transformed fungus structures and a large number of fungal structures exhibiting GUS expression was observed (221 ± 25 blue spots/cm2) (Figure 2C). After GUS assay, leaf surfaces were observed under scanning electron microscope. The analysis revealed that only germinating spores remained on the leaf surfaces (Figures 2D-E). Almost all spores observed were penetrating into leaf stomata. These phenotypes convincingly indicate that the silencing signal translocated across the germinated spores from tobacco into fungal cells. Recently, the translocation of RNAi molecules across the haustorium interface was demonstrated between a parasite and its host plants [13]. As a consequence, transgenic lettuce plants expressing a gus dsRNA induced specific gene silencing in their parasitic plant Triphysaria versicolor.
Figure 2.
Inoculations of GUS-RNAi tobacco lines with GUS expressing Fusarium verticillioides. (A) Mycelium cells exhibiting stable and high gus gene expression were used for the inoculation of plant leaves. (B) GUS assay carried out with GUS-RNAi tobacco leaves inoculated with the GUS+ fungus. Fungal mycelia interacting with the re-transformed plant did not show GUS expression; only a few germinating spores presented GUS expression. (C) GUS assay with non-transformed tobacco leaves inoculated with GUS+ fungus. Fungal mycelia showed high GUS expression level. (D, E) After GUS assay, leaf surfaces (presented in B and C) were observed under a scanning electron microscope in order to observe spores germinating and penetrating into stomata.
Infection by F. verticillioides can result in highly variable disease symptoms ranging from asymptomatic plants to severe rotting and wilting. In the more virulent pathogen F. oxysporum the whole process proceeds much faster and the transition from the initial symptomless phase to the necrotrophic phase occurs within a few days [39]. In contrast, the less aggressive pathogen F. verticillioides develops more slowly, leaving the plant more time to respond and restrict fungal growth [40]. In this study, the infection started from fungal conidia that initiated penetration to palisade and spongy mesophyll through stomata ~11 days after inoculation. We speculate that this fungus characteristic could be an important condition for plant-pathogenic fungi interaction-mediated gene silencing. These interaction types establish a long-term feeding relationship with a living host plant cell, allowing the nutrient-absorbing cells to uptake dsRNA molecules from their host. Indeed, this mode of plant colonization was confirmed when early stages of the F. verticillioides-maize interaction were characterized by using green fluorescent protein-expressing transgenic fungus isolates. Conidia were found only inside a cell in which an infection was established, while the surrounding cells appeared to be normal [40]. The differences in susceptibility could reflect the resistance of some cells or stages to the consequences of absorbed dsRNA.
In this work, transgene silencing in F. verticillioides was achieved by the interaction of fungal cells with a transgenic tobacco engineered for both transcribing and silencing the gus transgene expression, allowing the phenomenon of transitive silencing cycles of 'degradative-PCR' to take place in the plant cells. The 'degradative PCR' model proposes that siRNAs act as primers to transform the target mRNA into dsRNA. The nascent dsRNA is degraded by Dicer generating new siRNAs in a circle of dsRNA synthesis and degradation [41,42]. Further investigation should be carried out in order to determine whether this is important. It is known that gene silencing is dose-dependent and that more intact long hpRNAs could cause more pronounced silencing [43].
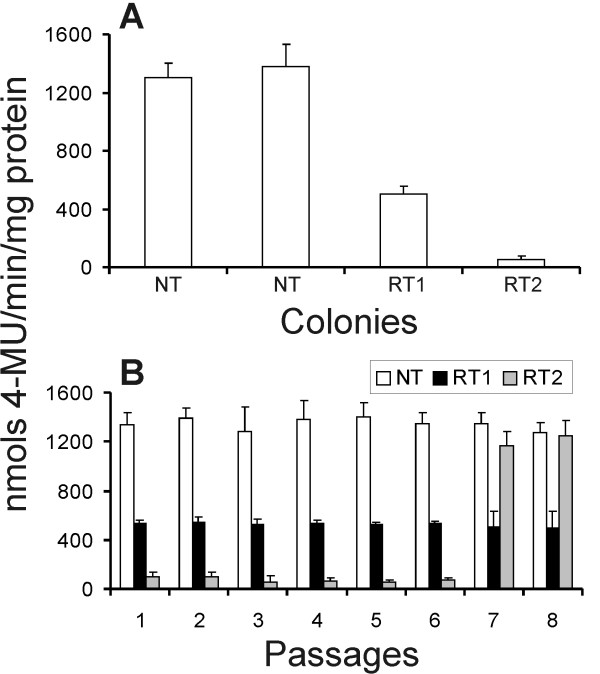
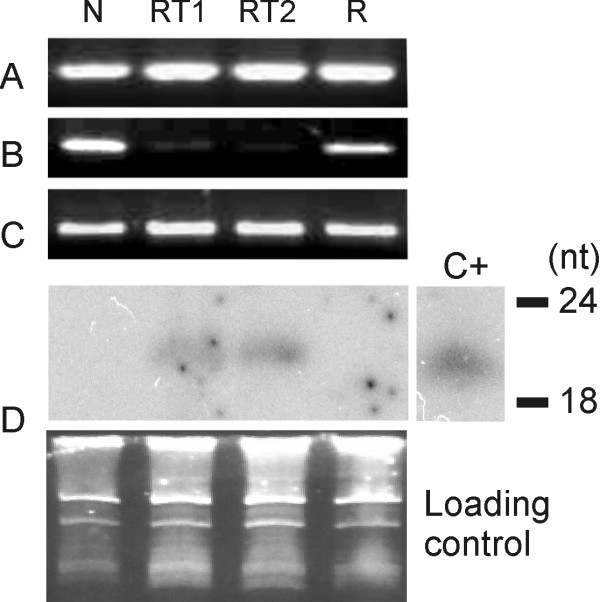
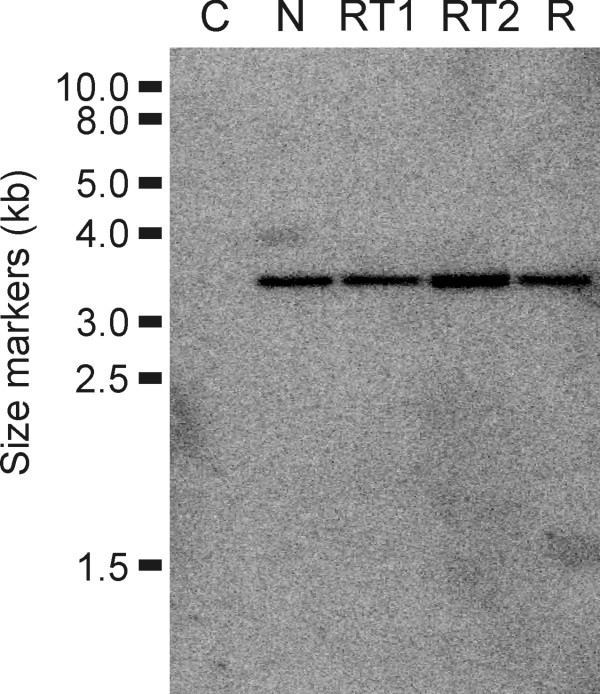
Ten fungal colonies were re-isolated from both GUS-RNAi and non-transformed tobacco lines and established in vitro. The gus gene expression was quantified in all re-isolated colonies and results revealed that two colonies isolated from GUS-RNAi plant lines presented a reduction of approximately 62% (RT1) and 96% (RT2) in the gus gene expression, compared with colonies isolated from non-transformed leaves (Figure 3A). All colonies isolated from non-transformed plants revealed normal GUS expression (activity raging from 1312 to 1378 nmol 4-MU.min-1.mg.protein-1). We further investigated the stability of gene silencing in mycelia cells over several passages in the absence of the RNAi trigger molecules from GUS-RNAi plants. The colonies showing a reduction of GUS expression were cultured in order to reach 70%-80% of confluence for eight passages (Figure 3B). GUS fluorimetric analyses revealed that the colony that presented 62% GUS expression reduction (RT1) remained expressing the gus gene in a similar pattern. However, the colony that presented 96% GUS expression (RT2) resumed normal GUS activity after the seventh passage. Steady-state levels of GUS mRNA in mycelia from the seventh-passage were estimated by semi-quantitative reverse transcription (RT)-PCR. gus-specific primers were used to distinguish cDNA from genomic GUS sequences. Results have shown a reduction in the steady-state level of gus transcript in colony mycelia of RT1 and RT2, which exhibit partial degrees of transgene silencing. In addition, after GUS-silencing phenotype reversion, the colony RT2 exhibits a gus transcript level similar to the control mycelia (which were not attached to hpGUS-tobacco leaves; Figure 4). Moreover, northern analyses showed siRNA bands of expected size range in both RT1 and RT2 silenced colonies and no signal was observed for the fungus colony isolated from non-transformed leaves (N) and RT2 colony that resumed normal GUS activity (R; Figure 4D).
Figure 3.
Isolation of Fusarium verticillioides colonies from both re-transformed and non-transformed tobacco lines. (A) The gus gene expression was quantified in fungal colonies isolated from non-transgenic (NT) and transgenic (RT1 and RT2) lines. (B) The two colonies isolated from GUS-RNA interference plant lines presented a reduction of the gus gene expression and were analysed for GUS expression over eight passages.
Figure 4.
Presence and expression of the gus gene in transformed Fusarium verticillioides isolated from small interfering (si)RNA-expressing tobacco plants. (A) Polymerase chain reaction (PCR) analysis for the presence of the gus gene in F. verticillioides isolates that were not inoculated in tobacco plants (N), which were isolated from inoculated GUS-RNA interference plants and exhibited a reduction of approximately 62% (RT1) and 96% (RT2) in the gus gene expression and RT2 isolate after reversion to the normal GUS+ phenotype after the 7th passage (R). (B) Reverse transcriptase (RT)-PCR analysis for the presence of transcripts from the endogenous gus gene in fungal cells. (C) RT-PCR analysis for the presence of transcripts from the fungal 5.8S rRNA housekeeping gene (internal control). (D) Detection of siRNA isolated from fungi isolated from transgenic (RT1, RT2 and R) and non-transgenic plants (N). The position corresponding to 18 and 24 nucleotides is indicated. C+: 100 pg of a gus gene-derived oligomer. Ethidium bromide-stained RNA serves as the loading control.
It has been shown that silencing phenotypes induced by RNAi can persist for generations. Despite numerous reports on RNA silencing in a variety of organisms, it is still not understood whether systemic smRNA signalling occurs in filamentous fungi. Nevertheless, silencing was shown to be a reversible dominant trait, operative in heterokaryotic strains containing a mixture of transgenic and non-transgenic nuclei, which suggests that a diffusable, trans-acting mobile RNA signal is involved [38,44]. In the filamentous fungus M. perniciosa, the gfp-silenced phenotype persisted for a period of 120 days after the transfection with dsRNAs [38]. In contrast, P. infestans treated with gfpdsRNA exhibited gene expression partially recovered after 4 days [45]. In Caenorhabditis elegans a single episode of RNAi in the nematode induced transgene silencing effects that were inherited over 80 generations in the absence of the original trigger [46]. In mice, microinjection of microRNAs into fertilized eggs also induced a heritable silencing phenotype, associated with the zygotic transfer of RNA molecules [47]. In vascular plants, recent studies suggest that smRNAs can move between cells (through plasmodesmata and endoplasmic reticulum) and can spread systemically via the long-distance transport systems (phloem) [48-52]. It was demonstrated that plant siRNAs do not only act at the site of synthesis, but are additionally mobile between cells [53]. There are indications that siRNAs can move 10-15 cells without amplification - probably as molecules of the 21-nt class - whereas movement over greater distances requires an amplification of the original signal [53]. In addition, certain cell types, such as neurons and sperm, are known to be more resistant to RNAi. Athough, so far, no cell type n fungi has been reported to be resistant to RNA silencing, this may possibly be the case with certain fungal cell types. Moreover, we can not exclude the possibility of mutation or an off-target effect on a gene related to silencing mechanism, or paramutation phenomenon associated with the reversion of gus gene normal expression.
In fungi, transgene gene silencing or instability of the silencing effect has been observed to associated with modifications in the organization of the transgene loci [31,44,54-56]. In order to investigate whether the gus gene silencing in fungal cells would be associated to changes in the status of the transgene in the genome, Southern analyses were carried out with fungal colonies that were re-isolated from both GUS-RNAi and non-transformed tobacco lines. Results have shown that all isolates presented a simple and unaffected integration pattern (Figure 5), demonstrating that the transgene locus was stably maintained in the fungal colonies re-isolated from GUS-RNAi plants and after several cultivation passages.
Figure 5.
Southern analysis of genomic DNA to detect the foreign gus gene in Fusarium verticillioides isolates. C = non-transformed; N = transgenic isolate that was not inoculated in tobacco plants; RT1 and RT2 = isolated from inoculated GUS-RNA interference plants; R = RT2 isolate after reversion to the normal GUS+ phenotype.
Conclusions
This work provides an example of RNA-mediated transfer of information between organisms and between plant and pathogen. Further studies should be carried out in order to access whether such RNA-mediated interference-transfer mechanisms participate in natural interactions. The use of siRNAs (RNAi) has become a powerful tool for specifically down-regulating gene expression and it has been demonstrated to be of great importance in basic and applied molecular plant-pathogen, mixotrophic and symbiotic interaction studies. From a biotechnological perspective, our results of in planta RNAi silencing of a gene expressed in a parasite fungus open new perspectives for the development of broad fungi-resistance strategies in plants and other organisms.
Methods
Plasmid construct and transgenic plants
Tobacco (cv Xanthi) was transformed as previously described by Horch et al. [57] with the vector pBI121 (Clontech) to express the gus gene. Transgenic lines presenting high level gus gene expression were re-transformed with the vector pC1302GUSi (Figure 1A). pC1302GUSi is an intron-spliced hpRNA vector constructed to promote gus gene silencing in re-transformed lines. In order to construct the pC1302GUSi, intron 3 from the malate synthase gene from Arabidopsis thaliana (ms-i3; GenBank accession number AB005235) was PCR-amplified from genomic DNA by using Platinum Taq DNA Polymerase High Fidelity (Invitrogen, CA, USA) and specific primers (5'-CTCTAGAGGCGCGCCGGTACCCCGGATCCATCAGCCTGTTCAAA-3' and 5'-TGAGCTCTCGCGATGGGGCCCACTAGTTTTATGGTCCATTTTC-3'), generating sites for XbaI, AscI, KpnI, BamHI and SpeI, ApaI, NruI, SacI, respectively. The 438-pb fragment was cloned into pCR2.1-TOPO (Invitrogen) and sequenced. The ms-i3 fragment was then removed with SacI and XbaI and inserted into the vector pUC19-35SAMVNOS [58] in order to generate the vector pSIU. A 627-bp fragment from gus gene coding sequence was removed with SspI and EcoRV from the vector pCambia3301 (Cambia, Brisbane, Australia) and cloned into the SmaI site from the pBlueScript KS+ (Fermentas, Ontario, Canada). The gus fragment was finally directionally cloned into the sites NruI and KpnI/XbaI from pSIU in order to generate sense and antisense arms, flanking the malate synthase gene intron 3 from Arabidopsis thaliana (GenBank accession No. AB005235). The gus inferring cassette (hpGUS) was removed with PvuII and cloned into SmaI from pCambia1302 (Cambia) generating the vector pC1302GUSi.
PCR analyses were carried out in order to detect the primary gus gene and the silencing cassette in the transformed and re-transformed (GUS-RNAi) lines. The primer pair GUS251 (5'-TTGGGCAGGCCAGCGATACGT-3') and GUS671 (5'-ATAACGCAGTTCAACGCTGAC-3') was used to amplify 420 bp from the functional gus gene and the primer pair GUSF (5'-TCAGGAAGTGATGGAGCATCAGG-3') and MSIR (5'-TAGTTGGTCTGGCAGGCTAGT-3') were used to amplify 627 bp from intro-spliced hpRNA vector (pC1302GUSi). PCR amplifications contained 0.4 μM of primer, 250 μM dNTP's, 1.5 μM MgCl2, 1.0 U Taq DNA polymerase (5 U/μL) and about 20 ng of genomic DNA. Temperature cycling was performed on a PCT-100 thermocycler (MJ Research, MA, USA) as follows: initial denaturation at 95°C for 5 min; 36 cycles of denaturation at 95°C for 1 min; primer annealing at 55°C for 1 min; extension at 72°C for 1 min; and a final extension at 72°C for 5 min.
Transgenic Fusarium
F. verticillioides strain S68 (a nia-mutant) was previously co-transformed with the vectors pNOM102 containing the gus gene under control of the gpd promoter from A. nidulans, and pNH24 carrying the F. oxysporum nitrate reductase gene (nia). The line TG8 exhibiting the nia+phenotype conferred by pNH24 was selected on MM with nitrate as the sole nitrogen source and purified by isolation of uninucleated conidia according to standard techniques. Only colonies harbouring the pNH24 vector (nia+) are able to grow on MM plates. The line TG8 was previously characterized and presented a single copy of the gus gene integrated into the genome and mitotic stability in respect of the GUS+ phenotype [59].
Inoculation of transgenic plants with gus+ Fusarium
One transgenic line of F. verticillioides over-expressing the gus gene was selected to inoculate both re-transformed and non-transformed tobacco lines. The fungus was grown on MM medium [59] at 26°C. Inoculation was carried out according to Dias et al. [58]. A mycelial agar plug 3 mm in diameter was cut from the growing margins of a 6-day-old F. verticillioides culture and applied to the adaxial surface of a leaf detached from 6-week-old in vitro plants (n = 12 per line; repeated twice). After 11 days at 26°C and under 90%-100% relative humidity, the agar plug was removed and leaves containing penetrative food-absorbing structures that remained attached to leaf surface were analysed for gus gene expression.
Fungal colonies were re-isolated from both GUS-RNAi and non-transformed tobacco lines. Small portions from mycelium were removed under stereomicroscope and cultured on MM medium (as described above) in order to reach 70%-80% of confluence for eight passages. GUS histochemical and fluorimetric assays were carried out in each passage. The qualitative β-glucuronidase activity assay was carried out according to Couteaudier et al. [60] using 4-methyl umbeliferyl β-D-glucuronide as a substrate. Fluorescence was measured on a TKO100 Minifluorimeter (Hoefer, CA, USA) at 365nm. Protein content was determined using a protein assay kit (BioRad, CA, USA).
GUS assays and scanning electron microscopy
The GUS histochemical assay was performed as described by Jefferson [61]. Stained materials were examined using a Zeiss Axiophot stereomicroscope and photographed using a Zeiss AxioCam ICC3 high-resolution digital camera system. Explants were also examined using a scanning microscope. For scanning microscopy, leaves were fixed with 3% glutaldeihyde in 0.1 M sodium cacodylate buffer at pH 7.2 for 15 h at 4°C and postfixed in 1% osmium tetroxide for 1 h. After washing in the same buffer, the specimens were dehydrated in a graded acetone series, critical-point dried with CO2, sputter-coated with a thin layer of gold and observed under a Zeiss DSM 962 SEM operating at 15 kV.
RT-PCR expression analysis
Mycelium cells were used for total RNA extraction as described, with Trizol (Invitrogen) as recommended by the manufacturer. Total RNA was used to produce cDNA using the reverse transcriptase Superscript III (Invitrogen), according to the protocol suggested by the manufacturer. PCRs were carried out as described [62], except that 20 ng of cDNA was used as a template, in reactions with 32 cycles of amplification. The number of amplification cycles was previously optimized in order to stop the reaction at the exponential stage, ensuring that amplification was semi-quantitative. Primers 5'-ATCACGCAGTTCAACGCTGAC-3' (GUS671) and 5'-TTGGGCAGGCCAGCGTATCGT-3' (GUS251) were used to amplify a 420 bp fragment from the gus gene. As an internal control, primers 5'-GGAAGTAAAAGTCGTAACAAGG-3' (ITS5) and 5'-TCCTCCGCTTATTGATATGC-3' (ITS4) were utilized to amplify a fragment of the 5.8S rRNA housekeeping gene. PCRs with total RNA presented no amplified fragments. Experiments were repeated three times.
RNAi analysis
The mycelia of the fungi were cultivated on static liquid MM medium for 14 days at 26°C. Total RNA was isolated by extraction with Micro-to-Midi Total RNA Purification System (Invitrogen) as recommended by the manufacturer. Total RNA from plant leaves was isolated according to Bonfim et al. [19]. siRNA analysis was carried out according to Bonfim et al. [19]. RNAs were hybridized with a DNA probe corresponding to the 627 pb-fragment from gus gene, excised with EcoRV and SspI from pCambia2301. Probes were labelled with α32P dCTP using a random primer DNA labelling kit (Amersham Pharmacia Biotech, NJ, USA) according to the manufacturer's instructions. Three oligomers (18, 24 and 38 nucleotides) were used as molecular size markers. A GUS-sequence-related 21-nucleotide oligomer was used as a positive control. The bands were visualized with a fluorescent image analyser (FLA-3000) (FUJIFILM, Tokyo, Japan).
Southern blot analysis
Genomic DNA was isolated as described by Chow et al. [63]. Southern blotting was carried out as described [64]. Genomic DNA (20 μg) was digested with SphI, separated on a 1% agarose gel, and transferred to a nylon membrane (Hybond-N+; Amersham Pharmacia Biotech). Hybridizations were carried out using the PCR amplified 420-bp fragment from the gus gene, labelled with α32P deoxycytidine triphosphate (3000 Ci mol-1) using a random primer DNA-labelling kit (Amersham Pharmacia Biotech) according to the manufacturer's instructions. The bands were visualized with a fluorescent image analyser (FLA-3000; FUJIFILM).
Abbreviations
dsRNA: double-stranded RNA; hpGUS: hairpin GUS; hpRNA: hairpin RNA; mRNA: messenger RNA; PCR: polymerase chain reaction; RISC: ribonucleoprotein complex; RNAi: RNA interference; RT-PCR: reverse transcription PCR; siRNA: small interfering RNA; smRNA: small RNA.
Authors' contributions
MLPT carried out the molecular studies and in vivo assays, analysed the data and drafted the manuscript. BBAD carried out the fungi inoculation assays. JAP performed fungi transformation. RCDA participated in the molecular analysis. FJLA is the research group leader, conceived the study and participated in its design, analysed the data and finalized the manuscript. All authors read and approved the final manuscript.
Contributor Information
Maria Laine P Tinoco, Email: mlaine@cenargen.embrapa.br.
Bárbara BA Dias, Email: barbara@cenargen.embrapa.br.
Rebeca C Dall'Astta, Email: rdallastta@gmail.com.
João A Pamphile, Email: japamphile@uem.br.
Francisco JL Aragão, Email: aragao@cenargen.embrapa.br.
Acknowledgements
We are grateful to Dr Leila Barros (Embrapa) for providing the tobacco plants and to Dr Guy de Capdeville for help with the electron microscopy. This work was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; Grant No. 500028/2006-0). MLPT was supported by a fellowship from CNPq.
References
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Tabara H, Grishok A, Mello CC. RNAi in C. elegans: soaking in the genome sequence. Science. 1998;282:430–431. doi: 10.1126/science.282.5388.430. [DOI] [PubMed] [Google Scholar]
- Huang G, Allen R, Davis EL, Baum TJ, Hussey RS. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc Natl Acad Sci USA. 2006;103:14302–14306. doi: 10.1073/pnas.0604698103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbairn DJ, Cavallaro AS, Bernard M, Mahalinga-Iyer J, Graham MW, Botella JR. Host-delivered RNAi: an effective strategy to silence genes in plant parasitic nematodes. Planta. 2007;226:1525–1533. doi: 10.1007/s00425-007-0588-x. [DOI] [PubMed] [Google Scholar]
- Steeves RM, Todd TC, Essig JS, Trick HN. Transgenic soybeans expressing siRNAs specific to a major sperm protein gene suppress Heterodera glycines reproduction. Funct Plant Biol. 2006;33:991–999. doi: 10.1071/FP06130. [DOI] [PubMed] [Google Scholar]
- Haywood V, Kragler F, Lucas WJ. Plasmodesmata: pathways for protein and ribonucleoprotein signaling. Plant Cell. 2002;14:S303–325. doi: 10.1105/tpc.000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U, Crete P, Leuenberger SA, Iglesias VA, Meins F Jr. High molecular weight RNAs and small interfering RNAs induce systemic posttranscriptional gene silencing in plants. Proc Natl Acad Sci USA. 2002;99:11981–11986. doi: 10.1073/pnas.182204199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B, Wang Y. Viroids: uniquely simple and tractable models to elucidate regulation of cell-to-cell trafficking of RNA. DNA Cell Biol. 2009;28:51–56. doi: 10.1089/dna.2008.0811. [DOI] [PubMed] [Google Scholar]
- Buhtz A, Springer F, Chappell L, Baulcombe DC, Kehr J. Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J. 2008;53:739–749. doi: 10.1111/j.1365-313X.2007.03368.x. [DOI] [PubMed] [Google Scholar]
- Palauqui J-C, Elmayan T, Pollien J-M, Vaucheret H. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 1997;16:4738–4745. doi: 10.1093/emboj/16.15.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Baulcombe DC. Systemic signalling in gene silencing. Nature. 1997;389:553. doi: 10.1038/39215. [DOI] [PubMed] [Google Scholar]
- Tomilov AA, Tomilova NB, Wroblewski T, Michelmore R, Yoder JI. Trans-specific gene silencing between host and parasitic plants. Plant J. 2008;56:389–397. doi: 10.1111/j.1365-313X.2008.03613.x. [DOI] [PubMed] [Google Scholar]
- Bakhetia M, Charlton WL, Urwin PE, McPherson MJ, Atkinson HJ. RNA interference and plant parasitic nematodes. Trends Plant Sci. 2008;10:362–367. doi: 10.1016/j.tplants.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, Johnson S, Plaetinck G, Munyikwa T, Pleau M, Vaughn T, Roberts J. Control of coleopteran insect pests through RNA interference. Nature Biotechnol. 2007;25:1322–1326. doi: 10.1038/nbt1359. [DOI] [PubMed] [Google Scholar]
- Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, Huang YP, Chen XY. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nature Biotechnol. 2007;25:1307–1313. doi: 10.1038/nbt1352. [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, Robinson SP, Gleave AP, Green AG, Waterhouse PM. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313X.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- Nunes ACS, Vianna GR, Cuneo F, Amaya-Farfan J, Capdeville G, Rech EL, Aragão FJL. RNAi-mediated silencing of the myo-inositol-1-phosphate synthase gene (GmMIPS1) in transgenic soybean inhibited seed development and reduced phytate content. Planta. 2006;224:125–132. doi: 10.1007/s00425-005-0201-0. [DOI] [PubMed] [Google Scholar]
- Bonfim K, Farias JC, Nogueira EOPL, Mendes EA, Aragão FJL. RNAi-mediated resistance to bean golden mosaic virus in genetically engineered common bean (Phaseolus vulgaris) Mol Plant Microbe Interact. 2007;20:717–726. doi: 10.1094/MPMI-20-6-0717. [DOI] [PubMed] [Google Scholar]
- Ketting R, Plasterk R. RNAi and its applications. Nature Rev Gen. 2005;6 http://www.nature.com/nrg/poster/rnai/nrg_rnai_poster.pdf poster. [Google Scholar]
- Kadotani N, Nakayashiki H, Tosa Y, Mayama S. RNA silencing in the phytopathogenic fungus Magnaporthe oryzae. Mol Plant Microbe Interact. 2003;16:769–776. doi: 10.1094/MPMI.2003.16.9.769. [DOI] [PubMed] [Google Scholar]
- Erental A, Harel A, Yarden O. Type 2A phosphoprotein phosphatase is required for asexual development and pathogenesis of Sclerotinia sclerotiorum. Mol Plant Microbe Interact. 2007;20:944–954. doi: 10.1094/MPMI-20-8-0944. [DOI] [PubMed] [Google Scholar]
- Graham L, Graham MY, Subramanian S, Yu O. RNAi silencing of genes for elicitation or biosynthesis of 5-deoxyisoflavonoids suppresses race-specific resistance and hypersensitive cell death in Phytophthora sojae infected tissues terrence. Plant Physiol. 2007;144:728–740. doi: 10.1104/pp.107.097865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond TM, Keller NP. RNA silencing in Aspergillus nidulans is independent of RNA-dependent RNA polymerases. Genetics. 2005;169:607–617. doi: 10.1534/genetics.104.035964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromley M, Gordon C, Rovira-Graells N, Oliver J. The Aspergillus fumigatus cellobiohydrolase B (cbhB) promoter is tightly regulated and can be exploited for controlled protein expression and RNAi. FEMS Microbiol Lett. 2006;264:246–254. doi: 10.1111/j.1574-6968.2006.00462.x. [DOI] [PubMed] [Google Scholar]
- Henry C, Mouyna I, Latge JP. Testing the efficacy of RNA interference constructs in Aspergillus fumigatus. Curr Genet. 2007;51:277–284. doi: 10.1007/s00294-007-0119-0. [DOI] [PubMed] [Google Scholar]
- Mouyna I, Henry C, Doering TL, Latge JP. Gene silencing with RNA interference in the human pathogenic fungus Aspergillus fumigatus. FEMS Microbiol Lett. 2004;237:317–324. doi: 10.1016/j.femsle.2004.06.048. [DOI] [PubMed] [Google Scholar]
- Khalaj V, Eslami H, Azizi M, Rovira-Graells N, Bromley M. Efficient downregulation of alb1 gene using an AMA1-based episomal expression of RNAi construct in Aspergillus fumigatus. FEMS Microbiol Lett. 2007;270:250–254. doi: 10.1111/j.1574-6968.2007.00680.x. [DOI] [PubMed] [Google Scholar]
- Yamada O, Ikeda R, Ohkita Y, Hayashi R, Sakamoto K, Akita O. Gene silencing by RNA interference in the koji mold Aspergillus oryzae. Biosc, Biotechnol, Biochem. 2007;71:138–144. doi: 10.1271/bbb.60405. [DOI] [PubMed] [Google Scholar]
- Moriwaki A, Ueno M, Arase S, Kihara J. RNA-mediated gene silencing in the phytopathogenic fungus Bipolaris oryzae. FEMS Microbiol Lett. 2007;269:85–89. doi: 10.1111/j.1574-6968.2006.00606.x. [DOI] [PubMed] [Google Scholar]
- Nakayashiki H, Hanada S, Nguyen BQ, Kadotani N, Tosa Y, Mayama S. RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet Biol. 2005;42:275–283. doi: 10.1016/j.fgb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Namekawa SH, Iwabata K, Sugawara H, Hamada FN, Koshiyama A, Chiku H, Kamada T, Sakaguchi K. Knockdown of LIM15/DMC1 in the mushroom Coprinus cinereus by double-stranded RNA-mediated gene silencing. Microbiology. 2005;151:3669–3678. doi: 10.1099/mic.0.28209-0. [DOI] [PubMed] [Google Scholar]
- Walti MA, Villalba C, Buser RM, Grunler A, Aebi M, Kunzler M. Targeted gene silencing in the model mushroom Coprinopsis cinerea (Coprinus cinereus) by expression of homologous hairpin RNAs. Eukaryotic Cell. 2006;5:732–744. doi: 10.1128/EC.5.4.732-744.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha YS, Covert SF, Momany M. FsFKS1, the 1,3-beta-glucan synthase from the caspofungin-resistant fungus Fusarium solani. Eukaryotic Cell. 2006;5:1036–1042. doi: 10.1128/EC.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas FE, de Haro JP, Torres-Martinez S, Ruiz-Vazquez RM. Mutants defective in a Mucor circinelloides dicer-like gene are not compromised in siRNA silencing but display developmental defects. Fungal Genet Biol. 2007;44:504–516. doi: 10.1016/j.fgb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Meyer V. Genetic engineering of filamentous fungi: progress, obstacles and future trends. Biotechnol Advanc. 2008;26:177–185. doi: 10.1016/j.biotechadv.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Khatri M, Rajam MV. Targeting polyamines of Aspergillus nidulans by siRNA specific to fungal ornithine decarboxylase gene. Med Mycol. 2007;45:211–220. doi: 10.1080/13693780601158779. [DOI] [PubMed] [Google Scholar]
- Santos ACC, Sena JAL, Santos SC, Dias CV, Pirovani CP, Pungartnik C, Valle RR, Cascardo JCM, Vincentz M. dsRNA-induced gene silencing in Moniliophthora perniciosa, the causal agent of witches' broom disease of cacao. Fungal Genet Biol. 2009;46:825–836. doi: 10.1016/j.fgb.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Lagopodi AL, Ram AF, Lamers GE, Punt PJ, Hondel CA Van den, Lugtenberg BJ, Bloemberg GV. Novel aspects of tomato root colonization and infection by Fusarium oxysporum f. sp. radicis-lycopersici revealed by confocal laser scanning microscopic analysis using the green fluorescent protein as a marker. Mol Plant Microbe Interact. 2002;15:172–179. doi: 10.1094/MPMI.2002.15.2.172. [DOI] [PubMed] [Google Scholar]
- Oren L, Ezrati S, Cohen D, Sharon A. Early events in the Fusarium verticillioides-maize interaction characterized by using a green fluorescent protein-expressing transgenic isolate. Appl Environ Microbiology. 2003;69:1695–1701. doi: 10.1128/AEM.69.3.1695-1701.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipardi C, Wei Q, Paterson BM. RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell. 2001;107:297–307. doi: 10.1016/S0092-8674(01)00537-2. [DOI] [PubMed] [Google Scholar]
- Hutvágner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Gordon KHJ, Waterhouse PM. RNAi for insect-proof plants. Nature Biotechnol. 2007;25:1231–1232. doi: 10.1038/nbt1107-1231. [DOI] [PubMed] [Google Scholar]
- Cogoni C, Irelan JT, Schumacher M, Schmidhauser TJ, Selker EU, Macino G. Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J. 1996;15:3153–3163. [PMC free article] [PubMed] [Google Scholar]
- Whisson SC, Avrova AO, West PV, Jones JT. A method for double-stranded RNA-mediated transient gene silencing in Phytophthora infestans. Mol Plant Pathol. 2005;6:153–163. doi: 10.1111/j.1364-3703.2005.00272.x. [DOI] [PubMed] [Google Scholar]
- Vastenhouw NL, Brunschwig K, Okihara KL, Muller F, Tijsterman M, Plasterk RH. Gene expression: long-term gene silencing by RNAi. Nature. 2006;442:882. doi: 10.1038/442882a. [DOI] [PubMed] [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-Mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- Chen X-Y, Kim J-Y. Transport of macromolecules through plasmodesmata and the phloem. Physiol Plant. 2006;126:560–571. [Google Scholar]
- Lough TJ, Lucas WJ. Integrative plant biology: role of phloem long-distance macromolecular trafficking. Ann Rev Plant Biol. 2006;57:203–232. doi: 10.1146/annurev.arplant.56.032604.144145. [DOI] [PubMed] [Google Scholar]
- Yoo B-C, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, Lee YM, Lough TJ, Lucas WJ. A systemic small RNA signaling system in plants. Plant Cell. 2004;16:1979–2000. doi: 10.1105/tpc.104.023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen RA. RNA traffics information systemically in plants. Proc Natl Acad Sci USA. 2002;99:11561–11563. doi: 10.1073/pnas.192449099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehr J, Buhtz A. Long distance transport and movement of RNA through the phloem. J Exp Bot. 2008;59:85–92. doi: 10.1093/jxb/erm176. [DOI] [PubMed] [Google Scholar]
- Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 2003;22:4523–4533. doi: 10.1093/emboj/cdg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogo MR, Vainstein MH, Aragão FJL, Rech E, Schrank A. High frequency gene conversion among benomyl resistant transformants in the entomopathogenic fungus Metarhizium anisopliae. FEMS Microbiol Lett. 1996;142:123–127. doi: 10.1111/j.1574-6968.1996.tb08418.x. [DOI] [PubMed] [Google Scholar]
- Barreto CC, Alves LC, Aragão FJL, Schrank A, Vainstein MH, Rech E. High frequency gene transfer by microprojectile bombardment of intact conidia from the entomopathogenic fungus Paecilomyces fumosoroseus. FEMS Microbiol Lett. 1997;156:95–99. doi: 10.1111/j.1574-6968.1997.tb12711.x. [DOI] [PubMed] [Google Scholar]
- Chicas A, Forrest EC, Sepich S, Cogoni C, Macino G. Small interfering RNAs that trigger posttranscriptional gene silencing are not required for the histone H3Lys9 methylation necessary for transgenic tandem repeat stabilization in Neurospora crassa. Mol Cell Biol. 2005;25:3793–3801. doi: 10.1128/MCB.25.9.3793-3801.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horch RB, Fry JE, Hoffmann NL, Wallroth M, Eichholtz D, Rogers SG, Fraley RT. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Dias BBA, Cunha WG, Morais LS, Vianna GR, Rech EL, de Capdeville G, Aragão FJL. Expression of an oxalate decarboxylase gene from Flammulina sp. in transgenic lettuce (Lactuca sativa) plants and resistance to Sclerotinia sclerotiorum. Plant Pathol. 2006;55:187–193. doi: 10.1111/j.1365-3059.2006.01342.x. [DOI] [Google Scholar]
- Pamphile JA, Rocha CLMSC, Azevedo JL. Co-transformation of a tropical maize endophytic isolate of Fusarium verticillioides (synonym F. moniliforme) with gusA and nia genes. Genet Mol Biol. 2004;27:253–258. doi: 10.1590/S1415-47572004000200021. [DOI] [Google Scholar]
- Couteaudier Y, Daboussi MJ, Eparvier A, Langin T, Orcival J. The GUS gene fusion system (Escherichia coli beta-D-glucuronidase gene), a useful tool in studies of root colonization by Fusarium oxysporum. Appl Environ Microbiol. 1993;59:1767–1773. doi: 10.1128/aem.59.6.1767-1773.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abreu EFM, Aragão FJL. Isolation and characterization of a myo-inositol-1-phosphate synthase gene from yellow passion fruit (Passiflora edulis f. flavicarpa) expressed during seed development and environmental stress. Ann Bot. 2007;99:285–292. doi: 10.1093/aob/mcl256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow TY-K, Kafer E. A rapid method for isolation of total nucleic acids from Aspergillus nidulans. Fungal Genet Newsl. 1993;40:25–27. [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3. Vol. 1. New York: Cold Spring Harbor Laboratory Press; 2001. Southern hybridization; pp. 6.33–6.64. [Google Scholar]